
An Information System for Training Assessment in Sports Analytics
Vanessa Meyer
a
, Lena Wiese
b
and Ahmed Al-Ghezi
c
Institute of Computer Science, Goethe University Frankfurt, Robert-Mayer-Str. 10, 60325 Frankfurt am Main, Germany
Keywords:
Sport Data Analytics, Human Activity Recognition, Data Visualization.
Abstract:
This paper presents an information system that analyzes and visualizes sports and human activity data. Clus-
tering is used to divide data into groups; however, the wide variation in methods for data preprocessing and
clustering makes it difficult to decide on appropriate methods. Thus, for the analysis of clustering methods, we
comparatively evaluate methods for preprocessing the data in addition to the different methods for clustering.
In addition, our sports analytics information system provides an approach that is able to assign athletes to a
cluster based on their individual features and hence provides an individual training assessment compared to
the clusters obtained on the data. The proposed visualization approach in comparison to a certain cluster offers
an intuitive solution for assessing the goodness of fit.
1 INTRODUCTION
Every day, a large amount of information is retrieved
and stored. To evaluate data, it has always been of
particular importance for people to compare things or
phenomena with each other based on their similarity,
to learn unknown patterns (Xu and Wunsch, 2005).
The similarity-based grouping of data plays a major
role in this kind of evaluation: the goal of clustering
methods is to find groups of similar data objects in
data sets (Bishop and Nasrabadi, 2006). As an unsu-
pervised learning methods, unlike supervised learning
methods, there is no need for labeled target variables
in clustering. Hence, we can conveniently base our
analysis only input variables.
Advances in modern communication technologies
(through portable, mobile devices and “wearables”)
have made the areas of “individualized training mon-
itoring” and “smart health” a central part of modern
life in order to improve athletic performance and in-
dividual health. In addition to these continuously col-
lected monitoring data, there are also conventional
data collected from laboratory tests (genomics, blood
values) or questionnaires (mental health). Very com-
plex data sets are therefore available in the area of
sports and health, covering different modalities and
granularities. From an economic perspective, the
sports sector is a key growth market: “The sport-
a
https://orcid.org/0009-0006-3394-6291
b
https://orcid.org/0000-0003-3515-9209
c
https://orcid.org/0000-0002-1683-0629
stech industry has experienced and is expected to ex-
perience exceptional growth with a compound an-
nual growth rate of more than 20% between 2018 and
2024” (Frevel et al., 2020). The ISPO (International
Trade Fair for Sporting Goods and Sports Fashion)
even speaks of “an increase of 60 percent to around
C82.3 million” in spending on fitness apps in Ger-
many in the year 2020
1
. The marketable goal of our
sports information system is to create personalized
recommendations for improvements in training and
health status. This is based on the analysis of clus-
ters/cohorts of similar athletes and the training rec-
ommendations that could be derived from them. As
a long-term vision such an information system may
also support commercial applications, for example for
manufacturers of fitness equipment – especially in
combination with mobile devices (smart watches or
fitness trackers connected to corresponding apps).
Contributions. Extending our previous work
(Meyer et al., 2023), we present an information sys-
tem to find prototypical features in sports data using
clustering techniques while optimizing associated
methods for preprocessing the data. Our use case
is a personalized assessment of the training status
of individuals in this sports information system. To
illustrate our system, a public data set is used for test-
ing: the Multilevel Monitoring of Activity and Sleep
in Healthy people (MMASH) data set (Rossi et al.,
1
https://www.ispo.com/en/topic/sportstech
Meyer, V., Wiese, L. and Al-Ghezi, A.
An Information System for Training Assessment in Sports Analytics.
DOI: 10.5220/0012724200003690
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 26th International Conference on Enterprise Information Systems (ICEIS 2024) - Volume 1, pages 149-160
ISBN: 978-989-758-692-7; ISSN: 2184-4992
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
149

2020a; Rossi et al., 2020b) available via Physionet
(Goldberger et al., 2000). The data set is prepro-
cessed accordingly before clustering, where different
variants of method combinations are tested here.
Furthermore, the following well-known clustering
methods are used: K-Means, Hierarchical Clustering,
Density-Based Spatial Clustering of Applications
with Noise (DBSCAN), Affinity propagation, Mean-
shift and Balanced Iterative Reducing and Clustering
using Hierarchies (BIRCH). By choosing different
clustering methods and preprocessing methods, the
differences between each method and the resulting
influence on the clustering results are clarified.
In addition, we present the user interface of our
system where users can create their visual profiles
based on selected features. For the self-assessment,
their own measurement values for selected features
can be entered. Based on the entered values an as-
signment to a group is made, as well as a presentation
of the differences between the own features and the
features of other individuals belonging to the cluster.
Outline. Section 2 includes related works with dif-
ferent applications of data mining and clustering in
sports. Section 3 presents the MMASH dataset; this
is followed by a description of the steps used to pre-
process the data. In Section 4 six different cluster-
ing methods as well as three cluster validation in-
dexes are described. Furthermore, the section com-
pares and interprets the quality of the clusters formed
by the various clustering methods. Afterwards, Sec-
tion 5 presents a web user interface. Here the user
can select a dataset to create a visual profile based on
selected features. In a final conclusion, Section 6 pro-
vides an outlook for future research.
2 RELATED WORK
Related works are surveyed in the following that deal
with clustering or other methods from the field of data
mining in sports. The related works show quite differ-
ent goals and applications of data mining techniques:
prediction of sports match results, performance im-
provement of individual athletes or teams, or determi-
nation of the market value of athletes or teams. Ac-
cording to (Cao, 2012), in the past, sports organiza-
tions relied on the experience of individuals such as
coaches or players. Over time, however, the amount
of data collected has increased, making the use of
methods such as data mining increasingly important.
In (Cao, 2012) a model is presented that focuses on
the prediction of scores of an NBA game; it is in-
tended to develop strategies in advance or can be con-
sidered for sports betting. Logistic Regression, SVM,
Artificial Neural Networks and Na
¨
ıve Bayes are used,
and the accuracy of prediction of the mentioned mod-
els in is considered most important.
In (D’Urso et al., 2022), a fuzzy cluster model is
proposed that can be applied to different types of vari-
ables, so-called mixed data from the field of sport.
