
UPCARE: User Privacy-Preserving Cancer Research Platform
Georg Bramm
1 a
, Melek
¨
Onen
2 b
, Martin Schanzenbach
1 c
, Ilya Komarov
3
, Frank Morgner
3
,
Christian Tiebel
4
and Juan Cadavid
5 d
1
Fraunhofer AISEC, Lichtenbergstraße 11, 85748 Garching bei M
¨
unchen, Germany
2
EURECOM, Campus Sophia Tech, 450 Route des Chappes, 06410 Biot, France
3
Bundesdruckerei GmbH, Kommandantenstraße 18, 10969 Berlin, Germany
4
IDG Institut f
¨
ur Digitale Gesundheitsdaten RLP, Große Bleiche 46, 55116 Mainz, Germany
5
Softeam Docaposte, 110 Esplanade du G
´
en
´
eral de Gaulle 92931 Paris, France
Keywords:
Distributed Analytics, Medical Data Management, Privacy-Preserving Computation, Cryptography,
Homomorphic Cryptography, Attribute Based Cryptography.
Abstract:
Cancer research has entered a new era with the advent of big data and advanced computational analytics.
However, the utilization of such medical data poses significant privacy and security challenges. This paper
presents a comprehensive examination of User Privacy-preserving Cancer Research Platform (UPCARE), a
research platform that enables the secure and ethical sharing of sensitive medical cancer data for collaborative
researchers, while safeguarding patient privacy. To our knowledge, only a few approaches have been pursued
so far in building a uniform cancer research platform protected by modern cryptography. We try to provide a
uniform platform for research UPCARE leverages cutting-edge cryptographic access methods, like attribute-
based encryption, as well as data anonymization techniques, like multiparty homomorphic encryption, to allow
secure data sharing with researchers, ensuring compliance with stringent regulatory requirements. This paper
discusses the architecture, methodologies, and applications of UPCARE, highlighting its potential to improve
cancer research and accelerate advancements in precision medicine while preserving user privacy.
1 INTRODUCTION
Cancer research has made remarkable strides in recent
years, driven by recent advancements in genomics,
computational biology, and data analytics. However,
these exciting developments come with a unique set
of challenges, particularly concerning the privacy and
security of sensitive medical data, especially in the
case of cancer data. Up until now, the data silos in
the different cancer registries are very heterogeneous
in nearly all aspects, for example like the utilized
software, and are therefore nearly impossible to com-
bine. The critical need to balance data sharing for
collaborative research and patient privacy protection
has never been more evident.
The conventional methods of data sharing and col-
laboration in the medical and research community are
a
https://orcid.org/0000-0002-9020-5856
b
https://orcid.org/0000-0003-0269-9495
c
https://orcid.org/0000-0001-6153-504X
d
https://orcid.org/0009-0002-3062-401X
often fraught with privacy risks, compliance issues,
and ethical dilemmas. Striking the right balance be-
tween unlocking the full potential of data-driven can-
cer research and safeguarding the privacy and rights
of individuals is a complex and pressing challenge.
Furthermore, the failure to address privacy and
security concerns can lead to data breaches, reputa-
tional damage, and legal consequences for research
institutions. In an interconnected world, where data
breaches are a constant threat, safeguarding sensitive
health data is paramount.
1.1 Regulatory Context
The significance of privacy in cancer research extends
far beyond legal and ethical obligations. While strict
regulatory frameworks such as the Health Insurance
Portability and Accountability Act (HIPAA) and the
General Data Protection Regulation (GDPR) enforce
the protection of patients’ privacy, the ethical and so-
cietal implications are equally profound. Addition-
52
Bramm, G., Önen, M., Schanzenbach, M., Komarov, I., Morgner, F., Tiebel, C. and Cadavid, J.
UPCARE: User Privacy-Preser ving Cancer Research Platform.
DOI: 10.5220/0012718100003767
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 21st International Conference on Security and Cryptography (SECRYPT 2024), pages 52-63
ISBN: 978-989-758-709-2; ISSN: 2184-7711
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

ally, use cases need to respect national regulations,
which specifies what kind of information needs to be
collected, how it should be made available to the pub-
lic for allowing cross-sectoral impact of the cancer
research as well as safeguarding the work split and
responsibilities between different cancer registries as
well as the entities communicating with the registries.
1.2 Research Objectives
The primary objective of this paper is to present
a holistic examination of UPCARE, which was de-
signed to address the privacy and security challenges
associated with cancer research. This platform aims
to facilitate the secure and ethical sharing of sensi-
tive cancer data among researchers while ensuring
compliance with stringent regulatory requirements.
Specifically, the research objectives include:
• Exploring the challenges and privacy risks in can-
cer research data sharing and computing.
• Assessing the potential use cases and applications
of the platform.
• Describing the architecture and functionalities of
UPCARE.
• Evaluating the cryptographic security and privacy
measurements taken, to ensure compliance with
stringent regulatory requirements .
• Outlining some future directions and challenges
in the field of privacy-preserving computation in
the area of cancer research.
The goal of UPCARE is to break down the restric-
tive data silos within the cancer registry landscape
and enable the combined usage of existing data sets
while preserving patient privacy. A user-centered ac-
cess decision and consent process should also allow
patients to make purpose-specific decisions and avoid
giving blanket data access. Instead of reinventing the
concept of a cancer registry from the ground up or
making unrealistic assumptions during the restructur-
ing of current processes and organizations, UPCARE
adopts a pragmatic perspective and respects the ex-
isting standards and processes. The focus is on re-
searching, developing, and demonstrating novel cryp-
tographic technologies that make personal data usable
while simultaneously protecting individuals’ privacy.
We support two different research scenarios, which
will be described in Section 3 in particular.
2 RELATED WORK
Projects related to the efforts undertaken in UPCARE
include the national Network Genomic Medicine
(nNGM) Lung Cancer, a collaborative research al-
liance of the German Oncology Centers
1
, and the
German Biobank Node (BBMRI)
2
. Such medical re-
search platforms usually face the same problem as
UPCARE: Heterogeneous data silos must be inte-
grated and a consolidated data pool must be provided
to researchers that want to perform any kind of data
analysis as part of their investigations. All platforms
fall under one or the other data protection regula-
tion whenever they are actually operated in the real
world. And while there exist tool boxes to support
data protection
3
, such efforts primarily address reg-
ulatory compliance and consent management. What
makes UPCARE unique is its use of state-of-the-art
cryptographic mechanisms to ensure data privacy for
patients, which also minimizes the attack surfaces.
One of the foundational works that introduced the
concept of privacy-preserving data mining and dis-
cussed methods for protecting individual privacy in
the context of data analysis was written by Agrawal
et al. (Agrawal and Srikant, 2000). Academic ap-
proaches in the literature are often based on the use
of a blockchain, like the general architecture that was
introduced by Yue et al. (Yue et al., 2016). It uti-
lizes a blockchain to enable patients to own, control
and share their data easily and securely without vio-
lating privacy, which provides a new potential way to
improve the intelligence of healthcare systems while
keeping patient data private. Some approaches try to
combine blockchain usage with attribute based en-
cryption (Pournaghi et al., 2020). Other approaches
only seem to be based on attribute based encryption
(Barua et al., 2011; Narayan et al., 2010).
