
Chaotic Convolutional Long Short-Term Memory Network for
Respiratory Motion Prediction
Narges Ghasemi
1,2 a
, Shahabedin Nabavi
1 b
, Mohsen Ebrahimi Moghaddam
1 c
and Yasser Shekofteh
1 d
1
Faculty of Computer Science and Engineering, Shahid Beheshti University, Tehran, Iran
2
Department of Computer Science, Viterbi School of Engineering, University of Southern California, U.S.A.
Keywords:
Convolutional Long Short-Term Memory, Deep Neural Network, Lung Motion, Radiotherapy, Respiratory
Motion Prediction.
Abstract:
One of the challenges of treating lung tumors in radiation therapy is the patient’s respiratory movements
during the treatment, which lead to tumor motion. The goal of respiratory motion prediction is to predict
the movements of lung tissues and lung tumors during the breathing cycle. Predicting respiratory movements
allows radiation to be directed only at the tumor, minimizing exposure to healthy tissue and reducing the risk
of side effects. Using 4D CT images, we can find the next position of the lung tumor and make a 4D radiation
therapy plan. As obtaining 4D CT scans is harmful to the patient due to radiation, the aim of this study is to
construct a 4D CT during a respiratory cycle using only a 3D image. In this paper, a Chaotic Convolutional
Long Short-Term Memory network is proposed, which utilizes chaotic features in respiratory signals to predict
pulmonary movements more accurately. The innovation of this method is paying attention to chaotic features
of respiratory signals, which leads to better interpretability of the presented model. The obtained results show
that the proposed method has a higher learning speed and better performance compared to previous models,
which generate 4D CT scans.
1 INTRODUCTION
Cancer Statistics 2022 reports that lung cancer re-
mains the foremost cause of cancer-related mortality
(Siegel et al., 2022), underlining the pressing need
for precise and effective treatments. Radiation ther-
apy, which employs high-energy rays to destroy can-
cer cells and shrink tumors, is a fundamental part of
lung cancer treatment. However, a significant chal-
lenge arises during the delivery of radiation therapy:
as patients breathe, the consequent movement of tu-
mors leads to inadvertent exposure of healthy tissue
to therapeutic rays. This not only increases the risk of
secondary cancers but also complicates the effective
targeting of lung tumors.
Respiratory motion prediction aims to predict how
lung tissues and lung tumors will move during breath-
ing. By integrating these predictions into treatment
a
https://orcid.org/0009-0006-6673-7760
b
https://orcid.org/0000-0001-7240-0239
c
https://orcid.org/0000-0002-7391-508X
d
https://orcid.org/0000-0002-6733-3702
planning, the precision in targeting tumors can be
significantly enhanced. Radiation therapists are then
able to tailor the treatment plan to synchronize with
the tumor’s movement, ensuring that the radiation is
delivered effectively to the intended target. The abil-
ity to concentrate radiation on the tumor, while limit-
ing exposure to healthy tissue, is crucial for reducing
side effects. This precision is especially important be-
cause radiation therapy, if not meticulously targeted,
can inadvertently harm nearby healthy tissues.
Efficient delivery of radiation, guided by accurate
prediction of pulmonary movements, can shorten the
duration of treatment sessions. This is of particular
importance for patients who face challenges enduring
lengthy treatments. Moreover, by precisely predicting
the movement of lung tissues, radiation therapists can
reduce the need for multiple treatment sessions that
may arise from initial targeting inaccuracies. This not
only improves the efficacy of the therapy but also sig-
nificantly enhances patient comfort by lessening the
overall treatment burden.
A series of three-dimensional Computed Tomog-
raphy (CT) scan images of the lung during the phases
Ghasemi, N., Nabavi, S., Moghaddam, M. and Shekofteh, Y.
Chaotic Convolutional Long Short-Term Memory Network for Respiratory Motion Prediction.