The mixed data are, among others, quantitative, nom-
inal and time series data. Suitable dissimilarity mea-
sures are calculated for each variable, which are given
weights during the clustering process, as each vari-
able has a different relevance for the results. Through
a simulation study, it was shown that the model can
handle outliers and assign correct weights to the dis-
tance matrices. The authors apply the proposed clus-
ter model to data of football players that include both
performance and positional characteristics.
A clustering algorithm is also used in (Narizuka
and Yamazaki, 2019). The focus of this work is
on team sports, specifically football matches. The
method divides formations of the games into sev-
eral average formations, which are again divided into
specific patterns, so that the formations are clustered
across several games. The methods used in this work
are hierarchical clustering and the Delauny method. .
The data used is from J1 league football matches. Ac-
cording to the authors, the method provides a tool for
formation analysis and characterizing and thus recog-
nizing team styles in the respective sport.
K-Means clustering was used in (Shelly et al.,
2020) to divide the data collected with wearables from
athletes in the sport of American football into train-
ing groups. The formation of the training groups
was based on the individual playing requirements of
the athletes. According to the authors, the results of
the analysis were confirmed when compared to tradi-
tional groupings for training in American football.
The focus in (Fister et al., 2020) is on individual
sports, such as running, cycling or triathlon, where
the time achieved is important for the quality of the
results. The authors review recent solutions regarding
post-hoc analysis. In this context, they mention per-
formance analysis, physical characteristics and ath-
letes’ behavior after a race. In their paper, the authors
achieve a robust solution based on heart rate data. It
could help athletic trainers advise their athletes to fur-
ther improve their performance and was tested on two
case studies of running athletes.
(Li et al., 2022) develop a model to assess the
physical fitness of athletes, in addition to a recom-
mendation model. Results from an experiment show
a higher classification accuracy compared to classical
methods. The authors estimate the application value,
research value and market application prospects for
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
150
monitoring human fitness using the presented meth-
ods to be high. In addition, the assessment method
obtained could be used to strengthen physical train-
ing in a targeted manner.
For prediction of the performance of trainings in
sports, (Li, 2022) use a generative adversarial neu-
ral network algorithm. Behavioral characteristics of
students are extracted; using a maximum pooling
method, the salient features of students are selected.
For prediction, the extracted features are subsequently
used as input to the neural network.
From an information system perspective, only few
related studies exist. From a physical education point
of view of pupils, (Feng, 2023) set up a web-based
system that applies a Decision Tree (DT) Classifier
for data mining.
More generally, (Herberger and Litke, 2021) pro-
vide a literature survey with a focus on professional
football; while not describing any technical applica-
tions in detail, the authors discuss pros and cons of
sports data analytics.
For the purpose of general athlete monitoring
(Thornton et al., 2019) survey some statistical ap-
proaches with a focus on individual’s data (so-called
“within-athlete changes”) with respect to training
load. Similarly, (Sarlis et al., 2023) analyze injury-
induced impact on performance in professional bas-
ketball; whereas (de Leeuw et al., 2023) discuss the
impact of internal and external training load on re-
cuperation and devise a specific heart rate model for
professional road cycling.
As opposed to these related works, in this paper
we particularly focus on clustering as a data mining
tool and present an intuitive user interface.
3 DATA PREPROCESSING
3.1 Data Set
For test purposes we use the publicly accessible Mul-
tilevel Monitoring of Activity and Sleep in Healthy
people (MMASH) data set, which is presented in
(Rossi et al., 2020b) and can be downloaded on
the PhysioNet page (Goldberger et al., 2000). The
MMASH data set can be assigned to the field of sports
and was published through the Open Database Li-
cense (ODbL). The authors of (Rossi et al., 2020b)
describe the MMASH data set as the first public data
set that offers psycho-physiological features on such
a scale. Accordingly, there are already other data sets
with data such as long-term heartbeat data, but not in
connection with actigraph or psychological data. The
MMASH data set contains information over a 24-hour
period of continuous measurements, including mea-
surements of time intervals between heart beats, heart
rate measurements, and others. The data, collected by
the company BioBeats and researchers from the Uni-
versity of Pisa, includes data from a total of 22 par-
ticipants, who are adults, male and mainly students at
the University of Pisa. According to the authors of
(Rossi et al., 2020b), this represents a sample that is
as homogeneous as possible. There are 7 files for each
participant with a total of 61 features.
3.2 Preprocessing
Prior to clustering, the MMASH data set is prepro-
cessed to obtain data in a suitable form for the chosen
clustering procedures. (Kirchner et al., 2016), point
out the importance of preprocessing data before ap-
plying clustering algorithms. According to the au-
thors, finding appropriate methods for preprocessing
and their execution order is challenging. Therefore,
in this paper, three different variants of preprocessing
data are used to later show the impact of preprocess-
ing on clustering results.
Table 1: Three versions of preprocessing pipelines for
MMASH data.
Version 1 Version 2 Version 3
Missing
Values
fill with
mean
fill with
mean
replace
with 0
Feature
Selection
Pearson
Correla-
tion
PCA Pearson
Corre-
lation
(different
treshold)
Scaling MinMax Standard MinMax
Before the preprocessing methods can be applied
to the MMASH data set, the data are first transformed
into a form suitable for the scikit-learn implementa-
tions
2
of the various clustering methods. Since time
series data are not considered in this paper, attributes
are transformed or aggregated accordingly. In addi-
tion, the original data (Rossi et al., 2020b; Rossi et al.,
2020a; Goldberger et al., 2000) are split into different
files for each participant. Finally, to put the data for
the clustering algorithms into a form where the values
of all participants are in one .csv file and in it each row
represents the attributes of an individual participant,
the files are merged into one remaining file.
The first step of data preprocessing is finding
missing values and dealing with them. In the first two
2
https://scikit-learn.org/stable/modules/clustering.html
An Information System for Training Assessment in Sports Analytics
151

versions, missing values are replaced with the corre-
sponding mean values of the attributes. In the third
version, missing values are replaced with the value
0. Outlier detection is not applied to the MMASH
data, since the data set contains very few data objects
anyway and without further domain knowledge it is
not possible to assess whether any outliers found are
really outliers. Subsequently, the dimension of the
data sets is reduced with respect to the number of at-
tributes. In the first version, a subset of the features is
created using the Pearson correlation and a set thresh-
old, in which features with higher correlation were
removed. In the second version, Principal Compo-
nent Analysis (PCA) is used. In the third version, sub-
sets are again created using Pearson correlation, this
time setting a different threshold. To scale the data, in
the first and third versions the MinMaxScaler, which
scales the values between 0 and 1 (0 is the minimum
of the values and 1 is the maximum of the values of an
attribute). In the second version, the StandardScaler
is used. Implementations
3
of scikit-learn were used
for the scaling methods.