Geva et al. (Geva et al., 2023) tried to access
cancer registry data in a privacy preserving man-
ner through the use of Homomorphic Encryption
(HE). The authors present a tool set for collaborative
privacy-preserving analysis of oncological data using
multiparty fully homomorphic encryption (FHE), al-
though most of the available solutions in literature de-
scribe very detailed and specific use cases (Son et al.,
2021). We designed UPCARE from the ground up in
such a way, that the utilized ABE scheme could be
easily swapped, in order to test different candidates
regarding performance and efficiency, like Bethen-
court et al, (Bethencourt et al., 2007), Sahai et al.
(Sahai and Waters, 2008), Lewko et al. (Lewko and
Waters, 2011), M
¨
uller et al. (M
¨
uller et al., 2009) and
Bramm et al. (Bramm et al., 2018). Because of its
stable performance and specific design decisions, like
its open world attribute setup, FAME in the KP-ABE
1
https://nngm.de/en/
2
https://www.bbmri.de/
3
https://www.tmf-ev.de/unsere-arbeit/projekte/magic
UPCARE: User Privacy-Preserving Cancer Research Platform
53

version by Agrawal et al. (Agrawal and Chase, 2017)
was chosen to be integrated into the platform. Thanks
to the foundational work of Gentry et al. (Gentry,
2009) and others (Van Dijk et al., 2010) schemes like
CKKS could be developed. This project vastly em-
ploys Lattigo to do HE computations, which imple-
ments a multi-party variant of the CKKS scheme by
Cheon et al. (Cheon et al., 2017).
Compared to all the other mentioned approaches
above, UPCARE tries to unify and simplify both
privacy preserving cryptographic techniques, namely
Attributed Based Encryption (ABE) and Homomor-
phic Encryption (HE) in an overall architecture. This
further improves privacy preserving data access to
cancer registries for eligible researchers.
3 USE CASES
The following two use cases exemplify UPCARE’s
commitment to cutting-edge privacy-preserving tech-
nologies and show how privacy friendly access to can-
cer registry data is possible. In the first scenario, UP-
CARE employs Multiparty Homomorphic Encryp-
tion (HE) to enable secure collaboration among multi-
ple cancer research institutions. This approach allows
researchers to analyze encrypted data without com-
promising individual patient details, fostering collec-
tive insights while preserving confidentiality. In the
second use case, Attributed Based Encryption (ABE),
coupled with a distinct ”patient disagreement” at-
tribute, empowers patients to exercise granular con-
trol over the sharing of their personal cancer data.
This user-centric approach allows patients to make
purpose-specific decisions by attaching conditions to
their data. This ensures that the access, which was
granted by implicit patient consent in the beginning
can be withdrawn by any patient on request. To-
gether, these use cases demonstrate UPCARE’s prag-
matic yet innovative approach to unlocking the po-
tential of cancer registry data without compromising
individual privacy.
3.1 Use-Case 1 (UC-1): Secure
Multi-Institutional Data
Collaboration
This use case ensures that sensitive patient informa-
tion remains confidential throughout the collaborative
research process, allowing institutions to glean valu-
able insights collectively without compromising indi-
vidual privacy.
Data Collaboration Setup. Each participating
institution encrypts its cancer registry data using
some customized public key. The encrypted data
is shared with a processing entity responsible for
performing aggregated analyses, the query platform.
Secure Data Analysis. The query platform can
perform computations on the encrypted data without
decrypting it, thanks to HE. By leveraging HE, it can
perform computations directly on the encrypted data,
eliminating the need for decryption and safeguarding
individual patient details. This approach enables
aggregated statistical analyses and patterns to be
derived without ever exposing sensitive information.
Consequently, the platform remains completely
data-blind, upholding both patients’ rights as data
subjects and cancer registry managers’ confidential-
ity obligations. Aggregated statistical analyses and
patterns can be derived without exposing individual
patients’ details.
Privacy-Preserving Results. The final results are
sent back to the requesting researchers in an en-
crypted form. Only authorized researchers with the
necessary decryption keys can access the detailed re-
sults and decrypt them.
3.2 Use-Case 2 (UC-2): Patient-Driven
Data Sharing Control
This use case empowers patients to actively partic-
ipate in and control the sharing of their health data
for research purposes, contributing to a more trans-
parent and patient-centric approach within the cancer
research ecosystem.
Attribute-Based Encryption Setup. Patient
data is encrypted using attribute-based encryption.
Attributes include medical information, research cat-
egories, and a specific ”patient agreement” attribute.
User-Centric Data Sharing Decision. Patients are
presented with requests for data access, specifying
the intended purpose (e.g., research study, clinical
trial). Patients can attach conditions to their data,
such as time limitations or restricted access to specific
set of attributes.
Selective Data Disclosure. The encrypted data
may only be decrypted by researchers that meet the
specified, policy based conditions, i.e. where the
attributes match the policy. A Patient disagreement
attribute acts as an additional layer of patient access
control, ensuring data is accessed only with implicit
patient consent, and not with explicit patient dis-
consent.
SECRYPT 2024 - 21st International Conference on Security and Cryptography
54
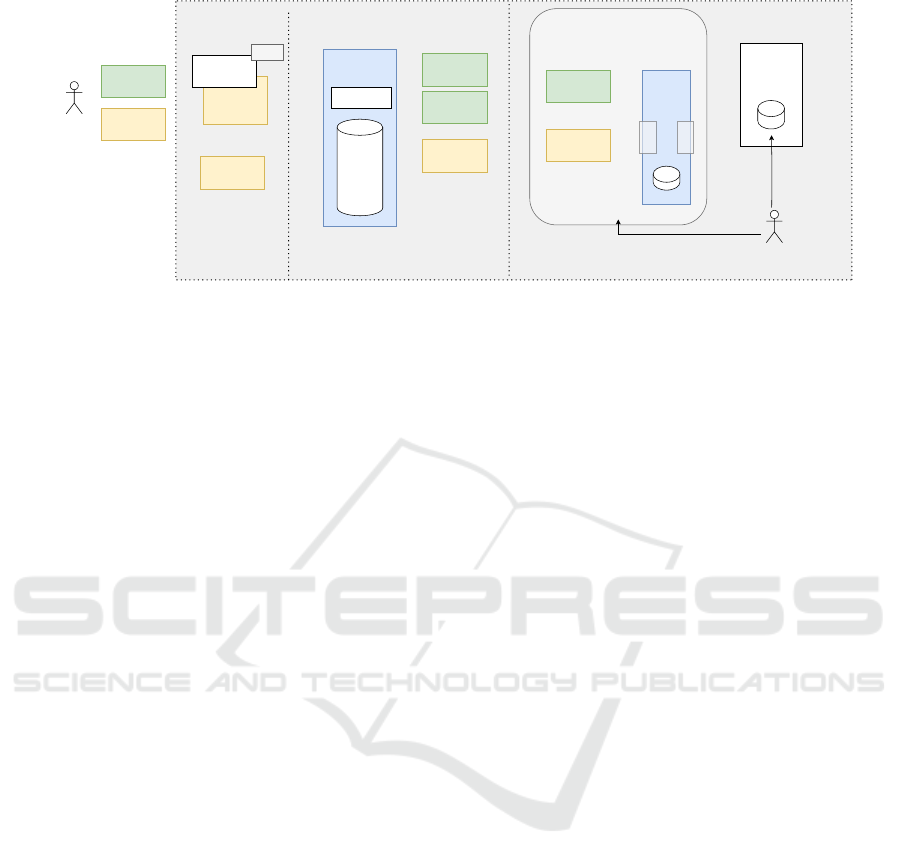
Users
Query result
cache (extra)
1. User Agent / Browser
2. Authorization layer
3. Query Platform
Privacy
Proxy
Existing
System
4. Privacy Proxies in Cancer Registries
Cancer Register
Data
management
Data Owner / Cancer Register
Operator
PP-Agg
Protection
Module
ABE encryption
module
PP-Agg
aggregation
module
Researcher
Portal, web front-
end
Key Server
ABE
ABE cache
(optional)
researchers
projects
queries
data owners
Ciphertext
computaion
and caching
PP-Agg Reveal
Module
ABE Decryption
Client
Management
System
Crypto
Backends
GUI
Auth* mechanism
with external IDP
FHIR Data
Storage
Module
Read API
Write API
Identity Provider
OpenID
Connect
Figure 1: The UPCARE platform components, composed of an authorization layer with a KS, a central QP and one or more
PP’s attached to existing cancer registries.
