
A Comparative Analysis of EfficientNet Architectures for Identifying
Anomalies in Endoscopic Images
Alexandre C. P. Pessoa
1 a
, Darlan B. P. Quintanilha
1 b
, Jo
˜
ao Dallyson Sousa de Almeida
1 c
,
Geraldo Braz Junior
1 d
, Anselmo C. de Paiva
1 e
and Ant
´
onio Cunha
2 f
1
N
´
ucleo de Computac
˜
ao Aplicada, Universidade Federal do Maranh
˜
ao (UFMA), S
˜
ao Lu
´
ıs, MA, Brazil
2
Universidade de Tr
´
as-os-Montes e Alto Douro (UTAD), Vila Real, Portugal
Keywords:
Endoscopy, Wireless Capsule Endoscopy, Deep Learning, EfficientNet.
Abstract:
The gastrointestinal tract is part of the digestive system, fundamental to digestion. Digestive problems can be
symptoms of chronic illnesses like cancer and should be treated seriously. Endoscopic exams in the tract make
detecting these diseases in their initial stages possible, enabling an effective treatment. Modern endoscopy
has evolved into the Wireless Capsule Endoscopy procedure, where patients ingest a capsule with a camera.
This type of exam usually exports videos up to 8 hours in length. Support systems for specialists to detect and
diagnose pathologies in this type of exam are desired. This work uses a rarely used dataset, the ERS dataset,
containing 121.399 labelled images, to evaluate three models from the EfficientNet family of architectures
for the binary classification of Endoscopic images. The models were evaluated in a 5-fold cross-validation
process. In the experiments, the best results were achieved by EfficientNetB0, achieving average accuracy and
F1-Score of, respectively, 77.29% and 84.67%.
1 INTRODUCTION
The gastrointestinal (GI) tract is part of the diges-
tive system, being fundamental in digestion, break-
ing down food, and absorbing nutrients. Digestive is-
sues such as bloating, constipation, or even diarrhoea
can be symptoms of chronic diseases such as cancer
and should be treated seriously. Cancers related to
the GI tract (esophageal, gastric, and colorectal, for
example) are some of the most common worldwide,
corresponding to 9.6% of cancer cases in the world,
with the second highest mortality rate (IARC/WHO,
2022b).
Worldwide, this type of cancer has the third high-
est incidence rate but has the second highest mortality
rate among cancer types, being more than 8%, with
lung cancer being the only one with a higher mortal-
ity rate. In Brazil specifically, 27% of the population
is afflicted with some disease in the gastrointestinal
a
https://orcid.org/0000-0003-4995-8909
b
https://orcid.org/0000-0001-8134-4873
c
https://orcid.org/0000-0001-7013-9700
d
https://orcid.org/0000-0003-3731-6431
e
https://orcid.org/0000-0003-4921-0626
f
https://orcid.org/0000-0002-3458-7693
tract. Colorectal cancer is among the three most com-
mon types of cancer among the entire Brazilian popu-
lation, surpassing the number of cases of lung cancer
(IARC/WHO, 2022a). The absence of specific symp-
toms in the initial stages results in delays in the diag-
nosis and treatment, with the prognosis of this disease
being strongly associated with the stage at which it
was diagnosed (Yeung et al., 2021).
By examining the interior of the GI tract, cancer
can be detected at an early stage, allowing for an ef-
fective treatment. The patient survival rate reaches
90% if it is diagnosed at an early stage. However, this
proportion drops to 14% in the case of an advanced-
stage cancer diagnosis (Siegel et al., 2020). Thus, en-
doscopy is one of the most used techniques for detect-
ing and analyzing anomalies in the GI tract.
However, traditional endoscopies are character-
ized by being invasive and somewhat painful, and
some complications, although rare, can include ex-
cessive sedation, perforations, hemorrhages, and in-
fections in general (Kavic and Basson, 2001). Con-
sequently, modern endoscopy has evolved into the
Wireless Capsule Endoscopy (WCE) procedure over
the past two decades. This exam consists of a pa-
tient ingesting a capsule a few millimetres in diame-
ter, coupled with a camera, a light source, a wireless
530
Pessoa, A., Quintanilha, D., Sousa de Almeida, J., Braz Junior, G., C. de Paiva, A. and Cunha, A.
A Comparative Analysis of EfficientNet Architectures for Identifying Anomalies in Endoscopic Images.
DOI: 10.5220/0012724900003690
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 26th International Conference on Enterprise Information Systems (ICEIS 2024) - Volume 1, pages 530-540
ISBN: 978-989-758-692-7; ISSN: 2184-4992
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

transmitter, and a battery. The patient uses a receiver
on their waist to receive the images captured by the
camera (Bao et al., 2015).
Despite being less uncomfortable for the patient,
the sheer amount of details contained in this type of
exam makes its analysis excessively time-consuming,
with a usual video of about 8 hours requiring around
2 hours to be analyzed by an expert while requiring
continuous focus (Hewett et al., 2010). As a result,
several details present in the videos may go unnoticed
by the expert, with up to 26% of polyps being un-
detected, depending on the endoscopist’s experience,
duration of the examination, patient’s level of prepa-
ration and size of the polyps (Ramsoekh et al., 2010).
Therefore, a support system for the expert to ana-
lyze this exam is desirable. Computer-aided detection
(CADe) and Diagnosis (CADx) Systems using Artifi-
cial Intelligence (AI) techniques have been proposed
and used in several medical areas in the last decades
(Litjens et al., 2017). Considering WCE images, sev-
eral works have been published for detecting and di-
agnosing pathologies such as polyps, hemorrhages,
colorectal tumours, and ulcers, among others (Zhuang
et al., 2021), using Deep Learning techniques (DL).
Thus, this work aims to evaluate convolutional neural
network models from the EfficientNet family of ar-
chitectures for the binary classification of Endoscopic
images, focusing on the Endoscopy Recommendation
System (ERS) dataset (Cychnerski et al., 2022).
The main contribution of this work is the eval-
uation of different versions of the EfficientNet ar-
chitecture for binary classification in endoscopy and
colonoscopy images using the ERS dataset, which
contains over 100 different kinds of pathologies
alongside images of healthy tissue from 6 distinct re-
gions of the GI tract. Unlike the work of Brzeski et al.
(Brzeski et al., 2023), which focused on classifying
areas with endoscopic bleeding in the ERS dataset,
this work considered all labels.
2 RELATED WORK
Recently, the focus of work related to endoscopic
image processing has been directed toward WCE
images. Muruganantham and Balakrishnan (Muru-
ganantham and Balakrishnan, 2022) presented a two-
step method that uses a convolutional network with a
self-attention mechanism to estimate the region where
a possible lesion would be located. This estimation,
used as an attention map, is fused with the processed
WCE image to refine the lesion classification process.
This work considered ulcers, bleeding, polyps, and
healthy classes, obtaining F
1
score values of 95.35%,
94.15%, 97.95%, and 93.55% for each class.
