
Fitting Tree Model with CNN and Geodesics to Track Blood Vessels in 2D
Medical Images and Application to Ultrasound Localization Microscopy
Data
Th
´
eo Bertrand
a
and Laurent D. Cohen
b
CEREMADE, UMR CNRS 7534, University Paris Dauphine, PSL, France
Keywords:
Geodesic Methods, CNN, Vessel Tracking, ULM Imaging, Eye Fundus.
Abstract:
Segmentation of tubular structures in vascular imaging is a well studied task, although it is rare that we try
to infuse knowledge of the tree-like structure of the regions to be detected. Our work focuses on detecting
the important landmarks in the vascular network (via CNN performing both localization and classification
of the points of interest) and representing vessels as the edges in some minimal distance tree graph. We
leverage geodesic methods relevant to the detection of vessels and their geometry, making use of the space of
positions and orientations so that 2D vessels can be accurately represented as trees. We build our model to
carry tracking on Ultrasound Localization Microscopy (ULM) data, proposing to build a good cost function
for tracking on this type of data. We also test our framework on synthetic and eye fundus data. Results show
that the Orientation Score built from ULM data yields good geodesics for tracking blood vessels but scarcity
of well annotated ULM data is an obstacle to the localization of vascular landmarks.
1 INTRODUCTION
Ultrasound Localization Microscopy is a quite recent
imaging technique that allows users to bypass the
compromise between precision and depth of penetra-
tion in ultrasound imaging.
It allows one to make highly resolved images of
the vascular network deeper in the skin tissues with
the help of micro bubbles used as contrast agents. We
refer to (Couture et al., 2018) for an overview of the
super resolution method.
In the present work, we introduce a new workflow
for complete end-to-end detection of vascular struc-
tures on ULM images, using deep learning to detect
landmarks (see Figure 1) allowing tracking of ves-
sels as edges in a tree graph with landmarks as ver-
tices. Our approach differs from classical Percep-
tual Grouping for blood vessel tracking as performed
in (Bekkers et al., 2018; Benmansour and Cohen,
2009). Indeed we are trying to take advantage of long
geodesics tracking blood vessels across an image, that
should behave well given the amount of literature on
the subject. While Perceptual Grouping usually fo-
cuses on computing short geodesics between close
a
https://orcid.org/0009-0005-7846-4985
b
https://orcid.org/0000-0002-3940-645X
points spread across the vessel network, we aim to
compute few long geodesics between key landmarks
of the vasculature. We try to take advandatage of the
information specifically given by ULM imaging, but
it must be noted that it is possible to adapt on other
types of images, for instance eye fundus images ob-
tained via direct photography. To do so, one may need
to evaluate local orientation information as we will
see in section 3. From a 2D image it can be done
using Orientation Scores (Duits et al., 2007) or sim-
ilar transforms such as the ones presented in (Zhang
et al., 2016) to lift a 2D image set in the plane to the 3-
dimensional space of positions and orientations. Raw
ULM data consists in ultrasound signals for imaging
blood vessels indirectly. Indeed, instead of directly
viewing the response of the tissues, it is the non-linear
response of microbubbles used as contrast agents that
is recovered and then treated to recover the position of
the bubbles. The set of the position of the microbub-
bles transported in the blood vessels allows one to
recover a highly-resolved image of the vascular tree
by projecting back on a grid as fine as needed (al-
though limited by the number of detected microbub-
bles). Some methods (see for instance (Song et al.,
2018)) then use the detected points to try and recover
trajectories of microbubbles along successive images,
thus allowing one to interpolate along those trajecto-
44
Bertrand, T. and Cohen, L.
Fitting Tree Model with CNN and Geodesics to Track Blood Vessels in 2D Medical Images and Application to Ultrasound Localization Microscopy Data.
DOI: 10.5220/0012723900003720
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 4th International Conference on Image Processing and Vision Engineering (IMPROVE 2024), pages 44-51
ISBN: 978-989-758-693-4; ISSN: 2795-4943
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
ries and infer even more points in order to provide
finer images. The data available and used in this work
is composed of detected trajectories of microbubbles.
We want to exploit previously recomposed trajecto-
ries by taking into account the additional information
contained in said trajectories, which are not only point
clouds but have velocity and temporal information.
Detection, segmentation or tracking tasks on med-
ical images are widely studied problems.
The contributions of our work include:
• working with ULM data, defining a Riemannian
metric in order to track vessels in ULM images,
• dealing with scarcity of data : 2 different high res-
olution images to make both the training and val-
idation set,
• carrying out detection of vascular landmarks in
such context,
• fitting a tree model with geodesics as edges to
take into account geometric and topological as-
pects into the tracking, thus investigating the effi-
ciency of using the tree-like nature of vasculature
to perform the tracking,
• comparing results on synthetic data (hand-made
black and white images to fit the framework used
for ULM data, i.e. few big images) and eye fundus
images (more images, but smaller).