Audit Trail and Transparency. A transparent au-
dit trail is maintained, detailing when and how patient
data is accessed. Patients have visibility into who ac-
cessed their data and for what purpose.
4 UPCARE PLATFORM
The success of the UPCARE platform hinges on the
synergy of multiple pivotal components: the Query
Platform (QP), Key Server (KS), and Privacy Prox-
ies (PP). The QP serves as central point for re-
searchers and institutions, providing a secure inter-
face for querying and analyzing cancer registry data
without compromising individual privacy. Comple-
menting this, the KS acts as the guardian of Attributed
Based Encryption (ABE) key material, ensuring se-
cure and authorized access to encrypted data. Mean-
while, the PP are the owners of the data requested by
the researchers. In specific applications like those ex-
plored in UPCARE, these privacy proxies (Privacy
Proxies (PP)s) would be deployed by cancer reg-
istries, at either national or regional level, within their
data infrastructure. Functioning as vigilant interme-
diaries, they act as safe gates to the registries’ data
management systems, as illustrated in Figure 1. To-
gether, these components form a robust and intercon-
nected framework, embodying UPCARE’s commit-
ment to fostering collaborative research while safe-
guarding the sensitive health information that propels
advancements in cancer research. A graphical repre-
sentation of the foundational architecture can be seen
in Figure 1.
4.1 Components
The UPCARE platform is composed of three main
components, namely a Query Platform (QP), a Key
Server (KS) and two or more Privacy Proxies (PP).
4.1.1 Query Platform (QP)
The central QP is responsible for accepting UC-1 or
UC-2 query requests by authorized researchers via
a web interface. It is also responsible for presenting
the query result to the researcher. Its web interface
needs to be able to run the required cryptographic
algorithms and protocols required for each use
case. UC-1 requires the web interface to be able to
decrypt ABE ciphertexts with a given secret key. In
comparison, UC-2 requires the web interface to be
able to generate HE key material, as well as decrypt
HE ciphertexts.
UC-1 Queries. are accompanied by ABE policies.
An ABE policy is dynamically generated and fetched
from the KS, in form of a secret key, on each reload
of or interaction with the web interface. The secret
key is used to decrypt the results of the UC-1 query
request. The request itself is based on plaintext JSON
and is defined by a selection, a projection, an upper
limit and a choice of PPs. On the left side of the
UI the selection, projection and choice of PP can be
configured for each query. On the bottom of the UI
the temporary generated key material is seaming-less
integrated into the web interface.
UC-2 Queries. are based on Multiparty Homomor-
phic Encryption (MHE) (Mouchet et al., 2021). A
temporary researcher key pair, composed of public
and private keys, is first generated by deriving it
from a common random polynomial, given as refer-
ence string. The common random polynomial is pro-
vided by the QP. The public key is used to re-encrypt
(through so called key-switching) the final result of
the UC-2 query request to the researcher, so that he
can, later on, decrypt the data with the corresponding
private key. The request itself is based on plain-text
UPCARE: User Privacy-Preserving Cancer Research Platform
55

JSON and is defined by a selection, a projection, a
computation method, and an upper limit of results. As
we want to protect the privacy of the patients, the re-
searcher has no information and no choice on which
PPs are going to be used in a UC-2 query. Again,
on the bottom of the UI the temporary generated key
material is seaming-less integrated into the web inter-
face.
4.1.2 Key Server (KS)
The central KS is responsible for creating, editing
and holding access policies, as well as deriving
ABE secret keys from those stored policies. Since
we regenerate the key material before each query
dynamically, it is in our interest to create the policy
at the time the query is formulated on the QP. This
approach also allows short-term changes to patient
consent to be included in the key material. Therefore
the decision was made to integrate a Key Policy At-
tributed Based Encryption (KP-ABE) scheme in our
platform. The corresponding derived secret keys give
the researchers the right to decrypt those ciphertexts,
where the attributes exactly match the policy. The
component is part of the authorization layer, which
allows internal or external Identity providers to be
attached to the system, via OpenID Connect. This
allows researchers to identify themselves via foreign
institutions.
ABE attribute universe. The attribute universe
is defined by all PPs in the network during the
initialization phase. In this phase all i PPs query their
external cancer registry database for all available field
descriptors d
j
. The field descriptors are collected on
the QP, where their union U =
j=i
S
j=0
d
j
is computed.
The set of all descriptors U, together with a special
additional patient-centric disagreement attribute d
dis
form the attribute universe U.
Patient Disagreement. Based on legal decisions
made in Germany, each German cancer patient gives
implicit agreement for the usage of the available med-
ical data. A patient can afterwards opt-out by giv-
ing explicit disagreement. This workflow is now in-
tegrated into the UPCARE platform by storing the at-
tribute d
dis
in the context of a registered patient, on
his disagreement. The central KS allows a patient to
store d
dis
for a specific research project or researcher
in an additional web front-end. The complete work-
flow can be seen in Figure 2.
4.1.3 Privacy Proxies (PP)
Two or more distributed PP are responsible for giving
access to the underlying cancer register DBMS in a
privacy preserving manner, according to the given
use cases UC-1 and UC-2. Each PP is composed of
three internal modules, an ABE module, a Privacy
Preserving Aggregation (PPAG) module and a data
module. The data module is responsible for importing
and converting all external data sources, provided
by the cancer registries, into a uniform JSON data
format. The data is not imported in a bulk, but rather
is accessed dynamically, on request by the ABE
module or the PPAG module.
The ABE Module. The abe module is responsible
for encrypting the plain-text input data from the data
module using ABE. It does so by first parsing the
key-value data sets for a specific patient ,defined by
the selection criteria of the query. Followed by the
parsing is the encryption of all values with the key as
corresponding attribute. The ABE module therefore
uses the master key (downloaded from the key server
on initialization) to calculate ciphertext values for all
given keys. The attribute set is extended by d
dis
, if
and only if the patient has expressed his disagreement
on the key server interface via opting out. More
details on the flow of data in the overall architecture
is given in section 4.4.1.
