
Towards the Standardization of Disease Registry Form Structure
Fatimetou Sidina, Hatem Bellaaj and Mohamed Jmaiel
ReDCAD Laboratory, University of Sfax, Sfax, Tunisia
Keywords: Disease Registry Form, Structure, Standardization.
Abstract: This paper presents a set of specifications for disease registry forms that vary from one registry to another,
emphasizing their standardization to ensure better interoperability and data analysis. After an in-depth review
of the state-of-the-art disease registry forms, we introduce a standardized structure adhering to the essential
data standards set by EPIRARE (Taruscio et al, 2014), a project funded by the European Union to improve
standardization and data comparability among patient registries, while respecting all question suggestions
provided by the Patient Registry Item Specifications and Metadata for Rare Disease PRISM project
(Richesson, Shereff and Andrews, 2012). This structure has been validated on several registries currently in
use, demonstrating a high level of accuracy.
1 INTRODUCTION
A disease registry (DR) includes information about
patients suffering from the same disease in order to
collect and track data related to their diagnoses,
treatments, outcomes, and demographics for research,
monitoring, and improving the understanding and
management of the condition. The information
collected by these registries becomes increasingly
meaningful depending on the protocols they follow.
It is necessary to establish standardized protocols for
diagnosis and treatment, which contributes to making
the collected data more reliable and comparable,
thereby enhancing the robustness of research
findings. Protocols can vary from one country to
another due to economic, demographic, and even
genetic differences.
Different national and international experiences
have been conducted. The latest report from Orphanet
(Orphanet Report Series, 2023) indicates a total of
827 registries, cohorts, and databases worldwide:
11% regional, 66.5% national, 11% European, and
11.5% global. Germany has the highest number (171
disease registries), followed by France (117 disease
registries).
There are many forms and structures which can be
included in disease registries. The main component is
the disease sheet (i.e. disease form), which is our
focus in this paper. Disease registry form includes
general data about the patient, circumstance of
discovery, clinical and analysis symptoms, treatment
and evolution. More information can be added
depending on physicians' needs.
The content of the disease registry form varies
across registries in terms of the data collected and the
structure, types, and presentation of that data.
There are several standardization efforts in the
creation of registries, defining essential data that
should be included in the registry form. (Aktaa et al,
2023) going further to specify the data type and how
it should be retrieved. There are also efforts to group
the questions/fields to be collected by registries
(Richesson, Shereff and Andrews, 2012), which can
be shared across various disease types. This is driven
by the fact that standardizing disease registry forms
will enhance the interoperability of health and
research data (Richesson, Shereff and Andrews,
2012). This, in turn, widens the scope of analyses and
research on diseases worldwide.
However, standardization efforts do not
encompass the standardization of the structures and
representation of registry forms, leading to multiple
implementation approaches for these registries. Each
registry has its unique way of implementing and
representing its forms. The Standardization of the
structure and representation of registries proposed in
this paper would not only reduce the design and
implementation efforts for registry forms but also
unify the structure of gathered data, even for registries
that do not adhere to a standard. For example, we
would no longer find registries with three levels of
organization alongside others with four levels, and we
would no longer encounter entire sections lacking
250
Sidina, F., Bellaaj, H. and Jmaiel, M.
Towards the Standardization of Disease Registry Form Structure.
DOI: 10.5220/0012708500003699
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2024), pages 250-257
ISBN: 978-989-758-700-9; ISSN: 2184-4984
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

fields specifying the collected data. These structuring
issues, often overlooked, can impact the automation
of disease registry implementations that comply with
global standards.
In this paper, we base our work on established
standards to introduce a well-defined structure and
representation for the sections of disease registry
forms. This is a crucial step for all who aim to
generate registry forms that comply with global
standards in terms of organization and structure. The
structure we have established has been validated
against seven resources, including standards and
currently used registries, and has yielded an average
accuracy of 0.88 and an accuracy of 1 for the two
standards used.
This paper is organized as follows: Section 2
delves into related work regarding the standardization
of disease registry forms, highlighting our specific
contributions in this area. Section 3 outlines the
skeleton and structure of disease registry forms.