Goel et al. (Goel et al., 2022) proposed an auto-
matic diagnostic method focused on angiodysplasia,
polyps, and ulcers on WCE images. The authors pre-
sented a dilated convolutional neural network archi-
tecture to classify between normal and anomaly im-
ages. The authors did not process complete videos of
the exams, but rather frames randomly selected and at
least 4 frames apart, eliminating redundancies. Some
regions of the edges of the images were removed to
remove black edges resulting from the capsule camera
capture process, which do not influence the presence
or absence of pathology. Finally, by using a dilated
convolutional neural network (i.e., with a particular
spacing between the pixels of the convolution win-
dows), it was possible to increase the receptive field
of the network without the need to increase the num-
ber of parameters. The authors obtained an accuracy
of 96%, sensitivity of 93%, and specificity of 97% us-
ing a private dataset.
The work of Yu et al. (Yu et al., 2022) presents
a multitask model for classification (treated as an in-
formation retrieval task) and segmentation of patholo-
gies in traditional gastroscopy images. The model
shares characteristics between tasks, including indi-
vidual characteristics for each one, aiming to improve
the performance of each task. The information re-
trieval task determines whether an image has the pres-
ence of cancer, esophagitis, or no abnormalities, us-
ing a deep retrieval module (Lin et al., 2015). This
module encodes image characteristics into a binary
sequence and then performs similarity queries to de-
termine the class of the analyzed image. The seg-
mentation task uses a segmentation architecture in-
spired by SegFormer (Xie et al., 2021), which is a seg-
mentation model based on Transformer (Dosovitskiy
et al., 2020). Considering the classification task, the
proposed method achieved 96.76% accuracy while
achieving 82.47% F
1
score for segmentation on a pri-
vate dataset.
Ma et al. (Ma et al., 2023) also proposed a method
to classify and segment pathologies in endoscopic im-
ages. The authors use their private dataset, which con-
tains standard gastroscopic images and the presence
of early-stage gastric cancer. The authors modified
the ResNet-50 (He et al., 2016) architecture based on
a guided attention inference network (Li et al., 2018)
for the classification task between these two classes.
On a private dataset, the authors achieved 98.84% ac-
curacy and 98.18% F1 Score for classification and a
Jaccard index value of 0.64 for segmentation.
Fonseca et al. (Fonseca et al., 2022) presented
binary classification experiments (healthy and abnor-
mal) on WCE images using three different convolu-
A Comparative Analysis of EfficientNet Architectures for Identifying Anomalies in Endoscopic Images
531

tional neural network architectures. In the tests car-
ried out, ResNet-50 obtained the best performance
among the used models, reaching 98% and 81% of F
1
values for healthy and abnormal images, respectively,
obtaining satisfactory results when working with a
relatively small dataset.
The work of Brzeski et al. (Brzeski et al., 2023),
the only other work in the literature that used the ERS
dataset for the classification task, proposed a method
for the binary classification of endoscopic bleeding.
The authors defined high-level visual features to in-
corporate domain knowledge into deep learning mod-
els. The extracted features generated by the pro-
posed feature descriptors were concatenated with the
respective images and provided as input to the convo-
lutional neural network architectures during the train-
ing and inference processes. The authors carried out
experiments with the VGG19 (Simonyan and Zisser-
man, 2014), ResNet-50, ResNet-152, and Inception-
V3 (Szegedy et al., 2016) architectures, with a per-
formance improvement when including the high-level
features in the first three architectures, reaching RO-
CAUC values of up to 0.963.
Work involving the classification of endoscopic
images in the literature tends to use datasets with
a limited number of pathologies, usually focusing
on images with the presence and absence of ulcers,
polyps and bleeding alongside healthy images. Fur-
thermore, the work by Brzeski et al., despite using
the ERS dataset, focused only on images with en-
doscopic bleeding. Therefore, the difference in this
work was the use of all pathologies present in the ERS
dataset, in addition to considering images from both
endoscopy and colonoscopy for the binary classifica-
tion between healthy images and those with anoma-
lies.
3 MATERIALS AND METHOD
3.1 Dataset
The ERS (Endoscopy Recommendation System)
dataset (Cychnerski et al., 2022) contains 5,970 im-
ages labelled by experts from 1,136 different patients.
This dataset was proposed to meet a need of the MAY-
DAY 2012 (Blokus et al., 2012) project, where an at-
tempt was made to create an ensemble of specialized
classifiers for endoscopic video images. As part of
a more extensive application, those classifiers were
trained for multi-class classification and ROI detec-
tion to detect locations where potential diseases could
occur.
Since this is a high-demand task for endoscopists
analyzing WCE videos, the authors tried to span nu-
merous sets of endoscopic diagnosis, using terminol-
ogy according to the Minimal Standard Terminology
(MST 3.0) (Aabakken et al., 2009). This resulted in
27 types of colonoscopic findings and 54 findings re-
garding upper endoscopy pathologies. The dataset
also included three miscellaneous terms applicable in
machine learning applications: healthy GI tract tis-
sues, image quality attributes, and images with endo-
scopic bleeding.
All terms collected were then separated into five
distinct categories, namely:
• Gastro: Anomalies categorized according to
MST 3.0 related to pathologies localizable by up-
per endoscopies, totalling 70 terms;
• Colono: Anomalies also categorized according to
MST 3.0, but concerning pathologies that colono-
scopies can detect, totalling 34 terms;
• Healthy: Labels of regions with no anomalies
detected, totalling seven terms (which identity
which region the image was captured from);
• Blood: Information indicating the presence of
blood in the image, totalling two terms (presence
and absence of bleeding);
• Quality: Categories referring to the quality of the
endoscopic image (blurring, lack of focus, exces-
sive light, etc.), as well as excess material in the
image (such as undigested food, bile, feces, etc.),
totalling ten terms.