The PPAG Module. is responsible for encrypting
the plain-text input data from the data module us-
ing a particular HE scheme named multi-party ho-
momorphic encryption (MHE) (Mouchet et al., 2021)
which allows the secure computation of their sum
and a collaborative decryption of this result encrypted
with a public key generated from multiple secret keys.
Thanks to the use of MHE, neither the researcher nor
any single privacy proxy can access the individual en-
crypted inputs before their aggregation. The aggre-
gation is performed collaboratively through the query
platform. Hence, the module allows a researcher the
dynamically generate a key pair in the browser and
use this key pair to generate the overall encryption key
and further aggregate encrypted data from the PPs in
a privacy preserving manner. More details on the flow
of data in the overall architecture is given in section
4.4.2.
4.2 Threat Model
The main threat model in the whole query process
is an honest-but-curious (HBC) QP who follows the
protocol, but carries out an insider attack on the data
flowing through itself. A successful insider attack on
the QP would compromise the data of all connected
SECRYPT 2024 - 21st International Conference on Security and Cryptography
56
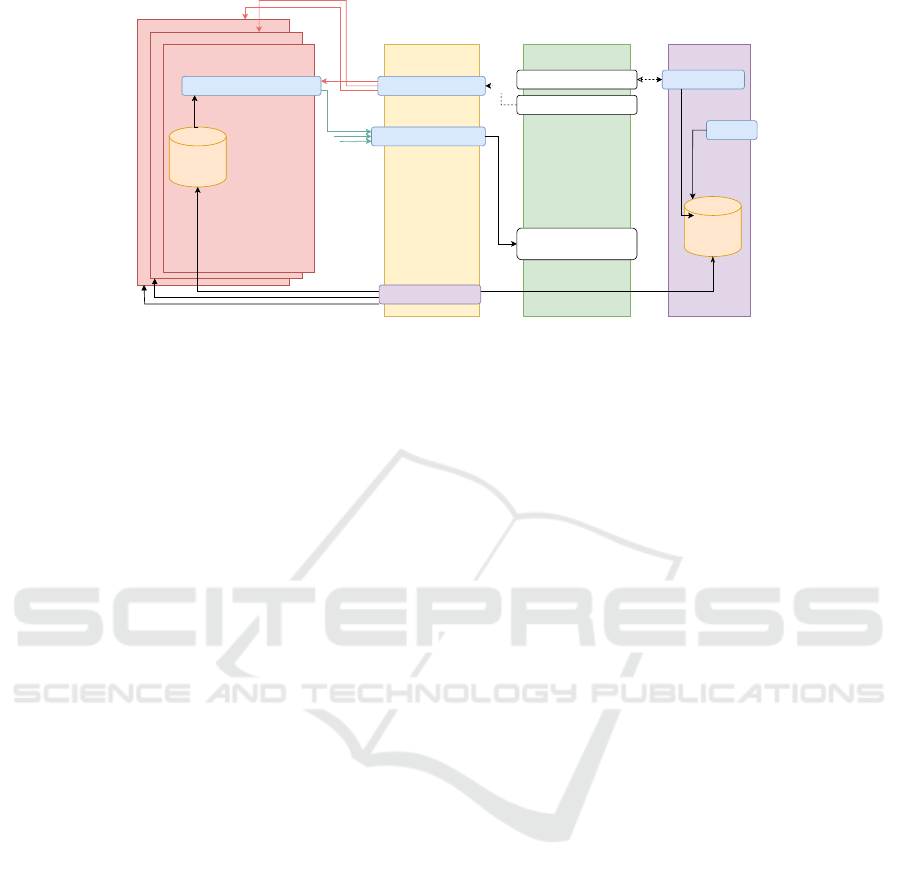
Privacy Proxy #n
UI Query InterfaceQuery PlatformPrivacy Proxy #1
forward Query
Cancer
Register
DBMS #1
[CTi] = Encrypt(PK, [key], [value])
Combine all [CTi]
Query
{"key": "value"}
Encrypted [CT
i
, ...]
Download SK
Query
WASM
Results ← Decrypt(SK ,[CTi])
Sample query: {
mode:
sum/avg/cnt,
selection:
(age > 50 ∧
BMI < 2.3) ∨
height > 1.75,
projection:
["blood-press",
"heart-rate"]
}
Privacy-Preserving ABE
Privacy Proxy #2
Keyserver
Attribute
DB
Disagree
getDatatypes
["key", "key2", ..., "key_n"]
get Datatypes
Query
Keygen(key1, ...keyn)
Figure 2: UC-1 in the Query phase. White boxes are coded in WebAssembly. Blue boxes are coded in Rust. Purple boxes are
coded in Go.
cancer registries, and thus would be the worst case.
PPs are responsible for ensuring the privacy protec-
tion of the cancer patients whose data they own. Be-
cause of this, they have no reason to deviate from the
protocol or exchange information with other actors.
Consequently, PPs are also considered HBC. The re-
searchers use the platform to perform some research
over the cancer data. We assume that researchers al-
ways act according to their best self-interest. This
means that researchers can deviate from the protocol
if this allows them to have access to more informa-
tion. For this reason, the researcher is considered ma-
licious. In addition to this setting, we consider that
PPs can not communicate with each other, privately,
and that the query platform can collude with the Re-
searcher.
4.3 Implementation
Our envisioned architecture ensures a secure, effi-
cient, and privacy-preserving environment for cancer
research, aligning with the goals of UPCARE. The
architecture leverages Rust’s performance, memory
safety, and concurrency features while incorporating
state of the art cryptographic techniques to protect
sensitive patient data. Its main features are:
Front-End. The user interface where researchers
and authorized patients interact with the system.
The UI was built using sveltekit, HTML, CSS, and
JavaScript, with additional Rust-generated (ABE) and
Go-generated (HE) browser-capable cryptographic
code (each compiled using WebAssembly).
Rocket Servers. The core back-end APIs (of
KS, QP and PP) are implemented in Rust using
the Rocket web framework. Each back-end API
manages incoming HTTP requests and orchestrates
communication from the front-end up until the PPs.
Every API handles a different, specific action from
authentication and query processing to response
generation.
Query Platform. A module within the Rocket
server responsible for processing and executing
queries on the cancer registry data. It allows either
personal ABE or statistical HE queries. The QP uti-
lizes secure multiparty homomorphic encryption to
perform computations on encrypted data or attribute
based encryption to allow encrypted data access.
The PPAG module implements privacy-preserving
algorithms for data analysis, currently sum, average
and count are available as options.
Key Server. The KS manages ABE keys used for
the decryption of sensitive, i.e. personal patient data.
It ensures the secure key distribution and thus allows
access control. The KS implements OAuth 2.0 for
secure authorization and data access.
Privacy Proxies. The PPs are intermediate compo-
nents between the cancer registry data sources and
the QP. They implement cryptographic techniques to
preserve individual patient privacy during data anal-
ysis or in the case of personal data access request by
a researcher. The ABE module was implemented in
Rust, while the HE module was implemented in Go.