4 CLUSTERING ANALYSIS
After the data has been preprocessed, clustering di-
vides the data into groups. Subsequently, the result-
ing clusters are visualized and evaluated using inter-
nal cluster indices. At the end, individual clusters are
described and interpreted.
4.1 Clustering Methods
• K-Means: A well-known partitioning method is
the K-means clustering method. In the K-means
method, parameter k is determined at the begin-
ning, where k is the number of groups to which
the individual data objects are to be assigned. To
determine the membership of the data objects in
the clusters, so-called centroids are determined
for each cluster and then each data object is as-
signed to the cluster whose centroid is closest
to the data object. The dispersion of the over-
all within-cluster is minimized by the iterative re-
distribution from the cluster members. In high-
dimensional spaces, however, these algorithms do
not show a good effect. This is because in high di-
mensional spaces almost all pairs of points are as
far away as the average. Thus, the distance con-
cept is poorly defined in high-dimensional spaces.
3
https://scikit-learn.org/stable/auto examples/preproce
ssing/plot all scaling.html
Another disadvantage is that the number of clus-
ters to be formed must be specified by the user
in advance. In addition, partitioning methods are
sensitive to initialization, as well as to noise and
outliers. Partitioning methods also often get en-
tangled in local optima. Non-convex clusters of
different sizes or densities cannot be handled by
partitioning methods like k-means.
• Hierarchical Clustering: In contrast to partition-
ing algorithms such as k-means, hierarchical clus-
tering combines or splits existing clusters. This
creates a hierarchical structure that contains the
order in which the clusters are combined or di-
vided. The combination of clusters takes place in
the agglomerative approach of hierarchical clus-
tering. In this approach, the individual data ob-
jects initially belong to their own cluster, each of
which contains only the one data object. Thus,
initially there are the clusters S
1
, S
2
, ...S
n
with n as
the number of data objects. Subsequently, a cost
function is to find pairs {S
i
, S
j
} that are to be com-
bined into a cluster at minimum cost. Thus S
i
and
S
j
are removed from the list of existing clusters
and a new cluster S
i
∪ S
j
is added. This proce-
dure is repeated until there is only one cluster that
contains all data objects. As for the cost function,
there are several variants such as complete linkage
(take maximum distance between members of two
clusters), average linkage (take average distance
between members of two clusters) and single link-
age (take minimum distance between members of
two clusters). Hierarchical procedures have the
disadvantages that once the decision to split or
merge groups has been made, no corrections can
be made, that there are poorly interpretable clus-
ter descriptors, and that the termination criterion
of this method is undefined. Hierarchical meth-
ods are less effective in high dimensional spaces
due to the curse of dimensionality.
• DBSCAN: In contrast to the k-means algorithm,
which assumes a convex shape of the clusters, in
DBSCAN all shapes of clusters can occur. This
is because in DBSCAN the clusters are formed in
such a way that points from a high-density area
are in one cluster. Points that are in high den-
sity areas are also called core samples. Thus, a
cluster is a set of these core samples. For the
DBSCAN algorithm, two parameters must be set.
One parameter is the minimum number of neigh-
bor points (min samples) to call a point a core
sample. The second parameter that must be set
is eps(ilon). With eps the distance is specified in
which the neighbors of a core sample are located.
For forming clusters with a larger minimum num-
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
152
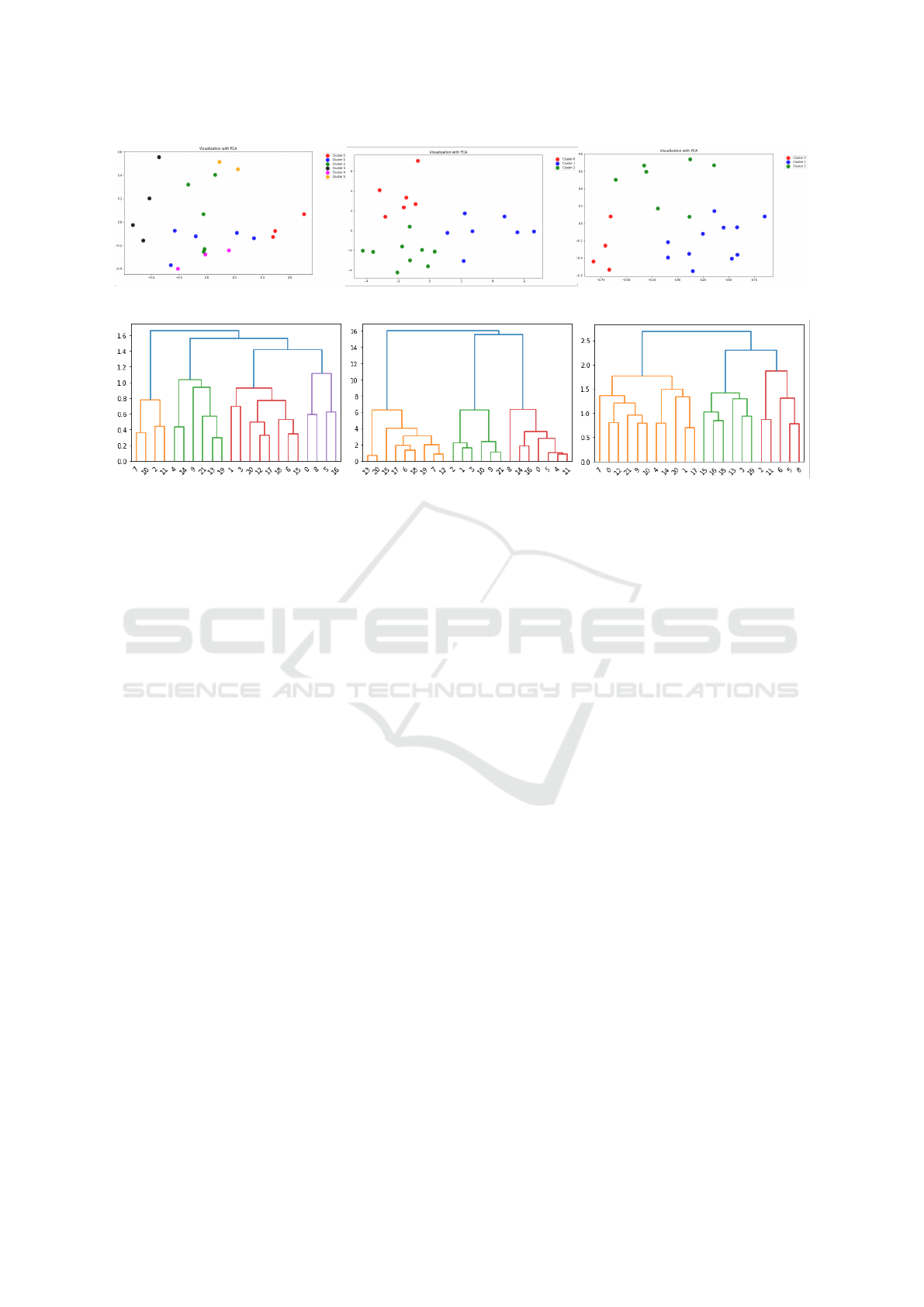
Figure 1: Resulting k-means clusters of the different data set variants (V1, V2, V3).