DOI: 10.5220/0012732600003720
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 4th International Conference on Image Processing and Vision Engineering (IMPROVE 2024), pages 99-106
ISBN: 978-989-758-693-4; ISSN: 2795-4943
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
99

of the respiratory cycle constitute a four-dimensional
(4D) CT scan. Essentially, a 4D CT scan compiles
multiple three-dimensional CT scans, each capturing
a different phase of respiration, to provide a time-
sequenced view of the lungs. This imaging technique
allows clinicians to observe the internal dynamics of
the body over time, which is invaluable in planning
treatment by considering the shape and movement of
the tumor and surrounding organs during the breath-
ing cycle. 4D CT scan images are a valuable tool for
estimating uncertainties related to respiratory move-
ments (Rehailia-Blanchard et al., 2019). In this pa-
per, we aim to propose a respiratory motion predic-
tion deep learning model that considers the inherent
characteristics of respiratory signals to predict future
slices of a lung CT volume based on the current slices
in the breathing cycle.
The contributions of this study are:
- A convolutional LSTM network architecture is
proposed in this study for respiratory motion predic-
tion to predict future slices in the breathing cycle us-
ing current slices. Using this method and 4D CT im-
ages as ground truth, the proposed model predicts the
slices of the next respiratory phase volume and com-
pares it with its corresponding slice.
- A chaotic feature extractor (CFE) and a chaotic
activation function (CAF) are used to improve the pre-
diction procedure and increase the accuracy of radia-
tion therapy. A chaotic convolutional LSTM model is
proposed to this end.
The rest of this article is organized as follows.
Section 2 reviews related studies. Section 3 describes
the proposed method. The results of the experiments
are presented in Section 4. Section 5 discusses the re-
sults obtained, followed by the conclusion in Section
6.
2 RELATED WORKS
Accurate respiratory motion prediction is crucial for
effective radiation treatment planning. Traditional
methods, such as those using external surrogates
marked on the chest or abdomen, have laid the
groundwork for capturing respiratory patterns. These
methods include predictive techniques based on the
autoregressive moving average method (McCall and
Jeraj, 2007), Kalman filter-based approaches (Lee
et al., 2011; Bukhari and Hong, 2014), and kernel es-
timation methods (Ruan, 2010). While valuable for
their non-invasiveness, they fall short in detailing the
complex internal organ motions that are vital for pre-
cision in modern medical imaging techniques.
With the advent of machine learning, more so-
phisticated approaches have emerged to surmount the
limitations of traditional methods. Techniques rang-
ing from Adaptive Neuro-Fuzzy Inference Systems
(Rostampour et al., 2018) to Artificial Neural Net-
works (ANNs) (Sun et al., 2017), and further to Re-
current Neural Networks (RNNs) (Kai et al., 2018),
have been explored for their capacity to factor in the
temporal dependencies missing from ANNs. The
evolution of RNNs into Long Short-Term Memory
(LSTMs) networks has specifically addressed the gra-
dient vanishing and explosion challenges characteris-
tic of earlier RNNs. In the domain of respiratory mo-
tion prediction, these LSTM networks, particularly
when trained on 4D CT imaging data, have shown
promise in enhancing prediction accuracy for treat-
ment planning (Lin et al., 2019). Additionally, re-
search has highlighted the potential of Conditional
Generative Adversarial Networks (CGANs), such as
the work of (Isola et al., 2017), originally designed
for image-to-image translation, in next frame predic-
tion tasks, including applications in respiratory mo-
tion prediction for medical imaging.
In the work of (Nabavi et al., 2020), the
ConvLSTM-based PredNet model, initially presented
in (Lotter et al., 2016) for predicting successive
frames in video sequences, has been adapted for fore-
casting subsequent slices in medical imaging. This
model leverages the existing slices as inputs to antic-
ipate future slices in a sequence. Additionally, work
of (Ghasemi and Samadi Miandoab, 2022) employs
parallelized ConvLSTM layers to enhance feature ex-
traction from the data slices, improving the predictive
capabilities of the model. Our study seeks to bridge
this gap by introducing a novel chaotic feature ex-
tractor (CFE) and a chaotic activation function (CAF)
within the ConvLSTM framework. These additions
are specifically designed to grapple with the unpre-
dictable, chaotic nature of respiratory movement. By
doing so, our model ventures beyond existing models,
leveraging the chaotic dynamics of respiratory mo-
tion to potentially elevate the accuracy of predictions
and adaptability to the unique breathing patterns of
patients.