They both act as guardians of sensitive information,
ensuring that only aggregated and privacy-preserving
HE encrypted statistical results or either attribute
based encrypted patient record data sets are exposed
to the query platform.
Logging and Auditing. The platform implements
logging and auditing mechanisms to track researcher
activities and data access on each of its components.
This facilitates transparency and compliance with
privacy regulations.
UPCARE: User Privacy-Preserving Cancer Research Platform
57
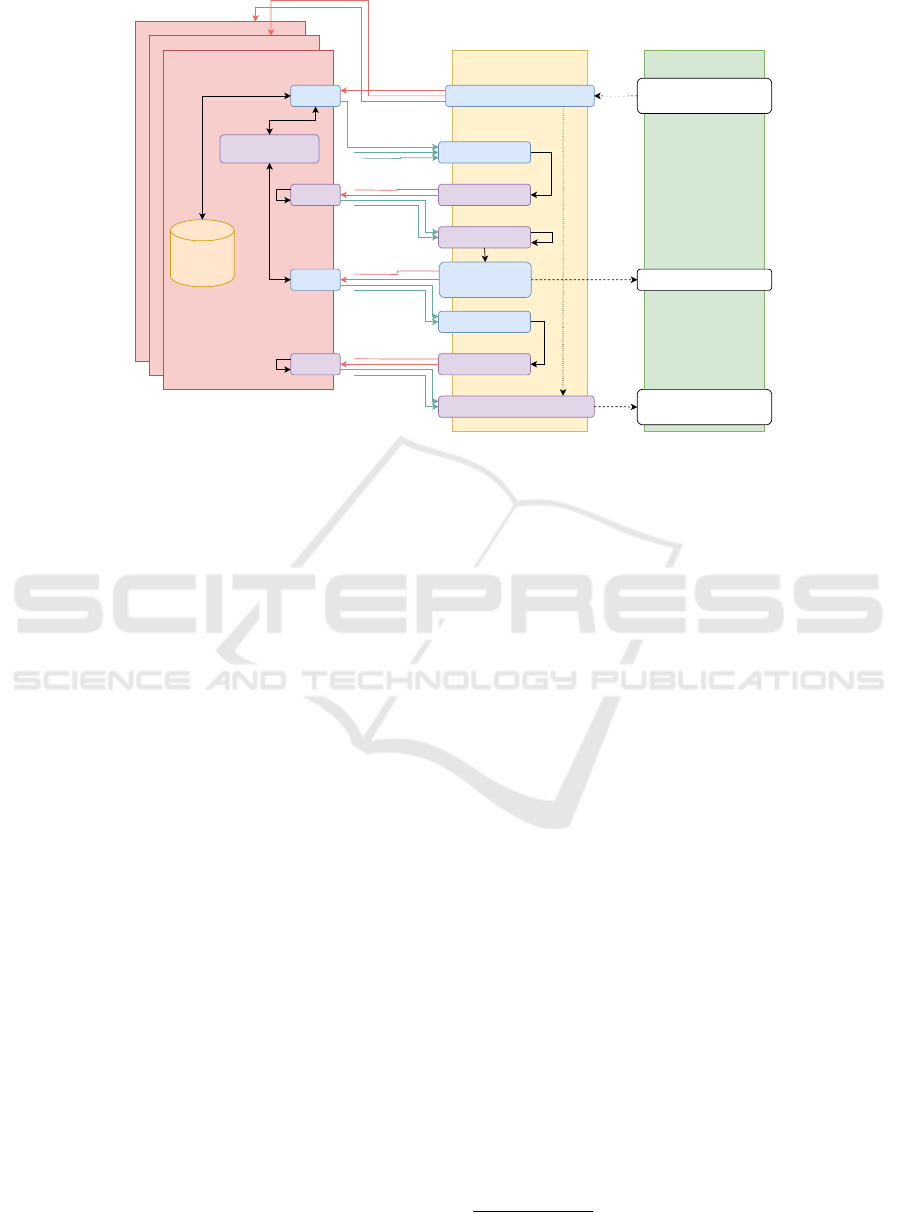
Privacy Proxy #n
UI Query InterfaceQuery PlatformPrivacy Proxy #1
store temporary PubK
k-anon Agg. of [CTi]
Cancer
Register
DBMS #1
store [sum
i
]
Fetch [CTi]
Query
[CTs] = Encrypt([sum
i
])
[CTi] = Encrypt([n
i
])
Query: [{sum
i
, n
i
}, ...]
Encrypted [CT
i
, ...]
KeySwitch
KeySwitch to
QP's tmp key
WASM
PrivK/PubK ← gen_keypair()
Query,
PubK
j = Decrypt(QPK,
[CTi]+... )
No Resultswith [CTs]
Continue .. Fetch sum
i
Fetch [CTs]
Encrypted [sum
i
, ...]
k-anon Agg. of [CTs]KeySwitch
KeySwitch to PubK
KeySwitch to temp. PubK of [CTs]
WASM
Results ← Decrypt(PrivK ,CTs)
j ≥ k .. continue,
j < k .. halt
Sample query: {
mode:
sum/avg/cnt,
selection:
(age > 50 ∧
BMI < 2.3) ∨
height > 1.75,
projection:
["blood-press",
"heart-rate"]
}
Privacy-Preserving HE
Privacy Proxy #2
PubK
[CTs]
[ ]
Figure 3: UC-2 in the Query phase. White boxes are coded in WebAssembly. Blue boxes are coded in Rust. Purple boxes are
coded in Go.
External Interfaces. Each component provides an
API for integration with external systems or research
applications. This procedure adheres to industry
standards and interoperability protocols.
Adaptability. The data import module allows to
import data from various sources like NoSQL, SQL,
CSV and so on. Therefore the system can be adopted
to various cancer databases, as long as a mapping
between input data format and the format standard,
based on FHIR, can be built. More details on FHIR
usage can be found in Section 5.1.
4.4 Integration
Both privacy-preserving components, the ABE and
PPAG modules, are seamlessly integrated into both
the individual PPs and the overall system architec-
ture through a standardized query protocol based
on JSON. Before responding to user queries, both
modules require an initialization phase upon system
startup, followed by a dedicated query phase where
they actively process queries.
4.4.1 ABE Flow
Initialization phase A master secret key and a
public key are generated on the KS based on all FHIR
data types available on the connected cancer registry
databases, which are queried using the corresponding
PP’s. In addition to the FHIR attributes, an attribute
”disagreement” is added to the set of attributes, in
order to represent patient disagreement. The public
key is distributed via a secure channel
4
to the PP’s,
where they are used to encrypt patient data from the
connected cancer registry database. A local database
is flushed and initialized on the KS, where user
disagreement can be stored permanently.
Query Phase. Once the key generation is finished,
the researcher sends a query to the query platform,
which forwards it to each of the PP’s. Each proxy pp
i
queries the corresponding data from the connected
cancer registry dbms and then encrypts the result us-
ing the public key PK, obtained from the KS and the
value of the current key as the current attribute. The
encrypted results are collected and combined on the
query platform before they are handed back to the UI
query interface, where a eligible researcher is able the
decrypt the results directly inside his browser, with
the help of his secret key SK and the decryption algo-
rithm compiled in Wasm. This exact flow can be seen
in figure 2.