Figure 2: Dendrograms of the different data set variants (V1, V2, V3).
ber of neighbors or small eps, a higher density is
needed. DBSCAN starts with a random core sam-
ple and finds all its neighbors that are also core
samples. The neighboring core samples are added
to the cluster and recursively all neighboring core
samples of these newly added core samples are
added to the cluster. Neighboring non-core sam-
ples that are in the neighborhood of core samples
are also added to the cluster, but no other neigh-
bors of non-core samples are added. These Non-
core samples that are near to core samples and
thus added to a cluster are border points of the
clusters. Non-core samples that are not in the
neighborhood of core-samples are outliers. All
core-samples, however, belong to a cluster. If eps
is too small, many points are marked as outliers
with the value -1. If eps is too large, it happens
that all points are assigned to just one cluster. So,
it is important to choose an appropriate value for
the parameter eps. Advantages of density-based
cluster algorithms are that arbitrarily shaped clus-
ters can be found, the clusters found can be of dif-
ferent sizes, and their resistance to noise and out-
liers. Disadvantages are the high sensitivity to the
parameters that must be set by the user.
• Affinity Propagation: In affinity propagation,
messages are exchanged between pairs of sam-
ples. These exchanged messages indicate whether
a sample is suitable as an exemplar for the other
sample. Based on the responses to values of
other pairs, the suitability is iteratively updated
until convergence is reached. Once convergence
is reached, the final clusters are formed. Based on
the given data, the number of clusters to be formed
is determined. The parameters that are important
for affinity propagation are firstly the preference
and secondly the attenuation factor. The prefer-
ence controls the number of copies to be used. To
avoid numerical fluctuations when updating the
responsibility and availability messages, the said
messages are damped with the damping factor.
Due to the complexity affinity propagation should
rather be used for small to medium sized data sets.
• Mean-Shift: Mean-Shift is a center-based algo-
rithm that aims to find blobs in a uniform density
of samples. In this process, candidate centers are
updated. This updating is done by having the can-
didate centers represent the mean of the points in a
region. Finally, the centers are formed by filtering
the candidates in a post-processing phase to re-
move near-duplicates. The number of clusters to
be formed is determined by the algorithm. Since
the Mean-shift algorithm requires multiple near-
est neighbor searches, it is not highly scalable. In
addition, Mean-shift stops iterating at only slight
changes in centroids, but is guaranteed to con-
verge. Labeling of the new samples is done ac-
cording to the nearest centroid.
• BIRCH: In BIRCH, a feature tree is created,
where the data is compressed into cluster fea-
ture (CF) nodes containing so-called feature sub-
clusters (CF subclusters). CF subclusters, which
belong to non-terminal CF nodes, can have CF
nodes as children. The CF subclusters contain
information such as the number of samples in a
subcluster, the linear sum, squared sum, centroids,
An Information System for Training Assessment in Sports Analytics
153

Figure 3: Resulting hierarchical clusters of the different data set variants (V1, V2, V3).
and the quadratic norm of the centroids. Thus, not
all input data need to be held in memory. The
input data is reduced to subclusters in BIRCH.
Therefore, this algorithm can also be used as an
instance or data reduction method before feeding
reduced data into a global clusterer.
4.2 Cluster Validation
After presenting some cluster algorithms, the next
step shows cluster validation indices (CVIs), which
are used and compared to measure the goodness of the
formed clusters. CVIs are used to combine compact-
ness and separability of clusters. Compactness mea-
sures the closeness of cluster elements within a clus-
ter (variance as a common measure) and separability
measures the distance between two different clusters.
In the following, some cluster validation indices are
presented based on (Rend
´
on et al., 2011).
• Silhouette Coefficient (SC)
The Silhouette coefficient is one of the evaluation
methods where the evaluation is performed us-
ing the model itself when the ground truth labels
are not present. The higher the Silhouette Score,
the better defined the clusters are considered. For
each sample of the dataset, the Silhouette Coeffi-
cient is defined by calculating the mean distance
between the sample and all other samples of the
same cluster (denoted as a in the formula below)
and the mean distance between a sample and all
other samples that are in a different, closest clus-
ter (denoted as b in the formula below). The fol-
lowing formula describes the SC s for a sample:
s =
b − a
max(a, b)
(1)
To obtain the SC for multiple samples, the aver-
age of the individual silhouette coefficients of the
samples is calculated. The application of the de-
scribed SC is usually done on the results of a clus-
ter analysis. The SC can take values between -1
and 1. If the SC is around zero, this indicates over-
lapping clusters. Higher values represent denser
and more distant clusters. A disadvantage is of the
SC is that higher values are calculated for convex
clusters. Non-convex clusters, which can occur in
DBSCAN, for example, have a lower SC.