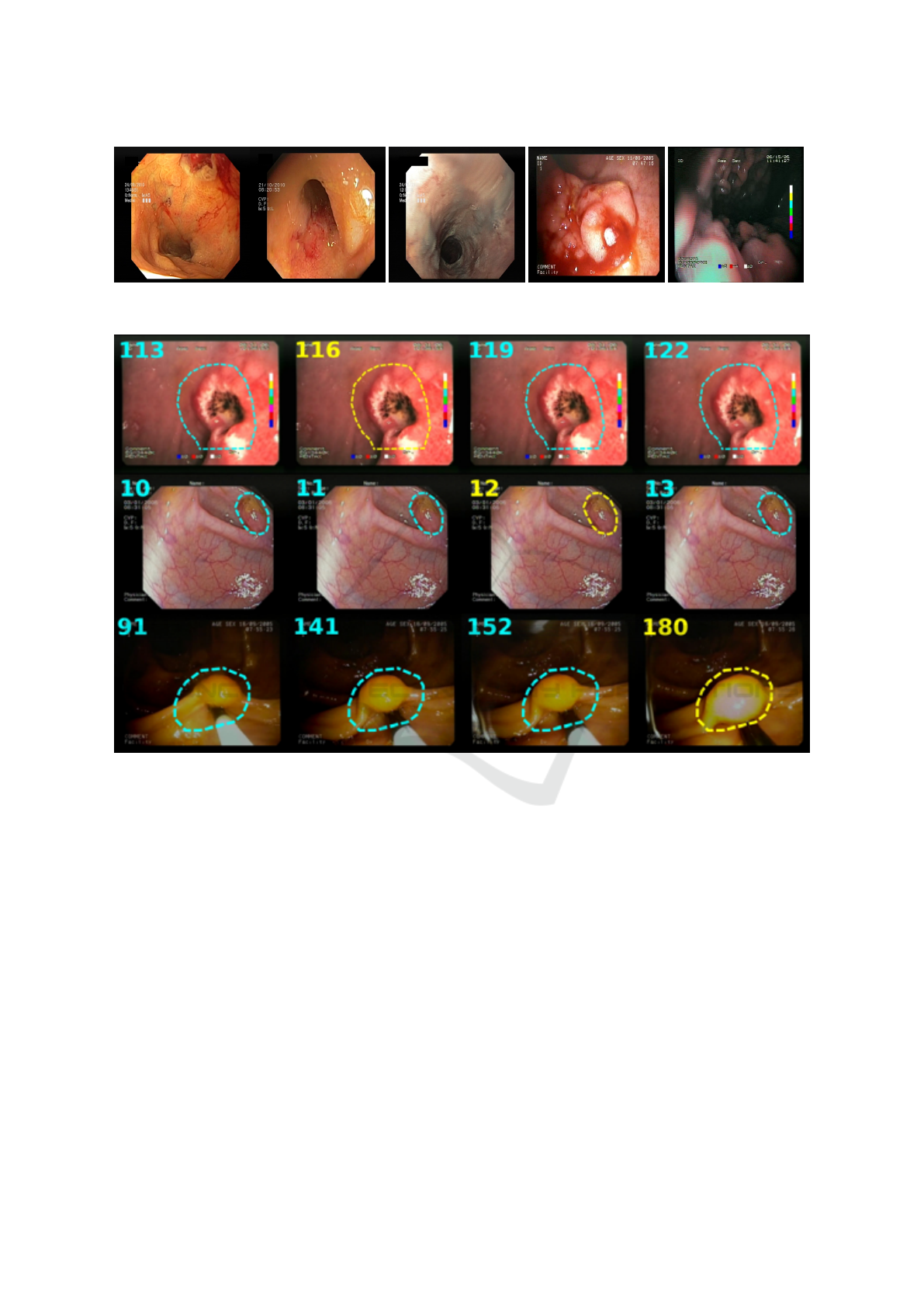
Figure 1 presents an example of each category.
The example of Gastro (1a) presents a frame of a case
of duodenal ulcer. The example of Colono (1b) illus-
trates a case of Chrohns disease. The Healthy exam-
ple (1c) is an esophagus image. An example of Blood
is presented in 1d. Notably, all images in this category
are labelled with a Gastro or Colono class. Finally, the
Quality example (1e) is an image with low lighting.
Figure 2 presents other image examples from this
dataset. The numbers in the upper left corner indicate
the frame number from the respective exam video.
The marked region shows the location of the anomaly
contained in the image. The colour of numbers and
markings indicate whether annotations are “Precise”
or “Imprecise”. Precise marks (in yellow, at frames
116, 12, and 180) were annotated by experts. The
Imprecise ones (in blue, composed of the remaining
frames) were defined using the neighbouring frames
to those marked by experts, in which the authors per-
formed a visual analysis and adjusted the binary mask
to match the region of interest visually.
The dataset contains 115,429 images with labels
categorized as Imprecise, significantly more than the
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
532
(a) Gastro. (b) Colono. (c) Healthy. (d) Blood. (e) Quality.
Figure 1: Examples of each category from ERS.
Figure 2: Example of images contained in the ERS dataset (Cychnerski et al., 2022).
number of images labelled by experts. Due to their
high abundance, however, imprecise images can be
instrumental in the training process of a deep learning
model, even with lower label accuracy. Furthermore,
the dataset has 866,612 images without any annota-
tions and 366,656 images from 7 WCE exams with
no labels.
In this dataset, images are split by patient, and
each patient can contain images from up to six differ-
ent exam videos. Each image may be associated with
0 or more labels (characterizing a multi-label dataset),
in addition to the possibility of having a binary mask
with the location of the finding for segmentation or
object detection applications. Finally, each image is
stored as a PNG file in the RGB colour space with an
original resolution of 720x576.
3.2 EfficientNet
EfficientNet is a family of CNN architectures pro-
posed by Tan and Le (Tan and Le, 2019), recog-
nized for its high performance in image classifica-
tion challenges such as ImageNet and ImageNet-V2
(Recht et al., 2019). The authors developed these
architectures by combining coefficients to scale the
network structure (the number of convolutional lay-
ers and their respective number of filters). This scal-
ing process, defined as Compound Scaling, was done
through a heuristic method based on Grid Search,
which uniformly adjusts the width and depth of the
network structure and regulates the feature maps from
a fixed set of scale coefficients. Such coefficients are
presented in the Equation 1:
A Comparative Analysis of EfficientNet Architectures for Identifying Anomalies in Endoscopic Images
533
d = α
φ
w = β
φ
r = γ
φ
(1)
Where:
• d: Depth;
• w: Width;
• r: Image resolution;
• φ: Coefficient that represents the amount of avail-
able computational resources, defined by the user;
The α, β, and γ coefficients define how the re-
sources will be assigned in relation to depth, width,
and resolution, respectively. These coefficients must
assume values greater than or equal to 1, subject to
the restriction presented in Equation 2:
α · β
2
· γ
2
≈ 2 (2)
These parameters can be estimated in 2 ways:
1. Assuming φ = 1 and estimating α, β and γ;
2. Assuming α, β and γ as constants and estimating
different values of φ.