Vessel segmentation is usually performed by comput-
ing scores of vesselness on the image, see the semi-
nal work (Frangi et al., 1998) and more recent work
(Jerman et al., 2016). The main idea in these works
being that high vesselness corresponds to regions in
the image where one orientation is dominant. Vessel-
ness is then defined as a function of the eigenvalues
of the Hessian (in dimension 2, one eigenvalue being
significantly higher than the other indicates a tubular
region). Modern methods of transposing the image in
a higher dimensional setting of Position-Orientation
space were used in (Zhang et al., 2016).
Other methods of vascular segmentation include
machine and deep learning methods that have become
accessible thanks to the availability of annotated data.
We can cite for instance (Oliveira et al., 2018) that
uses a fully-convolutional U-net for segmentation task
on eye fundus images.
A few works have already approached the prob-
lem of localizing vascular landmarks in eye fundus
images (Abbasi-Sureshjani et al., 2015; Pratt et al.,
2017; Wang et al., 2023; Calvo et al., 2011; Tetteh
et al., 2020) but they usually first focus on providing
a segmentation mask of the blood vessels in the im-
age before carrying post-processing on the segmenta-
tion to infer the positions of the landmarks (endpoints,
crossings of bifurcations of blood vessels). (Hervella
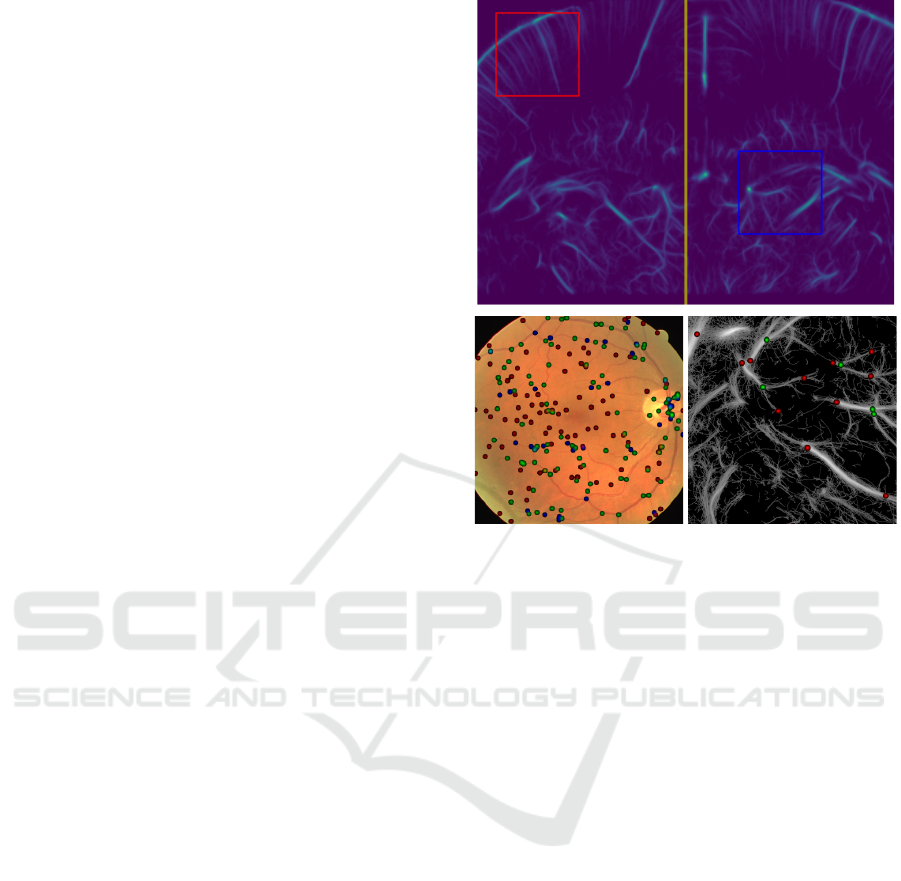
Figure 1: Top : Patches are made from a high resolution
image by cropping patches taken uniformly from each brain
half. Left : eye fundus image overlayed with heatmap of
vascular landmarks. Right : ULM image overlayed with
heatmap of vascular landmarks.
et al., 2019) tries to tackle the problem of finding
landmarks directly from input data, and we will be
building on this method here.