Moving to Section 4, we introduce a theoretical and
conceptual representation of disease registry forms,
discussing its validation. Finally, in Section 5, we
conclude our work.
2 RELATED WORK
Data collected from disease registries represent a
valuable source for clinical research. However,
despite the availability of a large amount of data from
registries worldwide, the utility of this data remains
limited due to the lack of interoperability and
consistency among these registries. For example, the
same question may be asked in multiple registries, but
it is formulated differently, and the types of responses
vary from one registry to another (Spisla and
Lundberg, 2012). Therefore, the standardization of
disease registries is a necessity to enhance the quality
of medical research and care globally (Computerized
Disease Registries | Digital Healthcare Research). This
underscores our commitment to this standardization
effort by establishing a uniform structure for all
registries, clearly defining the diverse components of
registry forms and their appropriate relationships.
The PRISM project, funded through an American
Recovery and Reinvestment Act (ARRA) grant
administered by the National Library of Medicine
(NIH), serves as a valuable resource for standardizing
questions in rare disease registries. It encompasses
over 2,200 questions (Richesson, Shereff and
Andrews, 2012). Each question is indexed by one or
more keywords that characterize its general content
category, such as demographic information,
medication details, medical history, and special
histories. Additionally, EPIRARE, funded by the
European Commission, presents a collection of
indicators and common data elements for the
European platform dedicated to the registration of
rare diseases (Taruscio et al, 2014). These are based
on the indicators identified by the EUROPLAN
project (Posada, Carroquino and Pérez, 2011) and the
EU Rare Disease Task Force (RDTF). The FHIR®
(Fast Healthcare Interoperability Resources)
standard, developed by HL7® (Health Level Seven),
facilitates easier and faster healthcare data exchange.
It defines a set of standardized formats, known as
resources, to represent various healthcare data types
such as medications, allergies, and diagnoses. These
standardized formats (FHIR® resources) enable
seamless exchange and sharing of data between
different healthcare systems and applications.
Similarly, SNOMED CT (Systematized
Nomenclature of Medicine Clinical Terms) provides
standardized terminology that can be utilized in
healthcare-related information systems, ensuring
consistency and interoperability across various
clinical specialties within healthcare systems.
However, despite this extensive collection, the
number of questions, indicators, and elements
provided by these initiatives remains limited when
compared to the diverse array of inquiries pertinent to
rare diseases. Consequently, disease registries
employing PRISM or common data elements for the
European platform, resources of FHIR®, or
standardized terminology of SNOMED CT may have
multiple sections lacking coverage by these
established questions, indicators, or elements,
resulting in an absence of complete standardization.
(Aktaa et al, 2023) undertook the standardization of
TAVI (Transcatheter Aortic Valve Implantation)
related data variables (i.e., data fields to be collected)
to address registry heterogeneity, facilitating
international comparative analyses and the
development of comprehensive valvular heart disease
registries, regardless of the treatment approach. These
variables were classified into two levels: Level 1 for
essential quality assessment data and Level 2 for
supplementary information useful in quality
evaluation and research but not universally required.
The selection of these variables was accomplished
through a modified Delphi method, with the Working
Group voting on a list of candidate variables
identified through a literature review. This effort
resulted in 93 Level 1 and 113 Level 2 variables
across ten TAVI care domains, including patient
characteristics, comorbidities, prior interventions,
and pre-procedural tests. This could be regarded as a
Towards the Standardization of Disease Registry Form Structure
251
general standardization of TAVI registries,
particularly as it takes into account demographic
differences. However, this applies only to a specific
procedure used for treating aortic valve disease,
Transcatheter Aortic Valve Implantation (TAVI).
Similarly, (Fulvio and Mantegazza, 2014) present the
European database for Myasthenia Gravis (EuroMG-
DB) as a model for an international disease registry.
The structure of EuroMG-DB follows a schematic
representation, including the Patient main page linked
to: (1) referring physicians and MG patients, (2)
diagnostic criteria, (3) thymus, (4) biological
samples, (5) other diseases, and (6) follow-up visits.
The RoPR project (Gliklich, Leavy and Dreyer,
2020) introduces an Outcome Measures Framework
that organizes disease registries into three
hierarchical levels: domains, subcategories of data
elements, and data elements. These domains
encompass: (1) Characteristics, which are further
divided into three main categories: Participants,
Diseases, and Providers. (2) Treatments, which can
be categorized into two main groups: Type and Intent.