The EfficientNet family of networks was defined
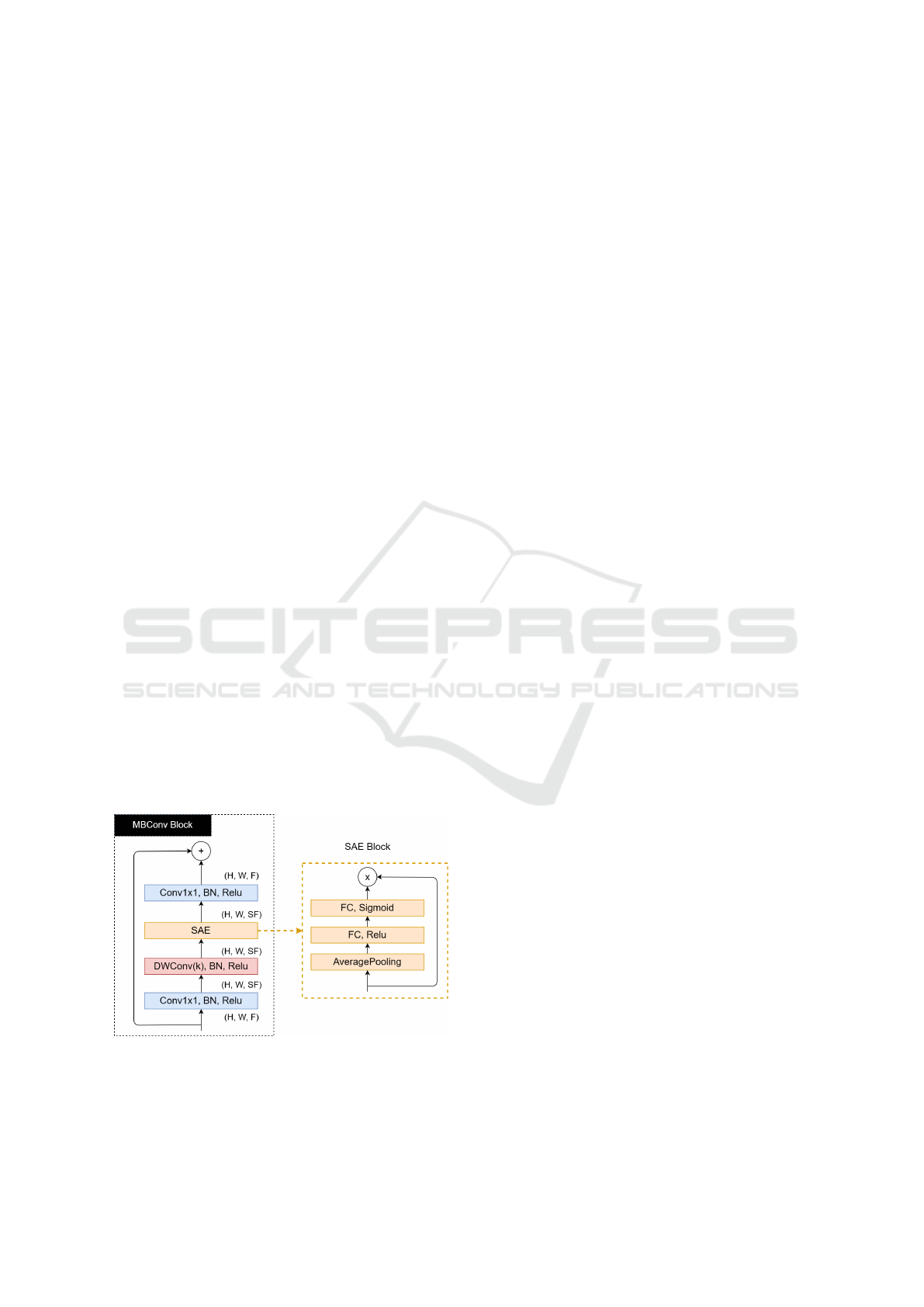
by this scaling method, being named from B0 to B7.
The structure of these networks is composed of blocks
called MBConv, being characterized by a combina-
tion between the Inverted BottleNecks (IB) (Sandler
et al., 2018) and the Squeeze-and-Excitation (SAE)
blocks (Hu et al., 2018). The IBs use depth-wise con-
volutions (DWConv) as alternatives to standard con-
volutions to reduce the computational cost of these
operations since DWConv operations have a smaller
amount of parameters to be adjusted (Howard et al.,
2017).
Figure 3: Structure of the MBConv and SAE blocks.
Figure 3 illustrates the structure of the MBConv
block and the SAE block. The input dimensions of
the MBConv blocks are H ×W × F, where H is the
height, W is the width, and F is the number of fea-
ture maps. More feature maps are generated after the
first 1x1 convolution (followed by Batch Normaliza-
tion and a Relu activation). This increases by a scaling
factor S that multiplies F. Afterwards, the DWConv
operations are applied, generating another increase in
the number of feature maps, which will be used as
input to the SAE block.
The SAE blocks, illustrated in Figure 3, assign
weights to feature maps that are more relevant to the
model’s objective and will have greater weights. Fi-
nally, a last 1x1 convolution decreases the number of
feature maps, assuming its initial value.
In this work, these models were chosen because
they have comparatively lower computational costs
(requiring fewer FLOPS in the inference process and
with fewer adjustable parameters) and have better per-
formances considering the top-1 and top-5 accuracy
metrics in ImageNet (Recht et al., 2019), compared
to other well-known CNN architectures. The experi-
ments conducted in this work involving EfficientNet
considered configurations from B0 to B3.
4 RESULTS AND DISCUSSION
4.1 Experiment Description
To conduct the experiments to evaluate the Efficient-
Net architectures in the binary classification of en-
doscopic images, the same approach as (Cychnerski
et al., 2022) was used to separate the images into
healthy and anomaly classes. In this approach, im-
ages were selected from the Healthy categories and
images without bleeding from the Blood category to
compose the set of healthy images, and images from
both the Gastro and Colono categories as well as
images with bleeding from the Blood category com-
posed the subset of images with pathologies.
A cross-validation method using five folds was
used. The folds were separated per patient to ensure
that exams from the same patient were not in separate
folds to avoid data leakage (Kaufman et al., 2012).
During each cross-validation stage, 20% of patients
from the training set were randomly selected for vali-
dation. As each patient has a different number of ex-
ams, the absolute number of images for each fold will
differ for each step. For the experiments, precise and
imprecise images were used to train and validate the
proposed models. For the evaluation, however, only
precise images were utilized.
Each model was trained for 25 epochs using Bi-
nary Cross-Entropy (Yi-de et al., 2004) as the loss
function, with the input resolutions ranging from
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
534

224x224 to 300x300, depending on which model was
being trained. The network weights were initial-
ized with the pre-trained weights on ImageNet (Recht
et al., 2019), available through Keras (Chollet et al.,
2015).
To evaluate the performance of each trained
model, the Binary Accuracy value was used, as well
as the F1-Score metric. This metric represents the
harmonic mean of precision, the ratio of all true pos-
itives to all positive values returned, and sensitivity,
which represents the ratio of true positives returned to
all positive values present. This metric can be calcu-
lated as in Equation 3:
F
1
= 2
precision ∗ recall
precision + recall
=
2t p
2t p + f p + f n
(3)
With t p being the true positives, that is, the images
with correctly classified anomalies; f p being the false
positives, with the healthy images being incorrectly
classified; and f n being the false negatives, these be-
ing the images with anomalies classified as healthy.
F
1
values vary between 0 and 1, with higher values
indicating better results for the model.
The only work in the literature that can be com-
pared to the obtained results is (Cychnerski et al.,
2022), in which the ERS dataset was published. In
that work, the authors conducted some experiments
to serve as a baseline for comparison. In the case
of binary classification, the authors tested several
deep neural network architectures, with MobileNet
v1 (Sandler et al., 2018) obtaining the best result.
The same cross-validation process was used for Mo-
bileNet v1, using the same five folds for a fair com-
parison.