The second part of our workflow makes use
of geodesic curves to track vascular structures in
the input images. Tracking vascular structures us-
ing geodesics has been done multiple times for in-
stance in (Deschamps and Cohen, 2000; Benmansour
and Cohen, 2011), those methods have had multi-
ple extensions, for instance taking advantage of roto-
translation group (Bekkers et al., 2014) or adding ves-
sel width information (Li and Yezzi, 2007).
2 DETECTING VASCULAR
LANDMARKS
The approach to detect the vascular landmarks is very
much like the one used in (Hervella et al., 2019). The
novelty of our approach is the scarcity of ULM data
we use for learning, the integration of endpoints in
the detection task, and the tracking described further
down.
Indeed, we want to generate heatmaps with multi-
ple channels indicating probable locations of vascular
Fitting Tree Model with CNN and Geodesics to Track Blood Vessels in 2D Medical Images and Application to Ultrasound Localization
Microscopy Data
45

landmarks.
We train a single U-net architecture to learn the lo-
calization and classification of interesting points in a
2D ULM image of brain vessels. The network outputs
4 channels : 3 for different types of landmarks (end-
points, bifurcations, crossings) and a last one to relax
classification. The heatmap predicted by the CNN is
then filtered to get the position of local maxima (after
thresholding at level r of output of network to reduce
noise).
2.1 ULM Data
Our data is composed of few highly resolved images
of rat brains obtained via ULM imaging. The scarcity
of data is a usual problem in medical images process-
ing. We were provided with two such images of rat
brains (two similar plans were imaged) by (Chavi-
gnon et al., 2020), we then proceeded to uniformly cut
those big (3210 × 2675) images into smaller square
patches. This way, we aggregate around 42 ULM im-
ages, making a training dataset of 21 and another set
for validation of 21 images. We make sure that there
are patches from both original images in both sets. We
also make sure that there is no overlapping between
the training and validation datasets by using different
brain halves to make those patches (see Figure 1 top).
As there is no available dataset annotation for seg-
mentation task of ULM data nor for the landmark lo-
calization and classification task, annotation for the
latter was produced by one of the authors. The dataset
annotation was made by selecting the point landmarks
by hand with the appropriate tool (Skalski, 2019).
One great difficulty of our approach is that we are
highly dependent on the accuracy of the initial anno-
tation of the data which can be hard given that there
are multiple visible vessel sizes on ULM images.
To prove the efficency of our method, we will
also train and evaluate our architecture on two other
datasets : one that consists in two synthetic images of
tubular structures arranged into a network, for train-
ing and validation, they are very big and are used to
imitate the case of ULM images (high resolution, few
images, thus cut into smaller patches); the other one
is simply a dataset using both images and groundtruth
from the DRIVE and IOSTAR datasets, much like in
the previously cited work (Hervella et al., 2019).
2.2 Training
The training loss is defined as L(θ,x, ˆy) = ∥ f
θ
(x) −
ˆy∥
2
2
the mean squared error (MSE), where ˆy is the
position of the labeled features in the input images
in our dataset convolved with a gaussian kernel ˆy =
∑
y∈D
k
σ
∗ δ
y
, f
θ
is our CNN architecture with param-
eters θ, applied on the x input image. k
σ
is a gaussian
kernel with chosen standard deviation σ.
Going through our data we minimize the loss eval-
uated on the training set over the space of parameters
θ ∈ Θ. We may recall that the U-net architecture is
an encoder-decoder architecture composed of multi-
ple convolution layers (3 × 3 filters and leaky ReLU
activation) and with skip-connections. (Ronneberger
et al., 2015) is the fundamental work introducing this
architecture.
To make up for the small size of our dataset,
we perform data augmentation via horizontal symme-
tries, translations, rotations, all randomly applied with
predefined parameters. It allows us to artificially ex-
pand our training dataset, leveraging equivariance of
our task by the action of those transformations.
3 FINDING APPROPRIATE
GEODESICS
Geodesics have been used for tracking vessels in vas-
cular images for a long time now. The works (De-
schamps and Cohen, 2000; Benmansour and Cohen,
2011) laid good basis for such work, and the book
(Peyr
´
e et al., 2010) that is a good introduction to the
use of geodesics for image analysis. These works
leverage our knowledge of geodesic curves and nu-
merical algorithms allowing us to compute them to
track tubular structures on medical images.
3.1 Geodesics for Vessel Tracking
Geodesics are curves that minimize a given energy
E(γ) =
R
1
0
P (γ(s),γ
′
(s))ds with γ ∈ Lip([0,1],Ω),
where P : Ω × R
d
−→ R
+
is some given feature po-
tential.
In fact, more than an energy, with a few hypothesis
on P , E may be seen as the length of the curve γ in
some geometry described by the potential P .