• Calinski Harabasz (CH) Index
The Calinski-Harabasz index, also called the Vari-
ance Ratio Criterion, can also be used for data sets
where there are no ground truth labels. Again, a
higher score indicates better defined clusters: If
the clusters are of higher density and well sep-
arated, the CH score is higher. The Calinski-
Harabasz index is used to represent the ratio of
the sum of the dispersion between clusters and the
dispersion within clusters:
s =
tr(B
k
)
tr(W
k
)
×
n
E
− k
k − 1
(2)
with n
E
as size of data set E, k as number of clus-
ters, and traces tr of between-group dispersion
matrix B
k
and of within-cluster dispersion matrix
W
k
where:
W
k
=
k
∑
q=1
∑
x∈C
q
(x − c
q
)(x − c
q
)
T
(3)
with C
q
set of points in cluster q and c
q
center of
cluster q and
B
k
=
k
∑
q=1
n
q
(c
q
− c
E
)(c
q
− c
E
)
T
(4)
with c
E
as the center of E and n
q
as the number
of points in cluster q. An advantage of this index
is that the values of the index can be calculated
quickly. A disadvantage is, as with the Silhouette
Coefficient, that the values of the index are gener-
ally higher for convex clusters.
• Davies Bouldin (DB) Index
Analogous to the previous two validation scores,
the Davies-Bouldin index is also used when no
ground-truth labels are present. Unlike the Sil-
houette Coefficient and the Calinski-Harabasz In-
dex, a lower Davies-Bouldin Index indicates that
the clusters of a model are better separated. The
average ‘similarity’ is indicated by the Davies-
Bouldin index, where this similarity is a compari-
son of the distance between clusters with the size
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
154
of the clusters. The closer the values of the index
are to zero, the better the partitioning is rated, with
zero being the smallest possible and therefore best
value. Advantages of the Davies-Bouldin index
are that a simpler calculation is performed than
with the Silhouette coefficient, and only pointwise
distances are used for the calculation and, conse-
quently, the index is based solely on sizes and fea-
tures inherent in the data set. A disadvantage of
this index is that the values are higher for convex
clusters than for non-convex clusters. In addition,
the distance metric is limited to Euclidean space
since the center distance is used.
DB =
1
k
k
∑
i=1
max
i̸= j
R
i j
(5)
with
R
i j
=
s
i
+ s
j
d
i j
(6)
where s
i
is the average distance between each
point of cluster i and the centroid of cluster i and
d
i j
is the distance between centroids i and j.
4.3 Clustering Comparison
Clustering was applied to the different variants of
the data set resulting from the three preprocessing
pipelines. The results are compared with respect to
the resulting clusters and cluster validation indices
Silhouette Score (SC), Calinski Harabasz Score (CH)
and the Davies Bouldin Score (DB). Based on the
results determined by the cluster validation indices,
the best possible clusters will be described and inter-
preted in more detail to perform cluster profiling.
• K-means: Before applying the k-means clustering
procedure to the MMASH dataset and the body
performance dataset, a suitable number of clus-
ters k is first determined using the elbow method.
First, k-means is applied to the MMASH dataset
with preprocessing version 1. For this version,
the elbow method results in a number k = 6 of
clusters. In contrast, for the other two prepro-
cessing versions of the MMASH dataset, only
three clusters were formed. This shows that al-
ready the results of the elbow method are influ-
enced by the way of preprocessing the data. In the
two-dimensional visualizations in Fig 1, which
are formed with the PCA method, it is also evi-
dent that depending on the version, the data points
are arranged differently in the coordinate system.
Hence, the components formed by PCA differ de-
pending on the version. The number of data ob-
jects in the respective cluster is shown in Table 2.
Table 2: Number of data points in the respective k-means
clusters for each data set variant.
Data Set Cl.
0
Cl.
1
Cl.
2
Cl.
3
Cl.
4
Cl.
5
MMASH V1 3 5 5 4 3 2
MMASH V2 6 7 9 - - -
MMASH V3 4 11 7 - - -
• Hierarchical Clustering: To find a suitable num-
ber of clusters for Hierarchical Clustering, den-
drograms were first considered. The groups high-
lighted in color in the dendrograms (Fig. 2) were
used as orientation for setting the number of clus-
ters. In this way, for the first MMASH version,
four groups were created in the hierarchical clus-
tering. However, for the other two versions of
the MMASH dataset, three groups were created
in each case. Again, the PCA method results in
different arrangements of the data points in the
coordinate system depending on the preprocess-
ing version: Fig. 3 again shows the visualizations
of the resulting clusters of the hierarchical clus-
tering for the MMASH dataset. The number of
data objects in the respective cluster is shown in
Table 3.
Table 3: Number of data points in the respective hierarchi-
cal clusters.
Data Set Cl. 0 Cl. 1 Cl. 2 Cl. 3
MMASH V1 4 8 6 4
MMASH V2 7 9 6 -
MMASH V3 5 11 6 -
• DBSCAN: In contrast to k-means and hierarchi-
cal clustering, DBSCAN does not initially specify
a number for the groups to be formed as a parame-
ter. Here, the groups are formed during the run of
the algorithm. Given eps = 0.6 and min samples
= 2, two clusters are created for the first version
of the MMASH dataset. For the second version
of the MMASH dataset, eps = 2 was set. The
value of min samples = 2 was also used here. This
created four clusters in the second version. The
parameters of the third preprocessing version of
the MMASH dataset were set to eps = 0.9 and
min samples = 2. Again, as in version 2, four
clusters were formed. The number of data objects
in the respective cluster is shown in Table 4.
• Affinity Propagation: As with DBSCAN, no pa-
rameter for the number of clusters is set for
affinity propagation. In the first version of the
MMASH dataset, five clusters were formed by
affinity propagation. In contrast, four clusters
An Information System for Training Assessment in Sports Analytics
155
Table 4: Number of data points in the DBSCAN clusters.
Data Set Noise Cl.
0
Cl.
1
Cl.
2
Cl.
3
MMASH V1 3 2 17 - -
MMASH V2 2 6 3 8 3
MMASH V3 6 4 5 5 2
were formed in each of the other two versions. In
version 1 and version 3, overlaps are also evident
in some of the clusters in the visualizations. Ta-
ble 5 shows the number of data points assigned to
each cluster.