(3) Outcomes, consisting of five main categories:
Survival, Clinical Response or Status, Events of
Interest, Patient-Reported, and Resource Utilization.
Finally, at the third level, you'll find subcategories of
data elements that are used to define an outcome
measure, including those that capture physical
findings and diagnoses. This project defines the
outcomes to be extracted from disease registries,
considering the variations among diseases and the
specific details they require. It goes further by
providing examples of these details for several
diseases, yet it does not specify the method or
structure for collecting this data.
All these standardization initiatives aim to establish
standardized disease registries. They do so by either
concentrating on specific aspects of disease registries,
emphasizing registry content and offering useful yet
limited collections, or by centering efforts on
standardizing the extracted outcomes. Nevertheless,
these approaches frequently result in an inability to
maintain a consistent standardized format for disease
registry forms. Alternatively, certain initiatives offer
a structured set of variables for collection, but this is
limited to a single disease.
The absence of standardization in the formats and
representation of disease registry forms renders
discussions on interoperability and comparability
between registries impossible and would necessitate
extensive reformatting of collected data (Fulvio and
Mantegazza, 2014)
. This drove our research efforts to
explore the standardized representation of disease
registry forms.
3 STRUCTURE AND CONTENT
OF A DISEASE REGISTRY
FORM
Disease registries can vary significantly based on
their intended purpose, context, and the overseeing
organization.The specific structure and content of a
registry depend on the registry's purpose (Gliklich,
Leavy and Dreyer, 2020). Regardless of the registry's
design, an electronic disease registry provides
healthcare professionals with a disease registry form
containing predefined options for its various sections.
The disease registry form acts as a key source of
input data, typically organized into sections or
domains (Item groups). The titles of these sections
may vary across registries. Each section consists of a
series of subsections (Item concepts), and within each
subsection, there is an array of fields (Questions) that
represent the data elements to be collected. These
fields may contain subfields (Content coding)
delineating the method or specifics of data collection,
as outlined in the Data Set for Rare Disease Patient
Registries Recommended for European Cooperation
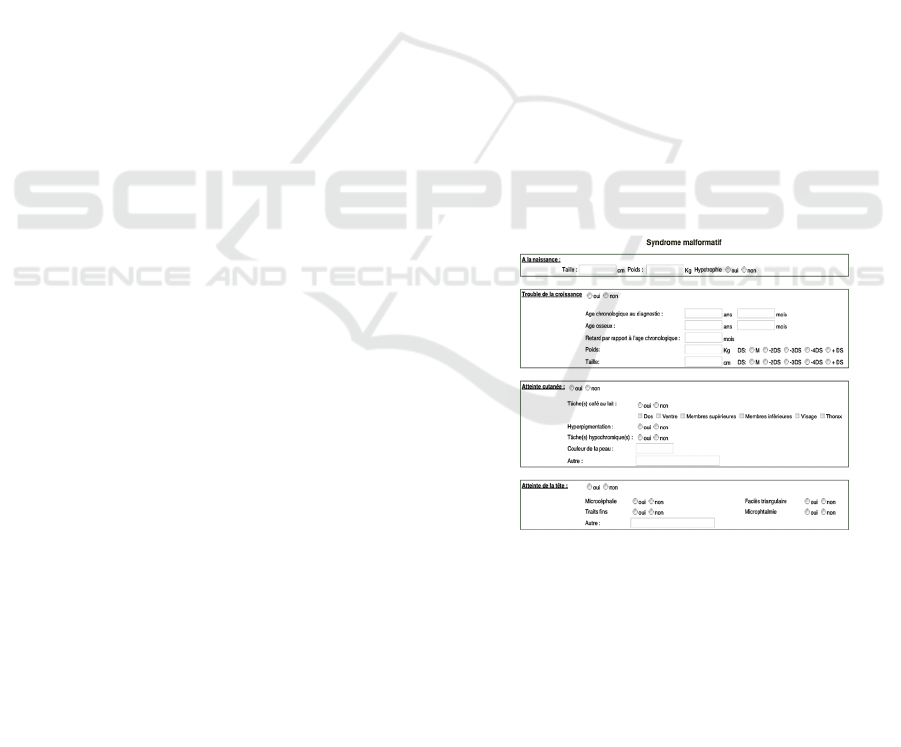
(Version 3.0) (Berger et al, 2021). “Figure 1”
illustrates an example of this structure within a
portion of the "Malformation Syndrome" section of
the TFAR registry (Bellaaj et al., 2017).