Given two points x
0
,x
1
∈ R
d
, a curve minimiz-
ing the energy E under the constraints γ(0) = x
0
and
γ(1) = x
1
is called a geodesic or minimal path (joining
x
0
and x
1
) according to the metric defined by P .
In general we will limit ourselves to the cases
where P is a Riemannian metric, i.e. the square root
of a quadratic form associated with a positive defi-
nite tensor field M : P (x,v) =
p
⟨
M(x)v, v
⟩
. It can
be interpreted as a local measure of the norm of some
velocity vector v in the neighbourhood of x.
The function E then defines a distance map:
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
46

d(x,y) = inf
γ(0)=x,γ(1)=y.
E(γ). (1)
Vessel tracking can be performed by finding
geodesics on an image of the vessels. To do so, we
simply need to define a metric that is well adapted.
For now, we will restrict ourselves to Riemmanian
metrics defined on the homogenous space of posi-
tions and orientations M
d
= R
d
× P
d−1
with P
d−1
≃
S
d−1
/{−1,1}, P
d−1
allows us to assimilate features
that have the same direction but not the same sign. we
will use the relaxed Reeds-Shepp metric that is well-
studied, Riemannian and penalizes curves that are not
planar.
The relaxed Reeds-Shepp metric is the one asso-
ciated with the metric tensor defined by:
P
ε
((x,θ),( ˙x,
˙
θ))
2
= (2)
C((x, θ))
2
(| ˙x · e
θ
|
2
+
1
ε
2
| ˙x ∧ e
θ
|
2
+ ξ
2
|
˙
θ|
2
),
with ξ,ε ∈ R , e
θ
the unit vector with orientation
θ. C is a cost function, in the following it will be
defined as C =
1
1+λW
2
with λ = 10
3
and W a [0, 1]-
valued score built from the image.
This vesselness score W is important because it
allows us to associate a θ coordinate to all the de-
tected landmarks by finding the orientation θ maxi-
mizing the score at its position.
We also look to enrich our orientation-dependent
score by adding information from the detection of
vascular landmarks by imposing ∀θ,W (x,θ) = 1 if a
bifurcation has been detected at position x, so that the
landmark point is accessible from any orientation.
Similarly, if a landmark has been found and clas-
sified as a crossing, we add a new point to our set of
detected points located at this position but with sec-
ond maximum intensity in the vesselness score.
Such geodesics are well-studied and have already
been used in previous works to accurately track blood
vessels (for instance in (Duits et al., 2018)). The main
asset of this model is that it helps avoid shortcuts in
the case where two different vessels cross in a 2D im-
age.
The geodesic distance can be computed efficiently
and fast using the Fast Marching Algorithm, we refer
to (Duits et al., 2018) and the attached library for ef-
ficient computational tools used in the present work.
3.2 Clustering Landmarks Using the
Geodesic Graph
Once we have defined a proper geodesic distance and
we are able to effectively compute it, we can build
a matrix D = (d(x
i
,x
j
))
1≤i, j≤n
l
of pairwise distance,
where the x
i
are the n
l
detected landmarks.
This step is the computational bottleneck as it re-
quires to compute n
l
(n
l
− 1)/2 coefficients, meaning
solving n
l
times the Fast Marching algorithm to iter-
atively fill the lines of the matrix, computing the map
d(x
i
,·) at every iteration i. Thus the complexity is
around O(n
l
N log(N)) with N the number of points.
The computed pairwise distance matrix thus de-
fines a complete weighted graph that we call the
geodesic graph. On a single image there may be
many different groups of vessels that appear, with the
computation of the pairwise distance we have already
computed the geodesic curves between each pair of
points. We then need to keep only the groups of points
that are relevant for representation of the vessels. To
cut the complete graph into smaller connected compo-
nents, we will simply perform hierarchical clustering
on the graph. Indeed, if the metric is well chosen to
make landmarks linked by the vascular network near
for the distance d, and landmarks not connected by
the vascular network far, we simply group aggregate
points that are near and separate them from the others
under some condition of threshold distance s
cluster
.
We may cite the work (M
¨
ullner, 2011) as a reli-
able source for theory and algorithms for hierarchical
clustering.
3.3 Linking Landmarks Through the
Geodesic Graph
Our landmarks are now separated into multiple
groups. Each of these group supposedly represents
one connected component of the visible vascular net-
work in the 2D image.
Those smaller groups represent smaller complete
graphs, but we still need to select which of the com-
puted geodesics represent the vessels.
Now the objective is to only keep the curves that
accurately represent blood vessels in the image. This
can be done by removing some of the edges in each
smaller graph.