Table 5: Number of data points in the affinity propagation
clusters.
Data Set Cl.
0
Cl.
1
Cl.
2
Cl.
3
Cl.
4
MMASH V1 2 5 6 4 5
MMASH V2 4 7 8 3 -
MMASH V3 9 4 4 5 -
• Mean-Shift: Mean-shift also does not require the
user to specify the number of clusters. In version
1 and version 2 of the MMASH dataset, five clus-
ters were formed using mean-shift. In the third
version of the MMASH dataset, however, only
three clusters were generated using mean-shift. It
can be seen that imbalanced clusters were formed
that included very many versus very few samples.
The number of samples in the respective cluster is
shown in Table 6.
Table 6: Number of data points in the mean-shift clusters.
Data Set Cl.
0
Cl.
1
Cl.
2
Cl.
3
Cl.
4
MMASH V1 15 3 2 1 1
MMASH V2 9 6 3 3 1
MMASH V3 20 1 1 - -
• BIRCH: With BIRCH, as with k-means and hier-
archical clustering, the number of clusters is cho-
sen by the user. For the MMASH dataset, four
clusters were selected as a parameter for each of
the first two preprocessing versions. For the third
version, the parameter for the number of clusters
is set to three. Table 7 shows the respective cluster
size based on the point assignment.
The individual results of the cluster validation in-
dices (CVI) are given in Table 8 for each cluster algo-
rithm and each version of the preprocessing.
Table 7: Number of data points in the BIRCH clusters.
Data Set Cl. 0 Cl. 1 Cl. 2 Cl. 3
MMASH V1 4 8 6 4
MMASH V2 6 9 6 1
MMASH V3 5 11 6 -
Table 8: Evaluation of different cluster algorithms that were
applied to three versions of preprocessed MMASH dataset.
Algorithm CVI V1 V2 V3
K-means
SC 0.2165 0.4483 0.1568
CH 5.4768 24.6601 4.6403
DB 1.1433 0.6833 1.6557
Hierarchical
SC 0.1868 0.4674 0.1410
CH 5.3219 24.4796 4.4898
DB 1.3433 0.6445 1.6918
DBSCAN
SC 0.0768 0.3822 0.0855
CH 2.2100 10.5337 2.7669
DB 1.8793 2.2796 1.8158
Aff. Prop.
SC 0.1732 0.4185 0.0857
CH 4.5213 22.2652 3.0880
DB 1.2958 0.7266 1.7820
Mean-Shift
SC 0.0068 0.4026 0.0264
CH 2.3804 21.4384 1.5655
DB 1.1292 0.5548 0.7846
BIRCH
SC 0.1868 0.4293 0.1410
CH 5.3219 20.9996 4.4898
DB 1.3433 0.5476 1.6918
Silhouette Score: The calculated silhouette scores
for the MMASH dataset are in the positive range for
all three preprocessing combinations and all cluster
algorithms. However, the scores for all three results
are closer to the 0 value rather than the 1 value, in-
dicating that there are partially overlapping clusters.
Especially the silhouette score of version 3 is very
low and closest to 0. This could be related to the
fact that more features were retained in this version
than in the other two versions and thus the dimen-
sion of the dataset is larger and affects the goodness
of clusters. Version 2 overall has the best silhouette
scores. If only version 1 is considered and the val-
ues of the different cluster algorithms are compared,
k-means has the highest and thus best silhouette score
for version 1. In the second version, the silhouette
score for hierarchical clustering is the highest com-
pared to the other cluster algorithms. As in version 1,
k-means is also in first place in version 3 with respect
to the silhouette score.
Calinski Harabasz Score: Overall, k-means pro-
vides the highest and thus best Calinski Harabasz
score for version 2. If only version 1 is considered,
k-means is also the algorithm that produces the high-
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
156

est score for this version of the MMASH dataset com-
pared to the other cluster algorithms. For the third ver-
sion, the k-means algorithm also produces the highest
Calinski Harabasz score, although this is somewhat
lower than for version 1.
Davies Bouldin Score: For the Davies Bouldin
score, smaller values represent better separated result-
ing clusters. The closer the value is to zero, the better.
The smallest and therefore best Davies Bouldin score
for the MMASH dataset was achieved by the BIRCH
algorithm applied to version 2. For version 1 and ver-
sion 3, mean-shift produced the smallest score.
5 IMPLEMENTATION
5.1 Description of the Streamlit App
Our Streamlit web application aims to analyze data
using the K-Means clustering algorithm. Users have
the option to upload a CSV file to our application. Af-
ter uploading a CSV file, various options are available
to pre-process the data. This includes in particular
measures such as dealing with missing values and the
selection of features. It is also possible to scale se-
lected features. In addition, users can enter their own
data in additional fields in order to be assigned to a
cluster based on the entered data after the clustering
algorithm has been executed.
The libraries used in our implementation are now
explained in more detail.
Streamlit: Streamlit
4
is a Python library for build-
ing web applications. It is used to create our user in-
terface and interact with users.
Pandas (Python data analysis): Pandas also be-
longs to the most used libraries in the field of Data
Science and offers among other things the possibility
to load CSV files and display them as a data frame. In
our Streamlit application it is used to read and manip-
ulate data from the CSV files uploaded by the user.
Matplotlib: Matplotlib is often used to visualize
data. We use this library to create 2D plots that visu-
alize the results of the k-means clustering algorithm.
4
https://docs.streamlit.io/
Scikit-learn: The scikit-learn library contains many
machine learning algorithms. Clustering and dimen-
sion reduction are among the applications of this li-
brary. In our case, it is used to perform K-Means clus-
tering and scaling data. To do this, we specifically use
sklearn.cluster.KMeans, sklearn.cluster.k means and
sklearn.preprocessing.MinMaxScaler.
NumPy (Numerical Python): NumPy is used for
numerical calculations and is considered a funda-
mental package for this. NumPy provides tools for
working with multidimensional objects (arrays) and is
widely used in data analysis. Within our application it
is used to work with arrays and numerical operations.
Plotly: The Plotly
5
library can be used to create in-
teractive graphs. In our streamlit application, plotly
express is used to create radar charts for cluster dis-
play with multiple features.
Seaborn: Seaborn
6
is based on matplotlib and is
also used to visualize data. We rely on this library
to provide a more attractive representation of data.