Figure 1: Example of Registry Form Section Structuring.
Different content of disease registry form experiences
are presented in the literature. For instance, (Salenius
et al, 1992) includes the “Patient's History”, “Clinic
Statistics” and “Progress Note”. This last consists of
four sections: (1) vital signs, allergies, average
weekly glucose measurements, and any point-of-care
values (2) eye, foot, and psychological screening
information, (3) smoking history, medication
compliance, and activity (minutes per week) and (4)
ICT4AWE 2024 - 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health
252

SOAP (subjective, objective, assessment, and plan)
note.
On the other hand, the Electronic Disease Form
(EDF) within the TFAR (Bellaaj et al, 2017)
encompasses 11 distinct sections: Register
Identification, Patient Identification, Family history,
Circumstances of discovery, Malformation
syndrome, Cytogenetic study, Hematological signs,
Molecular biology, Cell freezing, Clinical score and
Treatment. Each of these sections is further
subdivided into one or more subsections, totaling 37
subsections in the entirety of TFAR. For example, the
Malformation syndrome component comprises 11
subsections, many of which align with specific
medical specialties, such as Skin damage and
Urogenital malformation. Each subsection contains
various fields, including checkboxes and input fields.
Additionally, some fields are initially hidden and are
revealed only if the 'yes' option is selected.
The CASCADE FH Registry comprises four
domains (sections). 'Enrollment information'
encompasses the patient demographics section.
'Medical history' is divided into two subsections:
patient history and family history. 'Treatment,
laboratory, and examination' incorporates three
subsections: FH treatment, Examination/laboratory,
and Imaging/procedures (within 5 y). The 'Additional'
domain includes Patient-reported outcomes
subsection and an additional subsection for Clinical
trial participation and Provider contact information
(O’Brien et al, 2014).
The National Cardiovascular Data Registry
(NDCR)® ICD Registry ™ utilizes the ACC/AHA
Heart Failure Clinical Data Standards, clinical data
standards created by the American College of
Cardiology (ACC) for acute coronary syndromes,
heart failure, and atrial fibrillation (WRITING
COMMITTEE MEMBERS, Radford et al, 2005).
These standards categorize the collected data into 11
sections: Patient Demographics, Medical History,
Patient Assessment: Current Symptoms and Signs,
Patient Assessment: Summary Assessment,
Laboratory Tests, Diagnostic Procedures, Invasive
Therapeutic Procedures, Pharmacological Therapy,
End-of-Life Management, Patient Education:
Assessment of Status and Patient Education:
Intervention and Referral. For each of these sections,
the standards specify subsections and the requisite
data to be collected.
The structure of disease registry forms simplifies
data collection and analysis. The level of detail and
specialization, however, varies from one disease and
organization to another. For instance, in the case of a
Rare Disease Registry, more extensive and detailed
data may be necessary due to the rarity of the
conditions being studied. Conversely, for more
common diseases, less extensive data may suffice, as
a wealth of information about these conditions is
already available.
4 REPRESENTATION OF
DISEASE REGISTRIES FORMS
4.1 Definition
In our study, we investigated the contents of registry
forms, encompassing their individual sections, the
scope of data they cover, and the diversity in form
representation across each registry. However, upon
examining this structural representation, we observed
its near uniformity across the majority of international
and standardized registries. Nevertheless, not all
registries adhere to or adopt this common structure,
rendering the task of standardizing disease registries
increasingly challenging. This is why the definition
of formal representation is essential to guide the new
work of creating registries, especially for small
regional registry initiatives that generally do not
adhere to a well-defined standard. This
standardization of form format representation will
enable them to adopt a format that aligns with
international registries following standardization
guidelines. Hence, our aim is to introduce a
standardized theoretical and conceptual model for
registry forms, offering a universal representation.