4.2 Results
Table 1 compares the average results obtained with
MobileNetV1 and different EfficientNet configura-
tions (B0-B3) using the same cross-validation method
and folds for each architecture. Each EfficientNet
configuration uses a different input image resolution
(as shown in the table), while the MobileNetV1 model
was trained with 240x240 resolution images. Notably,
all the different trained EfficientNet configurations
obtained better results than the trained MobileNetV1
model, with differences of 8 to 15 percentage points
in the average Accuracy and 8 to 11 percentage points
in the average F1-Score. The standard deviation for
the Precision metric with MobileNet was also signifi-
cantly higher, resulting in a higher standard deviation
for the F1-Score.
When analyzing the results between the Efficient-
Net configurations, it is noticeable that the simplest
configuration (B0 with the lowest resolution) obtained
better results than the others regarding almost all met-
rics, with differences varying between 2 and 3 per-
centage points considering the F1-Score (with practi-
cally the same standard deviation).
In Table 2, we have the results for each individ-
ual fold of the cross-validation using EfficientNetB0,
which was the best-performing variation of the tested
architectures during the experiments. The Accuracy,
Precision, Recall, and F1-Score values for each test
fold and the mean and standard deviation for each in
this experiment are presented. The results are promis-
ing, reaching average values above 76% in all metrics.
However, the model’s difficulty in correctly classify-
ing healthy images is notable, as evidenced by the low
precision in fold 5, resulting in a relatively high stan-
dard deviation for this metric. This also resulted in
an accuracy value lower than the F1-Score achieved
due to more false positives and, consequently, a lower
number of true negatives (correctly classified healthy
images). However, the model performed better in cor-
rectly classifying images with anomalies, with F1-
Scores reaching 88.14% in fold 1 and obtaining an
average of 84.67%.
4.3 Case Study
Aiming to understand better the performance of Effi-
cientNet in classifying images from the ERS dataset,
we selected the model that performed best during the
experiments (EfficientNetB0 trained with folds 2-5)
and verified which images the model had the most
success and difficulties. To achieve this, we analyzed
the model outputs for the test set. We selected the im-
ages for which the model generated the highest and
lowest probabilities for correct and incorrect predic-
tions for both the positive and negative classes.
In Figure 4, we have examples of positive classi-
fications by the model. In Figure 4a and Figure 4c,
we have the predictions with the highest probability
for anomalies, for true positives and false positives,
respectively. Similarly, the lowest probability model
outputs for the positive class are presented in 4b and
4d. The positive case with the highest probability was
a polyp in the colon region. This was probably an
easy example of classification for the model because
it was a frame with a very visually evident polyp. The
case of the correct positive prediction with the low-
est probability is an image of a duodenal ulcer. It is
noticeable that there are bubbles in the image in this
particular frame, which can complicate the model’s
inference process. However, it was still able to clas-
sify this example correctly.
Both examples of false positives are images of the
A Comparative Analysis of EfficientNet Architectures for Identifying Anomalies in Endoscopic Images
535
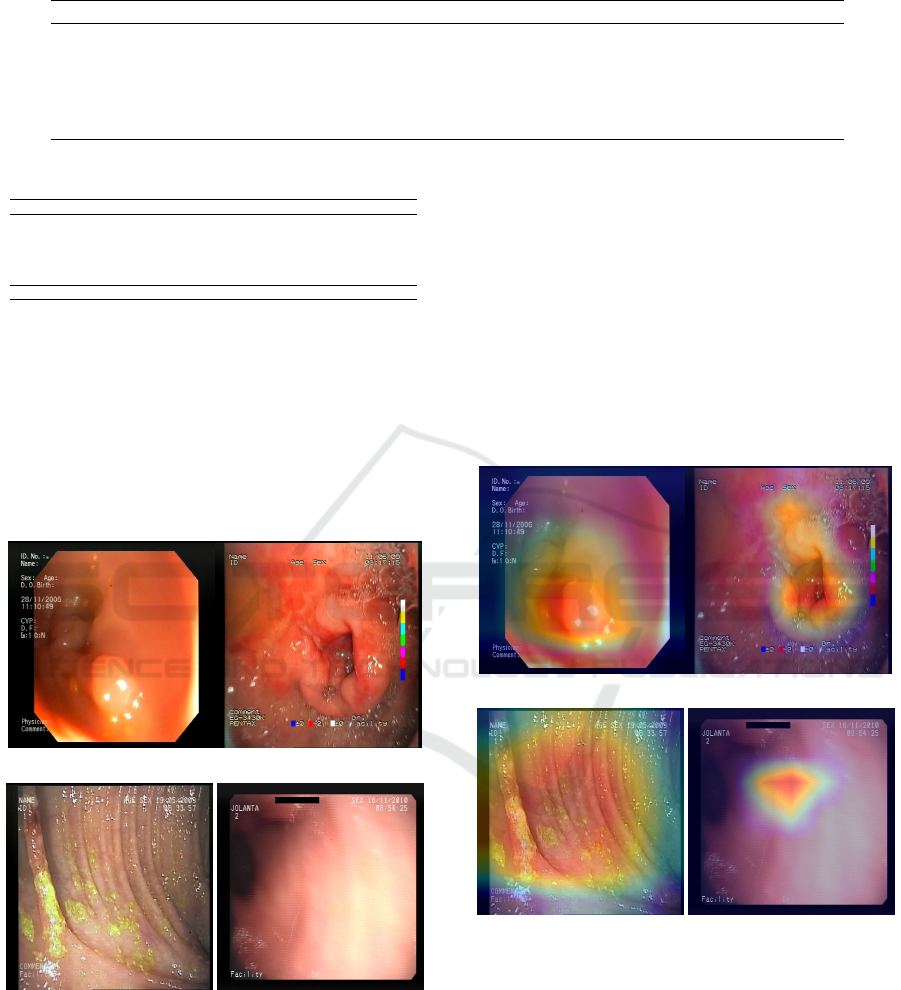
Table 1: Comparison between different EfficientNet architectures and MobileNetV1.
Model Resolution Accuracy (%) Precision (%) Recall (%) F1-Score (%)
EfficientNetB3 300x300 70.45 ± 4.64 69.84 ± 5.31 96.85 ± 0.94 81.04 ± 3.63
EfficientNetB2 260x260 74.32 ± 5.44 75.04 ± 5.88 91.71 ± 3.44 82.40 ± 3.95
EfficientNetB1 240x240 73.81 ± 5.62 74.38 ± 5.67 92.26 ± 2.63 82.23 ± 3.87
EfficientNetB0 224x224 77.29 ± 5.44 76.52 ± 6.08 95.21 ± 1.42 84.67 ± 3.51
MobileNetV1 240x240 61.93 ± 6.41 66.88 ± 6.85 83.88 ± 15.78 73.55 ± 7.79
Table 2: Results for cross-validation with EfficientNetB0.