5.2 Data Preprocessing
After a CSV file has been successfully uploaded, the
various options for pre-processing the data become
visible. To ensure that an appropriate error message
is displayed and the user can be informed if prepro-
cessing steps are necessary before cluster analysis, the
entire preprocessing is enclosed in a try-except block.
Handling Missing Values: Using the checkbox
with the label Handling missing values, users of our
application can choose how to handle missing values
in the data set they uploaded. There are two options
to deal with the corresponding missing values. On
the one hand, missing values can be ignored, on the
other hand, missing values can be replaced by the
mean value. If the Fill with mean option is selected,
the missing values in the entire data set will be re-
placed by the mean of the respective column. Another
checkbox with the label Replace 0 with mean can be
selected if cells that contain the value 0 should also be
replaced with the mean value.
Feature Selection: In the sidebar, below the Han-
dling missing values selection fields, the features can
be selected from a list of all available features that
5
https://plotly.com/python/
6
https://seaborn.pydata.org/
An Information System for Training Assessment in Sports Analytics
157

should be used for the cluster analysis. Selected fea-
tures are stored in a separate data frame selected data.
The user can use the sidebar to choose whether the
original data or the data with the selected features
should be displayed using checkboxes. This means
that users have access to the original data and the data
with selected features at any time.
Scaling the Data: To ensure that all data have the
same scale range, the user can choose to scale the
data. If the Scale data checkbox is activated, the data
is scaled using the min-max scaling from the Scikit-
learn library. If the data has been scaled, it can also be
displayed by activating the corresponding checkbox.
Entering Individual Data: Once the user has se-
lected features that should be considered for cluster
analysis, the user can enter their own values for the
features. These values are used for assignment to one
of the resulting clusters.
5.3 Cluster Assignment
After preprocessing the data and the user’s feature in-
put, the cluster analysis can be started. To do this,
the user enters the desired number of clusters k and
starts the cluster analysis by confirming the analysis
button. The K-Means algorithm from the Scikit-learn
library is run on the preprocessed data and the clus-
ter centers are determined. In addition, the values
entered by the user for the selected features are com-
pared with the descaled cluster centers (if the data was
previously scaled) so that the user can be assigned to
one of the resulting clusters. In order to compare the
data entered by the user with the calculated cluster
centers and determine a cluster assignment, the mini-
mum distance of the input vector to the cluster centers
is calculated. To do this, the np.linalg.norm method
is used, which calculates the Euclidean distance. The
following code snippet shows the cluster assignment
in more detail.
# Distances between input-vector and centers
dist = []
for center in centers_descaled:
dist.append
(np.linalg.norm(input_vector - center))
min_dist = min(dist)
assigned_Cluster = dist.index(min_dist)
The user’s entered feature values were previously
stored in an input vector array. The np.linalg.norm
function calculates the Euclidean distance between
the input vector and the descaled cluster centers (cen-
ters descaled). The array dist then contains the dis-
tances to each cluster center. Min(dist) is used to find
the index of the minimum distance in dist. This in-
dex corresponds to the cluster that has the minimum
distance to the input vector.
5.4 Visualization Techniques
The results of the K-Means cluster analysis should be
displayed visually in order to give the user an intuitive
insight into the assignment to one of the clusters and
to clarify the differences between the individual fea-
tures and the cluster center features of the assigned
cluster. This is intended to make the interpretation
and analysis of the results easier. Radar charts and
bar charts are used for the visualization, which illus-
trate the assignment to clusters and the differences to
the cluster centers. Each selected feature is also rep-
resented by a gauge plot showing the difference be-
tween the entered values and the cluster center values.
If more than two features have been selected by
the user, a radar chart is created using the plotly li-
brary. More specifically, go.Scatterpolar (where go
stands for plotly.graph objects) is also used to display
the cluster centers and the user’s input data as lines in
the radar chart. In order to be comparable, the data is
scaled to a common scale. In the case where only two
or fewer features have been selected, a 2D bar chart
is used to visualize the results. The bar chart is also
created using the plotly library. Here go.Bar is used
to display the cluster center bars and the user’s input
data. As with the radar chart, the data from the cluster
centers and the inputs are scaled to a common scale.
After visualizing the associated cluster, the dif-
ferences of each selected feature from those of the
cluster center are shown using gauge plots using
go.Indicator. In the following section, the visualiza-
tions are shown using a sports data set as an example.
To demonstrate the visualizations, we upload the
MMASH data, which was previously converted into
a suitable format, to our Streamlit application. As an
example, five of the features were selected and the
data was scaled. After starting the analysis, we get
the following visualizations. Figure 4 shows the radar
chart, in which the features of the cluster center (of the
cluster to which the user was assigned to, based on the
entered data) and the entered user data, which were
brought to the same scale, are plotted. This means
that several features can be viewed in direct compar-
ison. However, in this type of visualization it is im-
portant that the values of all features are scaled, since
otherwise features with generally much higher values
will always be visually more pronounced in the radar
chart and the features with generally lower values will
be less pronounced in the radar chart.
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
158

Figure 4: Visualization of a cluster and user characteristics
using a radar chart.
Figure 5: Visualization of cluster and user features using
gauge charts.
In order to be able to view the differences between
the user input and the cluster center with the actual
(unscaled) values for each feature, gauge charts as
shown in Figure 5 are used. The difference between
the user input and the value of the cluster center can
be read directly in the gauge charts. This makes it
possible to quickly assess whether a specific user fea-
ture is above or below the value of the cluster center.
6 CONCLUSIONS
Digitalization (supported by information systems) in
the fields of sports and healthcare is a strong growth
market (Schmidt, 2020) – especially through AI mod-
els, which make greater personalization and individ-
ualization possible in the first place. Individualiza-
tion is a paradigm shift in sports analytics that is
moving towards an individualized and fine-grained
evaluation of performance and health status based on
a cohort of similar athletes. The overall health of
the athletes may be analyzed through comprehensive
data analysis (laboratory tests, performance measure-
ments, blood values, but also psychological question-
naires). Individualization leads to better use of train-
ing and performance resources on the one hand and
reduces the negative side effects for athletes on the
other. In this way, our sports information system pro-
motes the innovation-driven use of data (taking mul-
timodal data types into account) by not only creating
a new business model, but also significantly improv-
ing existing approaches (such as existing training and
health apps). The application could be further im-
proved in the future. For example, the coloring of the
visualizations (especially the radar charts) could be
optimized to make them easier to read. In addition,
other clusters (not just the cluster to which the user
was assigned) could also be used for a comparison
and displayed visually. In particular, we aim to ana-
lyze recent clustering approaches that include a deep
learning component (Karim et al., 2021) as compared
to the conventional methods used here. Furthermore,
the application could be supplemented with more de-
tailed descriptions and explanations. The preprocess-
ing options can also be further expanded in the future
by adding additional preprocessing methods.