We need a few properties for the target graph, in-
ferred from the idea we have of the representation of
blood vessels:
• As it should represent vessels, it needs to be
”1−dimensional” i.e. it is represented by a pla-
nar graph.
• It does not have cycles.
• It is small for the geodesic distance (if previously
well chosen metric).
A good heuristic to have these properties is to look for
the minimal spanning tree in each smaller complete
graphs.
Fitting Tree Model with CNN and Geodesics to Track Blood Vessels in 2D Medical Images and Application to Ultrasound Localization
Microscopy Data
47

Figure 2: Geodesic graph on half of the synthetic validation
image, with N
θ
= 128. Left image shows detected land-
marks points and the geodesics linking them, right image
shows the input image.
A few algorithms are available to perform this
computation efficiently. For Kruskal’s algorithm,
time complexity is of the order of the sorting of
the edges’ weights O(e log(e)) with e the number of
edges in the graph.
4 RESULTS AND DISCUSSION
4.1 Synthetic Data
To test our framework we can execute our algorithm
on synthetic hand drawn data. The main difference
is that if we use 2D images to simulate our network,
there is no straightforward way to define orientation-
lifted images as we do with ULM microbubble tra-
jectories data. To generate such orientation-lifted im-
ages, we leverage our knowledge of Orientation score
techniques.
We define a simple transformation by convolution
with the help of anisotropic gaussian kernels : the
anisotropy in a direction θ of the kernel will allow
us to select only the parts where the local features are
aligned with this direction.
This kernel defines the lifting operator Φ :
∀u ∈ L
2
(Ω), ∀(x,θ) ∈ Ω × [0,π[, (3)
u
lifted
(x,θ) = (Φu)(x,θ) = (k
lift
θ
∗ u)(x),
To train the CNN to detect landmarks, we use two
synthetic images : one for the training set and the
other for the validation and apply the same approach
as in the case of ULM data.
Results: At the landmark-detection task level, we
were able to attain an F1 score of about 77% on the
validation dataset with hyper-parameters selected by
hand. During the training process, validation is made
on a set of images taken as random crops from the
original big image. The validation scores obtained on
those smaller (256 × 256) patches tend to give similar
scores on average along the dataset compared to eval-
uating on the whole validation image, although the
scores oscillate a lot along epochs. Figure 2 shows
the resulting tracking of synthetic vessels and the cor-
responding geodesic graph.
4.2 Eye Fundus Image
To reinforce our methodology, we apply our work-
flow to a classical dataset of eye fundus images, as a
middle ground between synthetic and ULM data.
The data came from the IOSTAR and DRIVE
dataset (Abbasi-Sureshjani et al., 2015), it was split
into a training set (30 images), a validation set (10
images) and a test set (21 images).
For the training, we perform random data augmen-
tation with translations and rotations, as to avoid over-
fitting and take advantage of equivariance properties
of the task at hand, and also random crops of fixed
size.
We built the vesselness score W for the compu-
tation of minimal paths by first applying a classical
Frangi filter (Frangi et al., 1998) on the input images
and then lifting the filtered images via Orientation
Score as described in the previous subsection.
Results: We were able to achieve satisifying re-
sults of about 60% in F1 score on both validation
dataset and test dataset (after hyper-parameter search-
ing on the validation data). These results on the land-
marks detection task is not as good as the ones pre-
sented in (Hervella et al., 2019) but they predict only
two classes of landmarks (crossings and bifurcations)
whereas we also added the additional endpoint class.
Figure 3 shows a sample of our tracking performed on
eye fundus data.
We are able to evaluate our approach end-to-end in
the case of eye fundus data. Indeed, there is no canon-
ical way to evaluate vascular tracking versus classic
Machine Learning methods generating a segmenta-
tion mask. The most straightforward approach is to
simply project the curves on a grid of the same size
as the Ground Truth segmentation. A more sophis-
ticated approach would be to carry out the tracking
using a model taking into account the width of the
vessels during the tracking such as the ones used in
(Chen et al., 2016), but it would be necessary to lift
once again the problem to a higher dimension and the
computations would be even heavier. We use here a
simple projection method where we define a single
width and define a mask by marking all pixels at a
distance from the curves below the set width.