4.4.2 PPAG flow
In this section, we describe the PPAG flow which
makes use of a dedicated multi-party homomorphic
encryption (MHE) scheme (Mouchet et al., 2021)
inspired by the work in (Bozdemir et al., 2023).
Initialization. This phase consists of the generation
of the common public key used for encryption by all
privacy proxies participating in the actual aggregation
operation. To collaboratively generate the public
4
Protected by the standard https protocol
SECRYPT 2024 - 21st International Conference on Security and Cryptography
58

key derived from MHE (Mouchet et al., 2021),
each party generates a public key from their secret
key (by encrypting a zero-message) and an initially
distributed/agreed common reference string. The
query platform performs the aggregation of these
public keys (a simple sum) in order to obtain the
common public key which is then distributed to
privacy proxies.
Query Phase. Once the key generation is finished,
the researcher sends the query to the query plat-
form which forwards it to the privacy proxies. Each
proxy pp
i
first determines how many instances of
its database it is gonna respond with. This num-
ber c
i
is encrypted with the public key and sent to
the query platform. The query platform aggregates
these numbers and the obtained total number is j de-
crypted and compared with a threshold value k. Only
if this threshold is exceeded, then the actual aggrega-
tion phase can start. Each proxy encrypts its data with
the common public key and sends it to the query plat-
form which aggregates these data similar to the previ-
ous aggregation operation. This sum is further resent
to the proxies for collaborative key switching so that
the result is encrypted with the researcher’s public key
at the end. This exact flow can be seen in figure 3.
4.4.3 Common Query Protocol
The queries of both uses cases are covered by a com-
mon, JSON-based query language. At first, a request
is generated on the UI, which is then added to either
a personal or a statistical query. Each request is trans-
formed into a Job on the QP and is stored locally in a
database for further procession by the QPs. As soon
as there is a new entry in the local database, the PP’s
start to work on the request. In a UC-1 query the
PP’s encrypt the corresponding query results using the
ABE key material. In a UC-2 query the PP’s encrypt
the corresponding query results using the HE key ma-
terial. After finishing their work, they write back the
result into the Job database. As soon as the job is fin-
ished, the results are streamed back to the researcher.
5 EVALUATION
The evaluation of the Privacy-Preserving Cancer Re-
search Platform (UPCARE) is paramount to ascertain
its effectiveness in addressing the complex challenges
inherent in collaborative medical research while safe-
guarding patient privacy. In this section, we present a
comprehensive evaluation of UPCARE, focusing on
its utilized dataset, its query performance, the sys-
tem scalability, a security analysis and additionally
compliance with regulatory frameworks. By scruti-
nizing the platform’s capabilities and limitations, we
aim to provide insights into its real-world applicabil-
ity, resilience to potential threats, and ability to meet
the evolving needs of cancer research stakeholders.
Through rigorous testing and analysis, we endeavor
to validate the integrity of UPCARE as a robust and
privacy-conscious solution poised to drive innovation
in the field of cancer research while maintaining the
highest standards of data privacy and security.
5.1 Dataset Description
To address a future compatibility question a Fast
Healthcare Interoperability Resource (FHIR) (Ayaz
et al., 2021) format of Health Level 7 Standards Or-
ganization (HL7) was used to store and request the
patient data. The Format is supported by FHIR Data
Storage module based on the FIDES System (Bun-
desdruckerei, 2018) as a native format such that no
additional conversion for data processing is required.
The patient data are high sensitive data. For a pur-
pose of test and proof of concept a generated patient
dataset of Synthea (Walonoski et al., 2018) was used
in the project. The dataset contains one million syn-
thetic patient medical records, encoded in HL7 FHIR
including demographic information, medical history,
medications, and lab results.
5.2 Security Analysis
Our purposed UPCARE solution solves the problem
of an (hbc) QP, by encrypting all plaintext data on
each PP, before arriving at the QP. Even when calcu-
lations are done on the QP, as in UC-2, QP is not able
to leak the corresponding, underlying plaintext data.
Both personal as well as statistical data are protected
from leaking at a central platform through means of
encryption. Indeed, in UC-2, thanks to MHE, nei-
ther the QP and the researcher nor any single PP can
access the encrypted data before aggregation. The ag-
gregation is performed collaboratively between PPs
through the QP. At the end of the protocol, a collab-
orative key switching is performed on the aggregated
data to enable its decryption by the actual researcher,
only. Regarding malicious insider attacks on the lo-
cal PP at the cancer registries, our proposed solution
does unfortunately not protect from data leakage, as
the medical plaintext data must be readable by the PP
for encryption. As previously mentioned these PP are
considered HBC, only.
UPCARE: User Privacy-Preserving Cancer Research Platform
59

5.3 Performance
In this section we evaluate the performance of both
Queries, i.e. UC-1 which involves secure data re-
trieval based on specific patient attributes and UC-2
which involves aggregated data analysis and statisti-
cal computations on encrypted data.
5.3.1 Methodology
The following methodology was used to measure
performance Initially, execute multiple UC-1 queries
across a range of test scenarios, varying the number of
attributes and the complexity of search criteria. Then
measure the query response time from the initiation of
the query to the retrieval of results, capturing the time
taken for data processing, encryption/decryption, and
transmission. Execute multiple UC-2 queries using
sample datasets of varying sizes, encompassing di-
verse research categories and demographic attributes.
Finally, Measure the query response time for the inte-
grated statistical operations.
5.3.2 Results
The following results where thereby gathered regard-
ing UC-1 queries: The performance of a UC-1 query
exhibits a positive correlation with the number of at-
tributes and the complexity of search criteria.On aver-
age, queries with fewer attributes and simpler search
criteria demonstrate faster response times, typically
ranging from 5 seconds to 25 seconds. This be-
haviour can be seen in figure 4 However, as the num-
ber of attributes and search complexity increases, the
query response time may exceed 25 seconds, particu-
larly in scenarios involving large datasets or complex
attribute-based access policies. The response time can
be optimized by an additional ABE ciphertext cache
on the QP.
Regarding UC-2 queries, the following results
were collected: This query demonstrates competitive
performance in conducting aggregated data analysis,
with response times typically ranging from 2000 mil-
liseconds to 15000 milliseconds. The query response
time exhibits scalability with dataset size, as larger
datasets incur marginally longer processing times due
to increased computational complexity. Complex
statistical computations, such as correlation analy-
sis across multiple attributes, may result in slightly
longer response times, particularly in datasets with
extensive attribute diversity.
Factors such as encryption/decryption overhead
and data transmission latency contribute to fluctu-
ations in query response time across different sce-
narios. Optimization strategies, such as indexing,
caching, and parallel processing, may be explored to
mitigate latency and enhance the efficiency of secure
data retrieval operations.
5.3.3 Conclusion
The performance of UC-2 queries underscores the
need for efficiency in homomorphic encryption opera-
tions. Despite the computational overhead associated
with encrypted data processing, UC-2 queries main-
tain satisfactory response times, facilitating timely in-
sights and decision-making in cancer research. Fur-
ther optimization opportunities, such as algorithmic
refinement and distributed computing strategies, may
be explored to enhance the scalability and efficiency
of aggregated data analysis operations. The evalua-
tion of query performance in UPCARE highlights the
platform’s capability to deliver secure and efficient
data retrieval and analysis functionalities. By sys-
tematically analyzing the response times of UC-1 and
UC-2 queries across diverse scenarios, we gain valu-
able insights into the platform’s performance charac-
teristics and identify opportunities for optimization.