CODE AVAILABILITY
The code of the Streamlit application is available at
https://github.com/VaneMeyer/CustomClusterVisual
izer.
ACKNOWLEDGEMENTS
This project was funded with research funds from
the Bundesinstitut f
¨
ur Sportwissenschaften based on
a decision of Deutscher Bundestag (Project Number:
ZMI4-081901/21-25).
REFERENCES
Bishop, C. M. and Nasrabadi, N. M. (2006). Pattern recog-
nition and machine learning, volume 4. Springer.
Cao, C. (2012). Sports Data Mining Technology Used in
Basketball Outcome Prediction. Master’s thesis, Tech-
nological University Dublin.
de Leeuw, A.-W., Heijboer, M., Verdonck, T., Knobbe, A.,
and Latr
´
e, S. (2023). Exploiting sensor data in pro-
fessional road cycling: personalized data-driven ap-
proach for frequent fitness monitoring. Data Mining
and Knowledge Discovery, 37(3):1125–1153.
D’Urso, P., De Giovanni, L., and Vitale, V. (2022). A robust
method for clustering football players with mixed at-
tributes. Annals of Operations Research, pages 1–28.
An Information System for Training Assessment in Sports Analytics
159

Feng, J. (2023). Designing an artificial intelligence-based
sport management system using big data. Soft Com-
puting, 27(21):16331–16352.
Fister, I., Fister, D., Deb, S., Mlakar, U., Brest, J., and Fis-
ter Jr., I. (2020). Post hoc analysis of sport perfor-
mance with differential evolution. Neural Computing
and Applications, 32(15):10799–10808.
Frevel, N., Schmidt, S. L., Beiderbeck, D., Penkert, B.,
and Subirana, B. (2020). Taxonomy of sportstech.
21st Century Sports: How Technologies Will Change
Sports in the Digital Age, pages 15–37.
Goldberger, A. L., Amaral, L. A., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000). Phys-
iobank, physiotoolkit, and physionet: components of
a new research resource for complex physiologic sig-
nals. circulation, 101(23):e215–e220.
Herberger, T. A. and Litke, C. (2021). The impact of big
data and sports analytics on professional football: A
systematic literature review. Digitalization, digital
transformation and sustainability in the global econ-
omy: risks and opportunities, pages 147–171.
Karim, M. R., Beyan, O., Zappa, A., Costa, I. G., Rebholz-
Schuhmann, D., Cochez, M., and Decker, S. (2021).
Deep learning-based clustering approaches for bioin-
formatics. Briefings in bioinformatics, 22(1):393–
415.
Kirchner, K., Zec, J., and Deliba
ˇ
si
´
c, B. (2016). Facili-
tating data preprocessing by a generic framework: a
proposal for clustering. Artificial Intelligence Review,
45(3):271–297.
Li, G. (2022). Construction of Sports Training Performance
Prediction Model Based on a Generative Adversarial
Deep Neural Network Algorithm. Computational In-
telligence and Neuroscience, 2022.
Li, X., Chen, X., Guo, L., and Rochester, C. A. (2022). Ap-
plication of Big Data Analysis Techniques in Sports
Training and Physical Fitness Analysis. Hindawi
Wireless Communications and Mobile Computing Vol-
ume 2022, Article ID 3741087.
Meyer, V., Al-Ghezi, A., and Wiese, L. (2023). Exploiting
clustering for sports data analysis: A study of public
and real-world datasets. In International Workshop on
Machine Learning and Data Mining for Sports Ana-
lytics, pages 191–201. Springer.
Narizuka, T. and Yamazaki, Y. (2019). Clustering algorithm
for formations in football games. Scientific Reports,
9(1):1–8.
Rend
´
on, E., Abundez, I. M., Gutierrez, C., Zagal, S. D.,
Arizmendi, A., Quiroz, E. M., and Arzate, H. E.
(2011). A comparison of internal and external cluster
validation indexes. In Proceedings of the 2011 Ameri-
can Conference, San Francisco, CA, USA, volume 29,
pages 1–10.
Rossi, A., Da Pozzo, E., Menicagli, D., Tremolanti, C.,
Priami, C., Sirbu, A., Clifton, D., Martini, C., and
Morelli, D. (2020a). Multilevel monitoring of activ-
ity and sleep in healthy people. PhysioNet.
Rossi, A., Da Pozzo, E., Menicagli, D., Tremolanti, C.,
Priami, C., S
ˆ
ırbu, A., Clifton, D. A., Martini, C.,
and Morelli, D. (2020b). A public dataset of 24-h
multi-levels psycho-physiological responses in young
healthy adults. Data, 5(4):91.
Sarlis, V., Papageorgiou, G., and Tjortjis, C. (2023). Sports
analytics and text mining nba data to assess recovery
from injuries and their economic impact. Computers,
12(12):261.
Schmidt, S. L. (2020). 21st Century Sports. Springer.
Shelly, Z., Burch, R. F., Tian, W., Strawderman, L., Piroli,
A., and Bichey, C. (2020). Using K-means Clustering
to Create Training Groups for Elite American Foot-
ball Student-athletes Based on Game Demands. Inter-
national Journal of Kinesiology and Sports Science,
8(2):47–63.
Thornton, H. R., Delaney, J. A., Duthie, G. M., and Das-
combe, B. J. (2019). Developing athlete monitoring
systems in team sports: Data analysis and visualiza-
tion. International journal of sports physiology and
performance, 14(6):698–705.
Xu, R. and Wunsch, D. (2005). Survey of Clustering Al-
gorithms. IEEE Transactions on Neural Networks,
16(3):645–678.
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
160