This model holds significant value as it furnishes an
all-encompassing framework, enabling automated
systems to consistently interpret the diverse
information types present within registries.
The disease registry form consists of multiple
sections, each containing one or more subsections.
Within each subsection, there exists a set of fields,
which can have some subfields if needed.
So we can represent a registry like 𝑆={𝑆
| 𝑖 ∈
1..𝑚} with 𝑆
is the section number i of registry
form, m is the number of sections in the form and each
section 𝑆
can be represented as shown in “Figure 2”.
with:
- Λ is the set of subsections of 𝑆
, Λ =
∪
𝜆
, n = |Λ|
- Γ is the set of titles of subsections, Γ =
∪
𝛾
, n = |Λ| = |Γ|
- Ω is the set of fields of 𝑆
,𝜴 =
{(𝝀
,𝛺
)| 𝑗𝜖1. . 𝑛} with: 𝑛 = |𝜦| =
Towards the Standardization of Disease Registry Form Structure
253
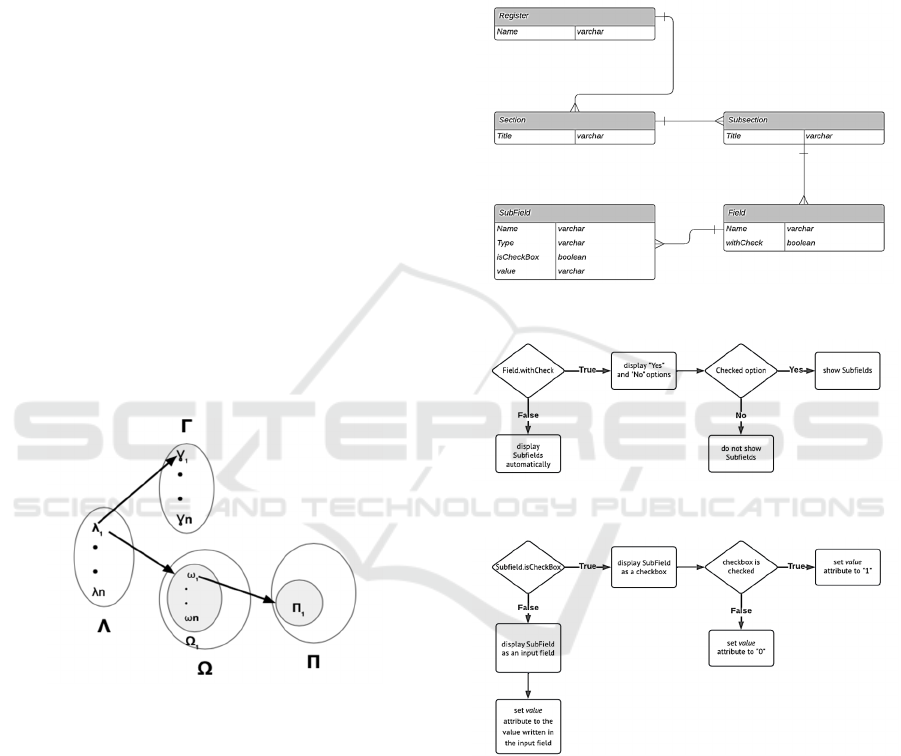
|𝜴| ; 𝛺
=∪
𝜔
set of fields of
subsection 𝜆
; 𝒑
𝒋
= |𝛺
| number of fields of
subsection 𝜆
; if 𝑝
= 0 , then 𝜆
is a blank
subsection
- Π is the set of subfields where Π=
∪
∗
𝛱
| 𝛱
=∪
𝞹
; kl= |𝛱
|
number of subfields of field 𝜔
; if 𝑘
= 0 , then
𝜔
is a blank field
- Π= 𝑰
𝟏
∪ 𝑰
𝟐
∪ 𝑰
𝟑
∪ 𝑰
𝟑
∪ 𝑰
𝟒
∪ 𝑰
𝟒
′ , with
- 𝑰
𝟏
={0,1} the set of checkboxes that
don’t require a condition
- 𝑰
𝟐
the set of input fields that don’t
require a condition
- 𝑰
𝟑
𝒂𝒏𝒅 𝑰
𝟑
set of checkboxes that
require verification of condition
respectively “if yes” and “if no”
- 𝑰
𝟒
𝒂𝒏𝒅 𝑰
𝟒
the set of input fields, that
require verification of condition
respectively “if yes” and “if no”
For each subsection 𝜆
∈ Λ, it is associated with a
title 𝛾
∈ Γ and a set of fields 𝛺
∈ Ω, where for
each field 𝜔
∈ 𝛺
, it is associated with a set of
subfields 𝛱
∈ Π.