Moving forward, continued refinement and optimiza-
tion efforts will be essential to ensure that UPCARE
maintains high-performance standards while fulfilling
the evolving demands of privacy-preserving cancer
research.
6 USE CASE EVALUATION
Cancer registration is an essential component of the
health care system and provides valuable insights into
the prevalence, treatment and outcomes of oncolog-
ical diseases. The comprehensive understanding of
cancer epidemiology and the evaluation of oncologi-
cal care heavily rely on the systematic collection and
analysis of data. In Germany, the data on oncolog-
ical care is reported to the German state cancer reg-
istries by the medical facilities involved in the treat-
ment according to the standardized oncological basic
data set (Tumorzentren eV, 2015). Various essential
pieces of information are recorded, including patient
details, diagnosis, surgical interventions, radiother-
apy, systemic therapy, and the course of the disease
leading to death (oBDS2021, 2021). Consequently,
the cancer registry is a systematic collection of infor-
mation in the form of a database on tumor diseases.
The evaluation of the use cases for the platform is es-
sentially based on the expertise of the Cancer Registry
Rhineland-Palatinate and the Rhineland-Palatinate le-
gal framework for the provision of personal patient
data from the cancer registry for research purposes.
SECRYPT 2024 - 21st International Conference on Security and Cryptography
60

2 4 6 8 10 2 4 6 8 10 2 4 6 8 10 2 4 6 8 10 2 4 6 8 10
0
5
10
15
20
25
30
10 Patients 20 Patients 30 Patients 40 Patients 50 Patients
Time in seconds
UC-1 Pat. query
UC-2 Stat. query
Figure 4: Performance, measured as query response time in seconds.
6.1 Current Situation
Cancer registries offer epidemiological and clinical
evaluations for all tumor entities, facility-related eval-
uations as well as individual evaluations for spe-
cific inquiries. For research purposes, aggregated,
pseudonymized or personal data can be provided for
precisely defined scientific oncological questions re-
lating to cancer prevention, cancer care and research
into the causes of cancer (KRRLP, 2012). Requesting
and providing data for research purposes is a man-
ual process, as there is currently no standardized, se-
cure request platform for cancer registry data for re-
searchers. The application is subject to an internal
formal and content-related review. In the case of a re-
quest for personal data, additional authorisations from
the Ministry of Science and Health, an ethics vote
and, if necessary, a hearing of the State Commissioner
for Data Protection and Information Security are re-
quired (LKRG215, 2015).
The possibility of requesting personal patient data for
research purposes has rarely been utilized to date. The
requested data is provided in a secure way. If personal
patient data is requested from several German can-
cer registries, each registry enables data access via its
own channel. A separate application must be submit-
ted to the respective registry for each cancer registry
from which data is requested. In such cases, several
ethics committees are involved, each overseeing the
ethical considerations for the registry in question.
As can be seen above, obtaining personal patient
data for a research project is a time-consuming pro-
cess for the applicant. Especially when data is re-
quested from several cancer registries.
6.2 Data Query via UPCARE
The use cases were evaluated from the perspective of
the cancer registry with regard to the following as-
pects:
Data Privacy and Security. Even with aggregated
data, there is a certain risk of drawing conclusions
about individual patients. Numerous factors con-
tribute to the creation of a high re-identification
potential. These factors may relate to data character-
istics, such as the uniqueness of feature expressions
and their combinations, as well as temporal and
spatial information. However, the likelihood of
successful re-identification depends on external
knowledge, whether publicly or non-publicly avail-
able, along with other influencing factors (Drechsler
and Pauly, 2024). By using HE, it is possible to
securely aggregate data without exposing individual
patient details. The encrypted aggregated data can
be analyzed to derive general patterns or statistics,
reducing the risk of re-identification of specific cases
based on unique characteristics. From a cancer
registry perspective, this is a big step towards patient
privacy. ABE leads the cancer registry to a standard-
ized data protection and data security concept for
the provision of personal patient data for research
purposes.
Accessibility. UPCARE provides researchers with
easy and secure access to a diverse range of cancer
data. The platform can be accessed at any time via a
web interface and enables secure data queries from
the connected cancer registries at a central hub.
Feasibility. The platform makes it possible to query
whether there is enough patient data for a study. The
applicant formulates the characteristics of the enquiry
and receives an aggregated result, for example the
number of patients, their gender and age. If the result
is sufficient for a cohort, the applicant can request the
patient data from the selected cancer registries via
the platform. In addition, by combining the cancer
registries via the platform, there is a good chance of
collecting sufficient data on rare tumors.
Interoperability. HL7 FHIR
5
is the chosen stan-
dard for communication between the platform and
5
https://www.hl7.org/fhir/
UPCARE: User Privacy-Preserving Cancer Research Platform
61

the cancer registries. The FHIR implementation
is currently based on FHIR resources generated
by Synthea (Walonoski et al., 2018). This is a
synthetic patient generator that produces realistic but
completely fictitious health data.
7 CONCLUSIONS
In conclusion, the Privacy-Preserving Cancer Re-
search Platform (UPCARE) represents a significant
advancement in the field of cancer research, empha-
sizing the paramount importance of preserving pa-
tient privacy while fostering collaborative data anal-
ysis. Through the integration of advanced crypto-
graphic techniques such as ABE and HE, UPCARE
ensures that sensitive health data remains confidential
and secure throughout the research process. The fine-
grained access control facilitated by ABE empowers
patients to maintain control over their data, while HE
enables secure and private computations on encrypted
data, safeguarding individual privacy. Moreover, the
automation of data processing and the use of cryptog-
raphy not only enhance privacy but also expedite the
speed of data requests and analysis, facilitating faster
insights and discoveries in cancer research. In this
way, data privacy-friendly analysis methods are de-
veloped and integrated into the architecture of cancer
registries, which, on the one hand, maintain a high
level of data protection and, on the other hand, pro-
vide meaningful findings for cancer research.
7.1 Future Work
Identifying avenues for future work is crucial for ad-
vancing the Privacy-Preserving Cancer Research Plat-
form (UPCARE) and addressing emerging challenges
in the field. Some potential directions for future work
include: Exploring advanced privacy-preserving tech-
niques, such as differential privacy or secure multi-
party computation, to further fortify data protection
and confidentiality mechanisms within UPCARE. In-
vestigate ways to integrate these techniques seam-
lessly into the existing UPCARE architecture without
compromising performance. Embrace emerging tech-
nologies such as blockchain and federated learning
to augment data security, transparency, and collabo-
ration within the UPCARE cancer research ecosys-
tem. Investigate opportunities to leverage these tech-
nologies to enhance data provenance, traceability, and
verifiability while preserving patient privacy. Con-
tinuously optimize UPCARE’s scalability and per-
formance to accommodate the growing volume and
complexity of cancer registry data. Explore meth-
ods to streamline data processing, improve query
response times, and reduce computational overhead
while maintaining privacy guarantees.