Fold Acc (%) Precision (%) Recall (%) F1-Score (%)
Fold 1 84.46 81.95 95.35 88.14
Fold 2 78.19 78.90 95.78 86.53
Fold 3 74.05 74.07 94.98 83.23
Fold 4 80.86 81.89 92.78 87.00
Fold 5 68.89 65.78 97.17 78.45
Mean 77.29 ± 5.40 76.52 ± 6.08 95.21 ± 1.42 84.67 ± 3.51
colon region, even though most healthy images in this
dataset are from colonoscopies. The case in 4c prob-
ably resulted in a high probability for the class of
pathologies due to the yellow spots along the tract tis-
sue, which the model may have identified as features
of some anomaly. The image presented in 4d gen-
erated a false positive, possibly because it is a low-
quality image with some visual artifacts that could
have been confused with pathology features.
(a) Maximum true positive. (b) Minimum true positive.
(c) Maximum false positive. (d) Minimum false positive.
Figure 4: Examples of predictions (correct and incorrect) as
pathologies.
To visualize the network’s decision process, the
Grad-CAM algorithm (Selvaraju et al., 2016) was
used to plot heatmaps that highlight the regions most
crucial for target class prediction. This algorithm was
applied to both the correct and incorrect cases pre-
sented previously to understand better the model’s
strengths and difficulties in detecting anomalies. Fig-
ure 5 shows the activation mappings of the last convo-
lutional layer of EfficientNetB0 plotted over the cor-
responding images of the positive class. In 5a and 5b,
it can be seen that the emphasis is around the lesion
region, and in the case of 5b, little focus was given
to the area with bubbles. For the case in 5c, it can
be seen that the entire region with the yellow marks
was highlighted, so, in fact, this was mistaken for a
lesion. Finally, in 5d, it is noted that a small region
in the middle of the image has been highlighted, but
nothing particularly notable occurs in this location.
(a) Maximum true positive. (b) Minimum true positive.
(c) Maximum false positive. (d) Minimum false positive.
Figure 5: Heatmaps mapping the activation for the positive
class.
Similarly to the previous figure, in Figure 6, we
have examples of the model’s negative classifications,
that is, predictions of healthy images. In 6a and 6b,
we have the frames that had the highest and lowest
probabilities for the negative class, respectively. This
probability was given as 1 − p, with p being the prob-
ability for the positive class. Both cases are from im-
ages of healthy tissue from the colon region. In con-
trast, the low probability of healthy tissue in Figure
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
536
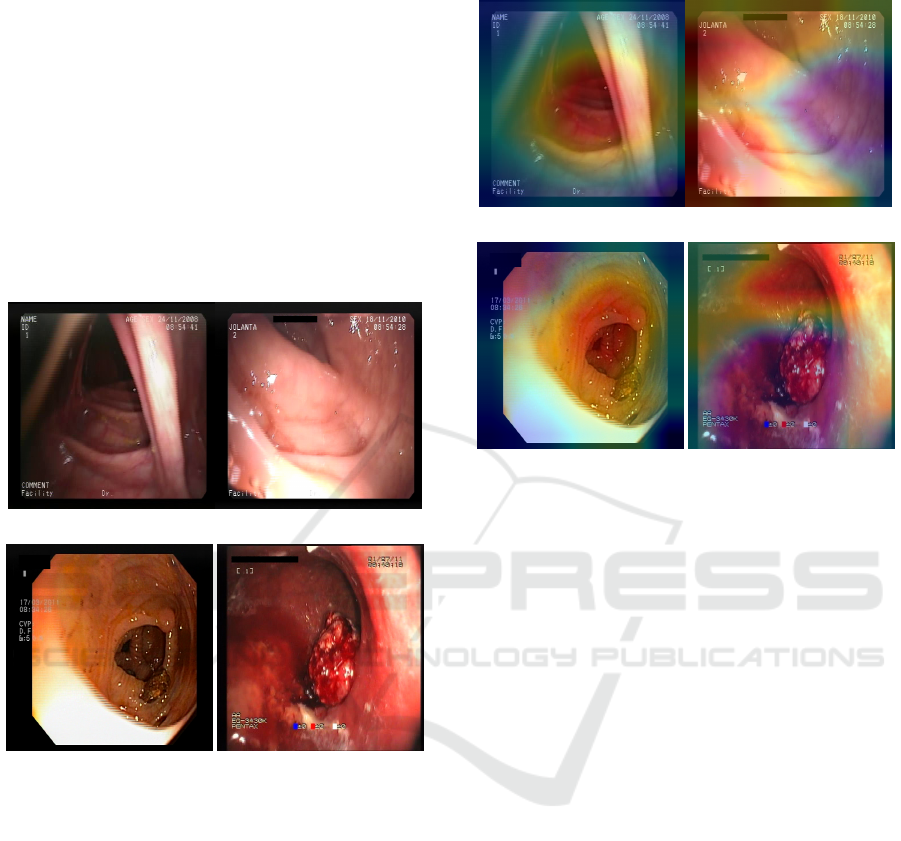
6b may be a consequence of the areas with high lu-
minosity in the lower part of the image. Figures 6c
and 6d, also parallel to the previous examples, are the
frames that had the highest and lowest probabilities
for the negative class, respectively, but were mistak-
enly classified as pathologies by the model. In Figure
6c, the model failed to recognize the region with a
polyp in the frame, probably attributing more signif-
icant importance to the areas of healthy tissue in the
image. In 6d, we have a severe case of false negative,
where the pathology is significantly visually evident.
Still, the probability of the positive class did not ex-
ceed the classification threshold and, therefore, was
erroneously classified as healthy.
(a) Maximum true negative. (b) Minimum true negative.
(c) Maximum false negative. (d) Minimum false negative.
Figure 6: Examples of predictions (correct and incorrect) as
healthy images.
As was done for positive examples, the Grad-
CAM algorithm was also used to visualize the de-
cision process in the classification examples for the
healthy class. To this end, the gradient of the out-
put of the network’s last convolutional layer was also
considered regarding the probability of the negative
class, that is, 1 − p, as previously described. Figure
7 presents the output of Grad-CAM for the presented
instances of classifications for the healthy class. In
7a, it can be seen that the activation for the healthy
class in this frame is in the center of the image, co-
inciding with the incidence of light from the colono-
scopic device and assigning less focus to the darker
parts of the image. Something similar happens in 7b,
and contrary to what was supposed, the regions with
the highest incidence of light were the most important
for the model’s decision in this classification.
(a) Maximum true negative. (b) Minimum true negative.
(c) Maximum false negative. (d) Minimum false negative.
Figure 7: Heatmaps mapping the activation for the negative
class.
For cases of incorrect classifications, 7c indicates
that the model assigned similar importance to a signif-
icant portion of the image, including the region con-
taining the polyp, pointing out that anomalies with
these features may be a weakness of this model. Fur-
thermore, the frame in 7d, despite pointing out that
little importance was given to the region where the
anomaly occurs for classification as a healthy im-
age, the rest of the image had enough healthy tissue
features to warrant a negative classification from the
model.