Figure 2: Relations between groups.
4.2 Entity-Relationship Disease
Registries Form Pattern
For a deeper and more accessible understanding of
the structure and easy interpretation of the
connections and interdependencies between entities,
as defined in the "Definition" section, we provide a
visual representation using the entity-relationship
diagram. This representation provides a clear and
concise representation of various entities, their
distinct attributes, and the connections between them,
making it easier to understand the interactions within
a given registry system (see “Figure 3”).
The diagram in “Figure 3” illustrates the essential
attributes for various entities. We use the attribute
'Name' for Register, Field, and SubField, while
employing the attribute 'Title' for Section and
Subsection. Additionally, Field and SubField have
additional attributes defining their representation and
data collection methods, as represented in the
algorithm outlined in “Figure 4” and “Figure 5”.
Figure 3: Diagram entity-relationship.
Figure 4: Field Attribute Constraints.
Figure 5: SubField Attribute Constraints.
4.3 Sample Structure of a Standardized
Registry Form
To illustrate the structured representation of data,
entities, and their connections, we present a sample
format of a standardized registry form. This
structured format is instrumental in ensuring
consistency, efficiency, and uniformity in the capture,
storage, and retrieval of information across the
diverse domains of registry forms.
ICT4AWE 2024 - 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health
254

The example showcased in “Figure 6” delineates
two sections within the registry. For instance, the first
section comprises two subsections: the first
subsection encompasses two fields, while the second
subsection includes one field. Within the first
subsection of the initial section, the first field has an
attribute of "withCheck" set to false, and all its
associated subfields possess the attribute
"isCheckBox" set to true. Conversely, the second
field has "withCheck" set to true, hence presenting
options for 'Yes' and 'No'. Subsequently, by selecting
“Yes”, the two subfields that appear have their
“isCheckBox” attributes set to false.
This detailed example (in “Figure 6”) exemplifies
how attributes like "withCheck" and "isCheckBox"
impact the presentation and behavior of fields and
subfields within the standardized registry structure,
demonstrating various conditional display settings
and attribute configurations.
Figure 6: Example of a standardized structure.
4.4 Universal Use of Proposed
Standard Structure
For our validation process, we examined the
structures of some registries actually in use alongside
a set of standard questions and responses, comparing
them to the standard structure proposed by our model
representation. We chose to use the accuracy metric
for this evaluation (see “Table 1”). In this simple
binary classification scenario, our goal is to determine
whether a given structure matches our model
representation or not. The choice to use the accuracy
metric is well-suited for this context. The
classification task is straightforward, dividing
structures into two categories: those that match (the
positive class) and those that do not (the negative
class), making accuracy a suitable measure.
Furthermore, we observed that the consequences and
costs associated with both false positives and false
negatives are similar, which further supports the use
of accuracy. Indeed, the accuracy metric measures the
proportion of correctly classified instances among the
total instances evaluated. It provides a
straightforward measure of how well a model is
performing overall in terms of classification
accuracy. The formula for calculating accuracy is as
follows:
Accuracy = (Number of Valid Sections) /
(Total Number of Sections)
(1)
Table 1: Validation Results: Comparison with proposed Structure.