By embarking on these future work initiatives,
UPCARE can evolve into a dynamic and adaptive
platform that not only advances cancer research but
also serves as a model for privacy-preserving data
sharing and collaborative innovation in healthcare.
ACKNOWLEDGEMENTS
This work was supported by the MESRI-BMBF
French-German joint project UPCARE (ANR-20-
CYAL-0003-01, 16KIS1383K).
REFERENCES
Agrawal, R. and Srikant, R. (2000). Privacy-preserving data
mining. In Proceedings of the 2000 ACM SIGMOD in-
ternational conference on Management of data, pages
439–450.
Agrawal, S. and Chase, M. (2017). Fame: fast attribute-
based message encryption. In Proceedings of the 2017
ACM SIGSAC Conference on Computer and Commu-
nications Security, pages 665–682.
Ayaz, M., Pasha, M. F., Alzahrani, M. Y., Budiarto, R., and
Stiawan, D. (2021). The fast health interoperability
resources (fhir) standard: systematic literature review
of implementations, applications, challenges and op-
portunities. JMIR medical informatics, 9(7):e21929.
Barua, M., Liang, X., Lu, R., and Shen, X. (2011). Espac:
Enabling security and patient-centric access control
for ehealth in cloud computing. International Jour-
nal of Security and Networks, 6(2-3):67–76.
Bethencourt, J., Sahai, A., and Waters, B. (2007).
Ciphertext-policy attribute-based encryption. In 2007
IEEE symposium on security and privacy (SP’07),
pages 321–334. IEEE.
Bozdemir, B.,
¨
Ozdemir, B. A., and
¨
Onen, M. (2023).
PRIDA: PRIvacy-preserving Data Aggregation with
multiple data customers. IACR cryptology e-print
archive, Paper 2024/074.
Bramm, G., Gall, M., and Sch
¨
utte, J. (2018). Blockchain-
based distributed attribute based encryption. In Pro-
ceedings of the 15th International Joint Conference on
e-Business and Telecommunications (ICETE 2018)-
Volume, volume 2, pages 99–110.
Bundesdruckerei (2018). From the almighty
administrator to the self-determined user.
https://www.bundesdruckerei.de/en/whitepaper/
download/2835/Whitepaper-Fides.pdf.
Cheon, J. H., Kim, A., Kim, M., and Song, Y. (2017). Ho-
momorphic encryption for arithmetic of approximate
numbers. In Advances in Cryptology–ASIACRYPT
2017: 23rd International Conference on the Theory
SECRYPT 2024 - 21st International Conference on Security and Cryptography
62

and Applications of Cryptology and Information Se-
curity, Hong Kong, China, December 3-7, 2017, Pro-
ceedings, Part I 23, pages 409–437. Springer.
Drechsler, J. and Pauly, H. (2024). Das reidentifika-
tionspotenzial von strukturierten gesundheitsdaten.
Bundesgesundheitsblatt - Gesundheitsforschung -
Gesundheitsschutz, 67(2):164–170.
Gentry, C. (2009). Fully homomorphic encryption using
ideal lattices. In Proceedings of the forty-first annual
ACM symposium on Theory of computing, pages 169–
178.
Geva, R., Gusev, A., Polyakov, Y., Liram, L., Roso-
lio, O., Alexandru, A., Genise, N., Blatt, M.,
Duchin, Z., Waissengrin, B., et al. (2023). Col-
laborative privacy-preserving analysis of oncologi-
cal data using multiparty homomorphic encryption.
Proceedings of the National Academy of Sciences,
120(33):e2304415120.
KRRLP (2012). Bericht des krebsregisters rheinland-
pfalz 2022/23. https://www.krebsregisterrlp.de/
fileadmin/user upload/dokumente/04 Ver%C3%
B6ffentlichungen/2023/KRB2022 Webversion 01.
pdf.
Lewko, A. and Waters, B. (2011). Decentralizing attribute-
based encryption. In Annual international conference
on the theory and applications of cryptographic tech-
niques, pages 568–588. Springer.
LKRG215 (2015). Landeskrebsregistergesetz - § 12 - abgle-
ichung und
¨
Ubermittlung personenidentifizierender
daten. https://www.landesrecht.rlp.de/bsrp/document/
jlr-KrebsRegGRP2015pP12.
Mouchet, C., Troncoso-Pastoriza, J., Bossuat, J., and
Hubaux, J. (2021). Multiparty Homomorphic Encryp-
tion from Ring-Learning-with-Errors. In Privacy En-
hancing Technologies Symposium (PETS).
M
¨
uller, S., Katzenbeisser, S., and Eckert, C. (2009). Dis-
tributed attribute-based encryption. In Information
Security and Cryptology–ICISC 2008: 11th Interna-
tional Conference, Seoul, Korea, December 3-5, 2008,
Revised Selected Papers 11, pages 20–36. Springer.
Narayan, S., Gagn
´
e, M., and Safavi-Naini, R. (2010). Pri-
vacy preserving ehr system using attribute-based in-
frastructure. In Proceedings of the 2010 ACM work-
shop on Cloud computing security workshop, pages
47–52.
oBDS2021 (2021). Einheitlicher onkologischer basisdaten-
satz 2021. https://basisdatensatz.de/basisdatensatz.
Pournaghi, S. M., Bayat, M., and Farjami, Y. (2020).
Medsba: a novel and secure scheme to share medi-
cal data based on blockchain technology and attribute-
based encryption. Journal of Ambient Intelligence and
Humanized Computing, 11:4613–4641.
Sahai, A. and Waters, B. (2008). Revocation systems with
very small private keys. Eprint2008.
Son, Y., Han, K., Lee, Y. S., Yu, J., Im, Y.-H., and Shin,
S.-Y. (2021). Privacy-preserving breast cancer re-
currence prediction based on homomorphic encryp-
tion and secure two party computation. Plos one,
16(12):e0260681.
Tumorzentren eV, A. D. (2015). Gesellschaft der epidemiol-
ogischen krebsregister in deutschland ev einheitlicher
onkologischer basisdatensatz. Verf
¨
ugbar: http://www.
tumorzentren. de/onkol-basisdatensatz. html [Stand:
19.06. 2015] View in Article.
Van Dijk, M., Gentry, C., Halevi, S., and Vaikuntanathan, V.
(2010). Fully homomorphic encryption over the inte-
gers. In Advances in Cryptology–EUROCRYPT 2010:
29th Annual International Conference on the The-
ory and Applications of Cryptographic Techniques,
French Riviera, May 30–June 3, 2010. Proceedings
29, pages 24–43. Springer.
Walonoski, J., Kramer, M., Nichols, J., Quina, A., Moe-
sel, C., Hall, D., Duffett, C., Dube, K., Gallagher, T.,
and McLachlan, S. (2018). Synthea: An approach,
method, and software mechanism for generating syn-
thetic patients and the synthetic electronic health care
record. Journal of the American Medical Informatics
Association, 25(3):230–238.
Yue, X., Wang, H., Jin, D., Li, M., and Jiang, W. (2016).
Healthcare data gateways: found healthcare intelli-
gence on blockchain with novel privacy risk control.
Journal of medical systems, 40:1–8.
UPCARE: User Privacy-Preserving Cancer Research Platform
63