4.4 Impact of Mislabeling
Finally, when carefully checking the labels assigned
to the images used in the experiments, a problem was
noticed in the separation chosen for healthy images
and those with pathologies. In the same way as the
binary classification experiments carried out in (Cy-
chnerski et al., 2022), images were selected from the
Healthy categories and images without bleeding from
the Blood category to compose the set of healthy im-
ages. However, it was noted that 3 images (frame 4
of patient 12, and frames 1 and 2 from patient 944, all
from the Precise subset) in the dataset were labelled
both as “No blood” and as an anomaly in the Gas-
tro or Colono category, which resulted in the same
image being labelled as both healthy and unhealthy.
Figure 8 shows the 3 frames where this occurred. The
A Comparative Analysis of EfficientNet Architectures for Identifying Anomalies in Endoscopic Images
537
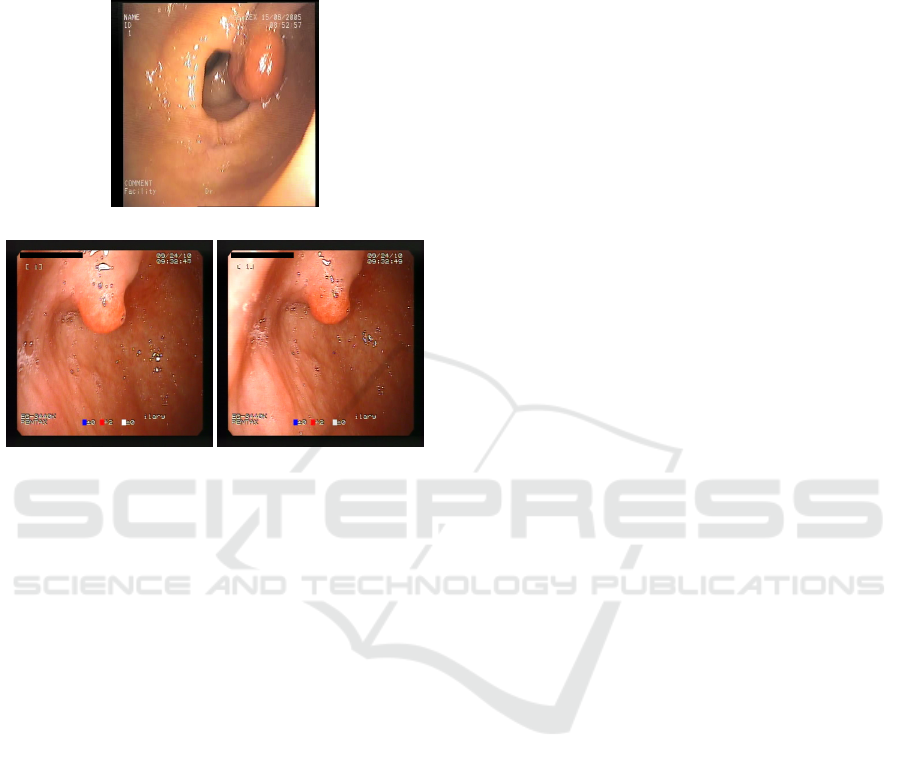
first frame is a colonoscopy exam, while the last two
frames occur one after the other in another upper en-
doscopy exam. Interestingly, the three frames present
cases of polyps.
(a) Frame 4 of patient 12.
(b) Frame 1 of patient 944. (c) Frame 2 of patient 944.
Figure 8: Frames labelled both positive and negative.
The EfficientNetB0 model trained with folds 2-5
of the cross-validation process presented previously
was also tested to evaluate the possible impact of this
mislabeling during the experiments. The probability
assigned to the positive class was verified. For the first
image, a probability of 0.4656 was assigned, which
would result in a wrong classification considering the
threshold of 0.5 used in this work. For the other
two images, the model assigned values of 0.6025 and
0.7797, respectively, correctly classifying the frames
as pathologies. It is assumed that these cases harmed
the learning of the model for pathology cases, in par-
ticular for polyp cases, even more so since the Keras
API dealt with the situation of the same instance with
multiple labels in a binary classification, keeping the
last assigned label, which in turn was “healthy” for
the 3 frames.
5 CONCLUSIONS
Various diseases in the gastrointestinal tract can be
detected and prevented through CADe and CADx sys-
tems applied to endoscopic exam images. The au-
tomatic early identification of pathologies present in
WCE exams can assist physicians in efficiently treat-
ing their patients. This work evaluated the perfor-
mance of networks from the EfficientNet family of ar-
chitectures for binary classification in WCE images.
The experiments were conducted using an extensive
dataset not used in the literature, and their results
were compared with the benchmark presented by the
dataset’s authors.
The results indicate that the simplest Efficient-
Net configurations obtained better results for binary
classification, but all the results were superior to the
best results obtained by the dataset authors. The val-
ues of the analyzed metrics indicate promising re-
sults for classifying anomalies in WCE images. Still,
it is essential to highlight that no studies involving
other possible tasks with this dataset, such as detect-
ing pathologies in images or multiclass classification.
ACKNOWLEDGMENTS
The authors acknowledge the Coordenac¸
˜
ao de
Aperfeic¸oamento de Pessoal de N
´
ıvel Superior
(CAPES), Brazil - Finance Code 001, Conselho
Nacional de Desenvolvimento Cient
´
ıfico e Tec-
nol
´
ogico (CNPq), Brazil, and Fundac¸
˜
ao de Am-
paro
`
a Pesquisa Desenvolvimento Cient
´
ıfico e Tec-
nol
´
ogico do Maranh
˜
ao (FAPEMA) (Brazil), Empresa
Brasileira de Servic¸os Hospitalares (Ebserh) Brazil
(Grant number 409593/2021-4) for the financial sup-
port.
REFERENCES
Aabakken, L., Rembacken, B., LeMoine, O., Kuznetsov,
K., Rey, J.-F., R
¨
osch, T., Eisen, G., Cotton, P., and
Fujino, M. (2009). Minimal standard terminology
for gastrointestinal endoscopy–mst 3.0. Endoscopy,
41(08):727–728.
Bao, G., Pahlavan, K., and Mi, L. (2015). Hybrid localiza-
tion of microrobotic endoscopic capsule inside small
intestine by data fusion of vision and rf sensors. IEEE
Sensors Journal, 15(5):2669–2678.
Blokus, A., Brzeski, A., Cychnerski, J., Dziubich, T.,
and Je¸drzejewski, M. (2012). Real-time gastroin-
testinal tract video analysis on a cluster supercom-
puter. In Zamojski, W., Mazurkiewicz, J., Sugier, J.,
Walkowiak, T., and Kacprzyk, J., editors, Complex
Systems and Dependability, pages 55–68, Berlin, Hei-
delberg. Springer Berlin Heidelberg.
Brzeski, A., Dziubich, T., and Krawczyk, H. (2023). Visual
features for improving endoscopic bleeding detection
using convolutional neural networks. Sensors, 23(24).
Chollet, F. et al. (2015). Keras. https://keras.io.
Cychnerski, J., Dziubich, T., and Brzeski, A. (2022). ERS:
a novel comprehensive endoscopy image dataset for
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
538

machine learning, compliant with the MST 3.0 speci-
fication. CoRR, abs/2201.08746.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Min-
derer, M., Heigold, G., Gelly, S., Uszkoreit, J., and
Houlsby, N. (2020). An image is worth 16x16 words:
Transformers for image recognition at scale. CoRR,
abs/2010.11929.