Register / standard Number of Valid
Sections
Total Number of
Sections
Accuracy
Tunisian registry GUELT 2013 36 44 0.82
Maghreb group for the evaluation of large B cell lymphomas
GEMLA
8 9 0.89
Tunisian registry of AMINOACIDOPATHIES 8 12 0.67
Tunisian registry of DIALYSIS 5 5 1
TFAR (Hadiji et al, 2012) 32 37 0.84
- (TARUSCIO ET AL, 2014) SET OF COMMON DATA
ELEMENTS FOR THE EUROPEAN RDR PLATFORM
5 5 1
- SAMPLE OF PRISM QUESTIONS AND SELECTED
METADATA (RICHESSON, SHEREFF AND ANDREWS,
2012). (224/2,200 QUESTIONS PRESENTED IN: [PDF FILE
(ADOBE PDF FILE), 318KB-MULTIMEDIA APPENDIX 1] )
22 22 1
Towards the Standardization of Disease Registry Form Structure
255

We've noted complete adherence to the standardized
structure model, reaching 100%, in the two standards
used. However, this varies between 100% and 67%
among other disease registries. Notably, even in cases
where sections deviate from our proposed
representation, there's potential to realign them with
our standardized model. Nevertheless, the lack of
standardization in section structures often leads
disease registry form developers to create sections
that diverge from the standardized form structure,
presenting the initial obstacle toward achieving
complete standardization of disease registries.
5 CONCLUSIONS
The paper proposes a standardized structuring of
disease registry forms, providing a clear definition of
various concepts and components within these forms,
as well as the relationships among these different
elements. Such structuring is crucial in progressing
towards the standardization of disease registries.
Adhering to this standard will result in a uniform
structural representation of disease registry forms, a
valuable uniformity for subsequent data analyses, and
a detailed guide for generating new disease registry
forms.
This work aims to simplify and unify the structure
of disease registry forms, establishing a standardized
representation that is universally applied. This
standardization represents the initial phase in a
broader effort to create a unified approach for data
collection and analysis across different disease
registries. By doing so, we not only enhance the
efficiency of this process but also facilitate the cross-
comparison of data and findings from various
sources.
The work represents the initial step towards
standardizing disease registries. A more generic
standardization will require further work on the
nature of different registry sections and their contents.
This will be our focus in future endeavors.
REFERENCES
Orphanet. (2023). Rare Disease Registries, cohorts and
databases. Orphanet Report Series, Rare Diseases.
Hadiji Mseddi, S., Kammoun, L., Bellaaj, H., Ben Youssef,
Y., Aissaoui, L., Torjemane, L., Telmoudi, F., Amouri,
A., Elghezal, H., Ouederni, M., Ben Abdennebi, Y.,
Hammemi, S., Ben Othmen, T., Ben Abid, H., Bejaoui,
M., Abdelhak, S., Hachicha, M., Dellagi, K., & Frikha,
M. (2012). Création et rapport du registre tunisien de
l’anémie de Fanconi (TFAR). Archives de Pédiatrie,
19(5), 467–475. https://doi.org/10.1016/j.arcped.20
12.02.017
Salenius, S. A., Margolese-Malin, L., Tepper, J. E.,
Rosenman, J., Varia, M., & Hodge, L. (1992). An
electronic medical record system with direct data-entry
and research capabilities. International Journal of
Radiation Oncology*Biology*Physics, 24(2), 369–376.
https://doi.org/10.1016/0360-3016(92)90693-C
Bellaaj, H., Mdhaffar, A., Jmaiel, M., & Freisleben, B.
(2017). An Adaptive Scrum Model for Developing
Disease Registries: Proceedings of the 10th
International Joint Conference on Biomedical
Engineering Systems and Technologies, 484–491.
https://doi.org/10.5220/0006297804840491
Fulvio, B., & Mantegazza, R. (2014). European Database
for Myasthenia Gravis: A model for an international
disease registry. Neurology, 83(2), 189–191.
https://doi.org/10.1212/WNL.0000000000000563
Computerized Disease Registries | Digital Healthcare
Research. (n.d.). Retrieved 12 October 2023, from
https://digital.ahrq.gov/computerized-disease-
registries
Spisla, C. M., & Lundberg, C. B. (2012). Standardization
of Patient Registries for Improved Data Collection and
Outcome Measurement. NI 2012 : 11th International
Congress on Nursing Informatics, June 23-27, 2012,
Montreal, Canada., 2012, 391.
Richesson, R. L., Shereff, D., & Andrews, J. E. (2012).