For the evaluation to be somewhat fair we evalu-
ate the F-score or Dice score on 24 DRIVE dataset
images and take the maximum score of the generated
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
48
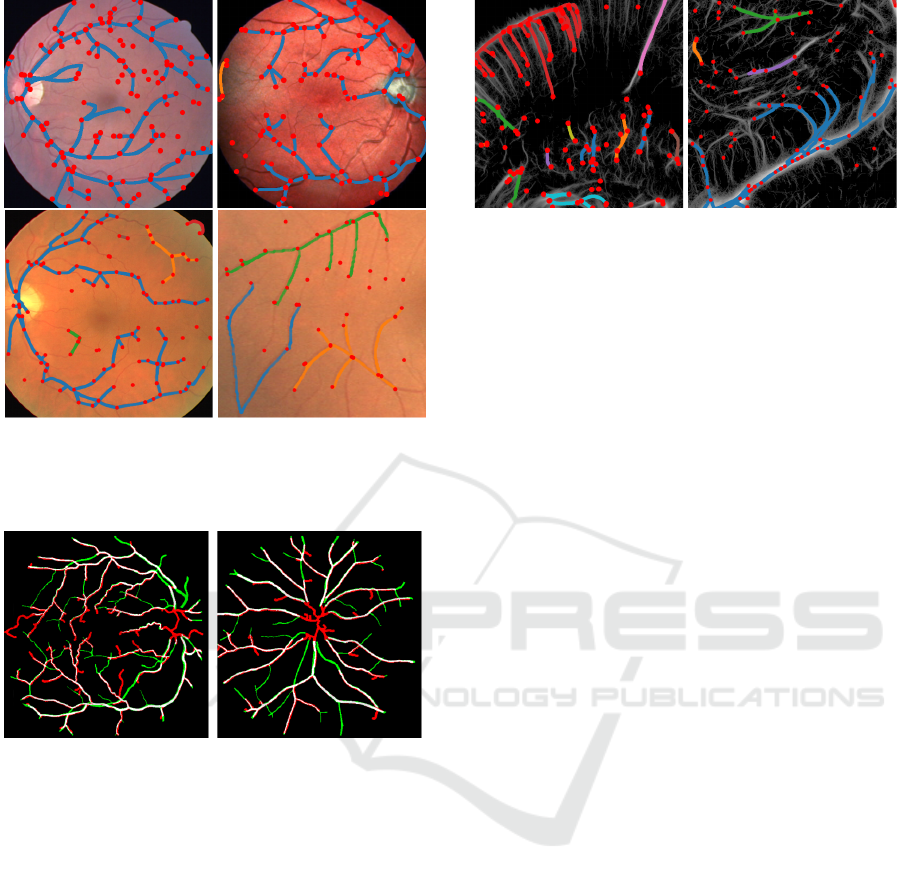
Figure 3: Geodesic tracking performed on two validation
images from the DRIVE and IOSTAR dataset, with N
θ
=
64. Big red points are the detected landmarks and curves
are the selected tree structures.
Figure 4: Example of naive segmentation mask generation
from our workflow. Green : Ground Truth. Red : proposed
segmentation. White : the intersection.
mask with multiple widths parameters and thresh-
old in the clustering step. Thus we evaluate in fact
max
σ,s
Dice where the maximum is taken over a uni-
form grid of values for both the width and the thresh-
old in region where the optimal parameters are likely
to be located. We report an average max
σ,s
Dice score
of around 56%, which is very low compared to the ex-
pected results of much more straightforward learning
methods such as (Ronneberger et al., 2015; Liu et al.,
2022) that range between 81 and 83%.
4.3 Rat Brain ULM Data
The main ideas of the data processing for ULM data
has already been described in Section 2.
We want to make full use of ULM data and use the
initial set of microbubbles path from the available data
(Chavignon et al., 2020) to construct the cost func-
Figure 5: Geodesic graph on patches cropped from the
ULM validation dataset (taken respectively from left and
right parts of rat brain), with N
θ
= 64. Big red points are the
detected landmarks and curves are the selected tree struc-
tures.
tion C in the relaxed Reeds-Shepp model as detailed
in Section 3.1. With this goal in mind we define W by
building directly the Orientation score from the his-
togram of microbubbles in the dataset just like it is
done for the input 2D image, but this time we add the
orientation of the given velocity vector for the orien-
tation coordinate. After renormalization it gives us a
function W (x,θ) with values beteween 0 and 1.
Results: With the described approach to learning the
detection of landmarks, we were only able to reach
low mean F1 scores of around 20 % (computed on
512 × 512 patches). Even with such a low score on
the detection-classification task, we are able to track
a few of the vessels in the image, as shown in Figure
5. Still, some big vessels remain untracked because
some points were not found at their tips.
4.4 Discussion
Application to real world data does not seem to work
in a very satisfying way.
We may note the following behaviours observed
after training with different hyper-parameters :
• The recovery of the geodesic tree structure is
highly sensitive to change in hyper-parameters (in
the definition of the metric tensor or the depen-
dence on the position of the detected points).