Fonseca, F., Nunes, B., Salgado, M., and Cunha, A. (2022).
Abnormality classification in small datasets of cap-
sule endoscopy images. Procedia Computer Science,
196:469–476. International Conference on ENTER-
prise Information Systems / ProjMAN - International
Conference on Project MANagement / HCist - Inter-
national Conference on Health and Social Care Infor-
mation Systems and Technologies 2021.
Goel, N., Kaur, S., Gunjan, D., and Mahapatra, S. (2022).
Dilated cnn for abnormality detection in wireless cap-
sule endoscopy images. Soft Computing, pages 1–17.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE Conference on Computer Vision and Pattern
Recognition (CVPR).
Hewett, D. G., Kahi, C. J., and Rex, D. K. (2010). Ef-
ficacy and effectiveness of colonoscopy: how do we
bridge the gap? Gastrointestinal Endoscopy Clinics,
20(4):673–684.
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D.,
Wang, W., Weyand, T., Andreetto, M., and Adam,
H. (2017). Mobilenets: Efficient convolutional neu-
ral networks for mobile vision applications. CoRR,
abs/1704.04861.
Hu, J., Shen, L., and Sun, G. (2018). Squeeze-and-
excitation networks. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR).
IARC/WHO (2022a). Cancer Today - Brazil Fact Sheet.
https://gco.iarc.who.int/media/globocan/factshee
ts/populations/76-brazil-fact-sheet.pdf. Accessed:
13/02/2024.
IARC/WHO (2022b). Cancer Today - World Fact Sheet.
https://gco.iarc.who.int/media/globocan/factsheets
/populations/900- world-fact-sheet.pdf. Accessed:
13/02/2024.
Kaufman, S., Rosset, S., Perlich, C., and Stitelman, O.
(2012). Leakage in data mining: Formulation, de-
tection, and avoidance. ACM Trans. Knowl. Discov.
Data, 6(4).
Kavic, S. M. and Basson, M. D. (2001). Complications
of endoscopy. The American Journal of Surgery,
181(4):319–332.
Li, K., Wu, Z., Peng, K.-C., Ernst, J., and Fu, Y. (2018).
Tell me where to look: Guided attention inference
network. In Proceedings of the IEEE Conference on
Computer Vision and Pattern Recognition (CVPR).
Lin, K., Yang, H.-F., Hsiao, J.-H., and Chen, C.-S. (2015).
Deep learning of binary hash codes for fast image
retrieval. In Proceedings of the IEEE Conference
on Computer Vision and Pattern Recognition (CVPR)
Workshops.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A.,
Ciompi, F., Ghafoorian, M., van der Laak, J. A., van
Ginneken, B., and S
´
anchez, C. I. (2017). A survey
on deep learning in medical image analysis. Medical
Image Analysis, 42:60–88.
Ma, L., Su, X., Ma, L., Gao, X., and Sun, M. (2023).
Deep learning for classification and localization of
early gastric cancer in endoscopic images. Biomed-
ical Signal Processing and Control, 79:104200.
Muruganantham, P. and Balakrishnan, S. M. (2022). At-
tention aware deep learning model for wireless cap-
sule endoscopy lesion classification and localiza-
tion. Journal of Medical and Biological Engineering,
42(2):157–168.
Ramsoekh, D., Haringsma, J., Poley, J. W., van Putten,
P., van Dekken, H., Steyerberg, E. W., van Leerdam,
M. E., and Kuipers, E. J. (2010). A back-to-back com-
parison of white light video endoscopy with autoflu-
orescence endoscopy for adenoma detection in high-
risk subjects. Gut, 59(6):785–793.
Recht, B., Roelofs, R., Schmidt, L., and Shankar, V. (2019).
Do ImageNet classifiers generalize to ImageNet? In
Chaudhuri, K. and Salakhutdinov, R., editors, Pro-
ceedings of the 36th International Conference on Ma-
chine Learning, volume 97 of Proceedings of Machine
Learning Research, pages 5389–5400. PMLR.
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and
Chen, L.-C. (2018). Mobilenetv2: Inverted residu-
als and linear bottlenecks. In Proceedings of the IEEE
Conference on Computer Vision and Pattern Recogni-
tion (CVPR).
Selvaraju, R. R., Das, A., Vedantam, R., Cogswell, M.,
Parikh, D., and Batra, D. (2016). Grad-cam: Why
did you say that? visual explanations from deep
networks via gradient-based localization. CoRR,
abs/1610.02391.
Siegel, R. L., Miller, K. D., Goding Sauer, A., Fedewa,
S. A., Butterly, L. F., Anderson, J. C., Cercek, A.,
Smith, R. A., and Jemal, A. (2020). Colorectal cancer
statistics, 2020. CA: A Cancer Journal for Clinicians,
70(3):145–164.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wo-
jna, Z. (2016). Rethinking the inception architecture
for computer vision. In Proceedings of the IEEE Con-
ference on Computer Vision and Pattern Recognition
(CVPR).
Tan, M. and Le, Q. (2019). EfficientNet: Rethinking model
scaling for convolutional neural networks. In Chaud-
huri, K. and Salakhutdinov, R., editors, Proceedings of
the 36th International Conference on Machine Learn-
ing, volume 97 of Proceedings of Machine Learning
Research, pages 6105–6114. PMLR.
Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M.,
and Luo, P. (2021). Segformer: Simple and efficient
design for semantic segmentation with transformers.
In Ranzato, M., Beygelzimer, A., Dauphin, Y., Liang,
P., and Vaughan, J. W., editors, Advances in Neural
A Comparative Analysis of EfficientNet Architectures for Identifying Anomalies in Endoscopic Images
539

Information Processing Systems, volume 34, pages
12077–12090. Curran Associates, Inc.
Yeung, M., Sala, E., Sch
¨
onlieb, C.-B., and Rundo, L.
(2021). Focus u-net: A novel dual attention-gated cnn
for polyp segmentation during colonoscopy. Comput-
ers in Biology and Medicine, 137:104815.
Yi-de, M., Qing, L., and Zhi-bai, Q. (2004). Automated im-
age segmentation using improved pcnn model based
on cross-entropy. In Proceedings of 2004 Interna-
tional Symposium on Intelligent Multimedia, Video
and Speech Processing, 2004., pages 743–746.
Yu, X., Tang, S., Cheang, C. F., Yu, H. H., and Choi, I. C.
(2022). Multi-task model for esophageal lesion anal-
ysis using endoscopic images: Classification with im-
age retrieval and segmentation with attention. Sen-
sors, 22(1).
Zhuang, H., Zhang, J., and Liao, F. (2021). A systematic re-
view on application of deep learning in digestive sys-
tem image processing. The Visual Computer, pages
1–16.
ICEIS 2024 - 26th International Conference on Enterprise Information Systems
540