Standardization of Questions in Rare Disease
Registries: The PRISM Library Project. Interactive
Journal of Medical Research, 1(2), e10.
https://doi.org/10.2196/ijmr.2107
Aktaa, S., Batra, G., James, S. K., Blackman, D. J.,
Ludman, P. F., Mamas, M. A., Abdel-Wahab, M.,
Angelini, G. D., Czerny, M., Delgado, V., De Luca, G.,
Agricola, E., Foldager, D., Hamm, C. W., Iung, B.,
Mangner, N., Mehilli, J., Murphy, G. J., Mylotte, D., …
Gale, C. P. (2023). Data standards for transcatheter
aortic valve implantation: The European Unified
Registries for Heart Care Evaluation and Randomised
Trials (EuroHeart). European Heart Journal - Quality
of Care and Clinical Outcomes, 9(5), 529–536.
https://doi.org/10.1093/ehjqcco/qcac063
Taruscio, D., Mollo, E., Gainotti, S., Posada de la Paz, M.,
Bianchi, F., & Vittozzi, L. (2014). The EPIRARE
proposal of a set of indicators and common data
elements for the European platform for rare disease
registration. Archives of Public Health, 72(1), Article 1.
https://doi.org/10.1186/2049-3258-72-35
Posada, M., Carroquino, M. J., & Pérez, H.
(2011).European Project for Rare Diseases National
Plans Development (EUROPLAN): Selecting
indicators to evaluate the achievements of RD
initiatives. Retrieved 16 October 2023, from
http://www.europlanproject.eu/_europlanproject/Reso
urces/docs/2008-2011_3.EuroplanIndicators.pdf
Gliklich, R. E., Leavy, M. B., & Dreyer, N. A. (2020).
Registries for Evaluating Patient Outcomes: A User’s
Guide(Fourth edition). Agency for Healthcare Research
ICT4AWE 2024 - 10th International Conference on Information and Communication Technologies for Ageing Well and e-Health
256

and Quality (AHRQ). https://doi.org/10.23970/AHRQ
EPCREGISTRIES4
O’Brien, E. C., Roe, M. T., Fraulo, E. S., Peterson, E. D.,
Ballantyne, C. M., Genest, J., Gidding, S. S.,
Hammond, E., Hemphill, L. C., Hudgins, L. C., Kindt,
I., Moriarty, P. M., Ross, J., Underberg, J. A., Watson,
K., Pickhardt, D., Rader, D. J., Wilemon, K., &
Knowles, J. W. (2014). Rationale and design of the
familial hypercholesterolemia foundation CAscade
SCreening for Awareness and DEtection of Familial
Hypercholesterolemia registry. American Heart
Journal, 167(3), 342-349.e17. https://doi.org/10.10
16/j.ahj.2013.12.008
WRITING COMMITTEE MEMBERS, Radford, M. J.,
Arnold, J. M. O., Bennett, S. J., Cinquegrani, M. P.,
Cleland, J. G. F., Havranek, E. P., Heidenreich, P. A.,
Rutherford, J. D., Spertus, J. A., Stevenson, L. W.,
Heidenreich, P. A., Goff, D. C., Grover, F. L., Malenka,
D. J., Peterson, E. D., Radford, M. J., & Redberg, R. F.
(2005). ACC/AHA Key Data Elements and Definitions
for Measuring the Clinical Management and Outcomes
of Patients With Chronic Heart Failure: A Report of the
American College of Cardiology/American Heart
Association Task Force on Clinical Data Standards
(Writing Committee to Develop Heart Failure Clinical
Data Standards): Developed in Collaboration With the
American College of Chest Physicians and the
International Society for Heart and Lung
Transplantation: Endorsed by the Heart Failure Society
of America. Circulation, 112(12), 1888–1916. https://
doi.org/10.1161/CIRCULATIONAHA.105.170073
Berger, A., Rustemeier, A.-K., Göbel, J., Kadioglu, D.,
Britz, V., Schubert, K., Mohnike, K., Storf, H., &
Wagner, T. O. F. (2021). How to design a registry for
undiagnosed patients in the framework of rare disease
diagnosis: Suggestions on software, data set and coding
system. Orphanet Journal of Rare Diseases, 16(1), 198.
https://doi.org/10.1186/s13023-021-01831-3
Towards the Standardization of Disease Registry Form Structure
257