• Our framework is thought for ULM images and
does not necessarily adapt well to the eye fundus
images dataset considered in the tracking step, al-
though results might get better if one can tune
the Orientation Score well enough such that ori-
entation are well separated and landmarks can be
linked i.e. reasonably close for the geodesic dis-
tance.
• Defining the Orientation-dependent cost function
from the position of the microbubbles and their
estimated velocity vector seems to be a good ap-
proach to perform tracking on ULM data as we
Fitting Tree Model with CNN and Geodesics to Track Blood Vessels in 2D Medical Images and Application to Ultrasound Localization
Microscopy Data
49

can see from recovered geodesics in Figure 5
• The results on the synthetic images tend to show
that if we can provide a good enough segmen-
tation it would be relatively easy to provide a
good detection of landmarks and retrieve a good
geodesic tree tracking.
5 CONCLUSION AND FURTHER
WORKS
In this work we have investigated the possibility to
recover a complete tracking of the vessels in 2D im-
ages of vascular networks. It was done using CNN
techniques from the literature to extract vascular land-
marks that define the main points of interest defining
the network. Our method is interesting because it fits
a length-minimizing tree model to the image (using
geodesics in a certain geometry to represent vessels)
and thus includes both topological (tree-like struc-
ture) and geometrical (fitting geodesics) information
to our tracking.
Although results on real world data are not satis-
fying for a complete recovery of the vasculature, nor
for segmenting the vascular network, we have shown
the potential of using ULM data and the information
they carry can be used to accurately track vessels.
Further research prospects include incorporating
scale information or scale equivariance to distinguish
vessels and help the localization process and also pro-
vide width information
ACKNOWLEDGEMENTS
This work was funded in part by the French govern-
ment under management of Agence Nationale de la
Recherche as part of the ”Investissements d’avenir”
program, reference ANR-19-P3IA-0001 (PRAIRIE
3IA Institute). The authors would to thank Dr Olivier
Couture and his team for the support on ULM data,
and Dr Erik Bekkers and Dr Jiong Zhang for the ac-
cess to the DRIVE and IOSTAR datasets.
REFERENCES
Abbasi-Sureshjani, S., Smit-Ockeloen, I., Zhang, J., and
Romeny, B. (2015). Biologically-inspired supervised
vasculature segmentation in SLO retinal fundus im-
ages. pages 325–334.
Bekkers, E., Duits, R., Berendschot, T., and ter
Haar Romeny, B. (2014). A multi-orientation anal-
ysis approach to retinal vessel tracking. Journal of
Mathematical Imaging and Vision, 49(3):583–610.
Bekkers, E. J., Chen, D., and Portegies, J. M. (2018). Nilpo-
tent Approximations of Sub-Riemannian Distances
for Fast Perceptual Grouping of Blood Vessels in 2D
and 3D. Journal of Mathematical Imaging and Vision,
60(6):882–899.
Benmansour, F. and Cohen, L. (2011). Tubular struc-
ture segmentation based on minimal path method and
anisotropic enhancement. International Journal of
Computer Vision, 92:192–210.
Benmansour, F. and Cohen, L. D. (2009). Fast Object Seg-
mentation by Growing Minimal Paths from a Single
Point on 2D or 3D Images. Journal of Mathematical
Imaging and Vision, 33(2):209–221.
Calvo, D., Ortega, M., Penedo, M. G., and Rouco, J.
(2011). Automatic detection and characterisation of
retinal vessel tree bifurcations and crossovers in eye
fundus images. Computer Methods and Programs in
Biomedicine, 103(1):28–38.
Chavignon, A., Baptiste, H., Hingot, V., Lopez, P., Eliott,
T., and Couture, O. (2020). Opulm pala.
Chen, D., Mirebeau, J.-M., and Cohen, L. D. (2016). Vessel
tree extraction using radius-lifted keypoints search-
ing scheme and anisotropic fast marching method.
Journal of Algorithms & Computational Technology,
10(4):224–234.
Couture, O., Hingot, V., Heiles, B., Muleki-Seya, P., and
Tanter, M. (2018). Ultrasound Localization Mi-
croscopy and Super-Resolution: A State of the Art.
IEEE Transactions on Ultrasonics, Ferroelectrics,
and Frequency Control, 65(8):1304–1320.
Deschamps, T. and Cohen, L. D. (2000). Minimal paths in
3D images and application to virtual endoscopy. In
Vernon, D., editor, Computer Vision — ECCV 2000,
pages 543–557, Berlin, Heidelberg. Springer Berlin
Heidelberg.
Duits, R., Felsberg, M., Granlund, G., and ter Haar Romeny,
B. (2007). Image analysis and reconstruction using a
wavelet transform constructed from a reducible rep-
resentation of the Euclidean motion group. Interna-
tional Journal of Computer Vision, 72:79–102.
Duits, R., Meesters, S. P., Mirebeau, J.-M., and Portegies,
J. M. (2018). Optimal paths for variants of the 2D and
3D reeds–shepp car with applications in image anal-
ysis. Journal of Mathematical Imaging and Vision,
60(6):816–848.
Frangi, A. F., Niessen, W. J., Vincken, K. L., and Viergever,
M. A. (1998). Multiscale vessel enhancement fil-
tering. In Wells, W. M., Colchester, A., and Delp,
S., editors, Medical Image Computing and Computer-
Assisted Intervention — MICCAI’98, pages 130–137,
Berlin, Heidelberg. Springer Berlin Heidelberg.
Hervella, A., Rouco, J., Novo, J., Gonzalez, M., and Ortega,
M. (2019). Deep multi-instance heatmap regression
for the detection of retinal vessel crossings and bifur-
cations in eye fundus images. Computer Methods and
Programs in Biomedicine, 186:105201.
Jerman, T., Pernus, F., Likar, B., and Spiclin, Z. (2016). En-
hancement of Vascular Structures in 3D and 2D An-
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
50

giographic Images. IEEE Transactions on Medical
Imaging, 35(9):2107–2118.
Li, H. and Yezzi, A. (2007). Vessels as 4-D curves: Global
minimal 4-D paths to extract 3-D tubular surfaces and
centerlines. IEEE transactions on medical imaging,
26(9):1213–1223.
Liu, W., Yang, H., Tian, T., Cao, Z., Pan, X., Xu, W., Jin,
Y., and Gao, F. (2022). Full-resolution network and
dual-threshold iteration for retinal vessel and coronary
angiograph segmentation. IEEE journal of biomedical
and health informatics, 26(9):4623–4634.
M
¨
ullner, D. (2011). Modern hierarchical, agglomerative
clustering algorithms. CoRR, abs/1109.2378.
Oliveira, A., Pereira, S., and Silva, C. A. (2018). Reti-
nal vessel segmentation based on fully convolutional
neural networks. Expert Systems with Applications,
112:229–242.
Peyr
´
e, G., P
´
echaud, M., Keriven, R., and Cohen, L. D.
(2010). Geodesic Methods in Computer Vision and
Graphics. Arxiv.
Pratt, H., Williams, B., Ku, J., Vas, C., McCann, E., Al-
Bander, B., Zhao, Y., Coenen, F., and Zheng, Y.
(2017). Automatic Detection and Distinction of Reti-
nal Vessel Bifurcations and Crossings in Colour Fun-
dus Photography. Journal of Imaging, 4(1):4.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Navab, N., Hornegger, J., Wells,
W. M., and Frangi, A. F., editors, Medical Image Com-
puting and Computer-Assisted Intervention – MICCAI
2015, pages 234–241, Cham. Springer International
Publishing.
Skalski, P. (2019). Make Sense. https://github.com/
SkalskiP/make-sense/.
Song, P., Trzasko, J. D., Manduca, A., Huang, R., Kadirvel,
R., Kallmes, D. F., and Chen, S. (2018). Improved
super-resolution ultrasound microvessel imaging with
spatiotemporal nonlocal means filtering and bipartite
graph-based microbubble tracking. IEEE Transac-
tions on Ultrasonics, Ferroelectrics, and Frequency
Control, 65(2):149–167.
Tetteh, G., Efremov, V., Forkert, N. D., Schneider, M.,
Kirschke, J., Weber, B., Zimmer, C., Piraud, M., and
Menze, B. H. (2020). DeepVesselNet: Vessel Seg-
mentation, Centerline Prediction, and Bifurcation De-
tection in 3-D Angiographic Volumes. Frontiers in
Neuroscience, 14:592352.
Wang, G., Huang, Y., Ma, K., Duan, Z., Luo, Z., Xiao, P.,
and Yuan, J. (2023). Automatic vessel crossing and bi-
furcation detection based on multi-attention network
vessel segmentation and directed graph search. Com-
puters in Biology and Medicine, 155:106647.
Zhang, J., Dashtbozorg, B., Bekkers, E., Pluim, J. P. W.,
Duits, R., and ter Haar Romeny, B. M. (2016). Ro-
bust retinal vessel segmentation via locally adaptive
derivative frames in orientation scores. IEEE Trans-
actions on Medical Imaging, 35(12):2631–2644.
Fitting Tree Model with CNN and Geodesics to Track Blood Vessels in 2D Medical Images and Application to Ultrasound Localization
Microscopy Data
51