3 METHOD
The Method section is organized into three primary
parts. First, we lay out the problem formulation. The
second part details the proposed method, and the fi-
nal part describes the implementation details and set-
tings, offering insight into how the model is applied
and tested.
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
100

3.1 Problem Formulation
In this study, we approach the prediction of respira-
tory motion using 4D CT data to infer the future state
of lung CT volumes. We seek to calculate the lung
CT volume at a future time point t + ∆t based on the
current volume at time t. Our model works by pre-
dicting each corresponding slice of the future CT vol-
ume, ˆs
i
t+∆t
, from the current CT volume slice, s
i
t
, and
then reconstructing the complete future volume
ˆ
V
t+∆t
.
The prediction for each slice at a given time step t and
slice s
i
t
is formulated as:
ˆs
i
t
+∆t
= f (s
i
t
) (1)
where f denotes the motion prediction model.
3.2 Proposed Method
The input of the model is the current CT volume, and
we aim to predict the CT volume at the next time step.
These volumes are sliced, and the input of the model
is a slice of the current volume, aiming to predict the
corresponding slice in the next time step. Initially,
a chaotic feature extractor generates a feature map
with dimensions similar to the input image. This fea-
ture map is then concatenated with the input image,
followed by a convolution operation that reduces the
channel size to one, merging the information of the
image and the chaotic features. ConvLSTM blocks,
interlaced with chaotic activation functions, further
extract features, and a final convolution block gener-
ates the predicted future slice. Batch Normalization is
applied between ConvLSTM blocks to enhance train-
ing speed. The architecture of the proposed frame-
work is illustrated in Figure 1.
Chaos is characterized by seemingly irregular,
unpredictable long-term, and non-periodic behavior
within a deterministic system. Research of (Michalski
et al., 2014) has demonstrated that respiratory signals
exhibit complex nonlinear dynamics and chaotic be-
havior. To explore this chaotic behavior in lung CT
scans, a 2D discrete cosine transform (2D DCT) is
computed on the input image. The image is then tra-
versed in a zigzag pattern from the bottom right to the
top left, forming a 1D vector. This vector is subjected
to a chaos test (Gottwald and Melbourne, 2004), ver-
ifying its chaotic properties.
3.2.1 Chaotic Feature Extractor
Artificial intelligence has achieved remarkable suc-
cess in practical applications, yet it often lacks a
close representation of the chaotic firing properties
observed in biological neurons. This limitation has
spurred the development of neuron architectures that
embody intrinsic chaos, as seen in biological systems.
In (Harikrishnan and Nagaraj, 2019; Balakrishnan
et al., 2019), a single-layer chaos-inspired neuronal
architecture for classification problems is proposed.
In our research, the Generalized Lur
¨
oth Series (GLS)
neurons form the input layer, consisting of n neurons
G
1
, G
2
, ..., G
n
, where n is the length of the vectorized
input image, and the pixel values are normalized to
the interval [0,1].
The GLS neuron is described by a 1-D piecewise
linear chaotic map T : [0, 1) → [0, 1), given by:
T (x) =
(
x
b
if 0 ≤ x < b
1−x
1−b
if b ≤ x < 1
(2)
where x lies in the range [0,1). The parameter b is a
crucial component of the GLS map, influencing the
neuron’s chaotic behavior. In this study, the value of
b was determined through empirical testing. Various
values were trialed, and the optimal b was selected
based on the performance of the model.
Each GLS neuron starts with an initial neural ac-
tivity q, representing the initial value for the chaotic
map. These neurons then fire chaotically in response
to a stimulus, a real number between 0 and 1. The
firing time—or the number of iterations required for
the neuron’s output to fall within an epsilon neighbor-
hood of the pixel’s initial value—is used as the chaotic
feature for that particular pixel. The concept of Topo-
logical Transitivity (TT) ensures that this process will
converge, meaning that firing will eventually cease,
as demonstrated in (Harikrishnan and Nagaraj, 2019;
Balakrishnan et al., 2019).
The features extracted by this method, now in vec-
tor form, are then reshaped to match the input image’s
dimensions and concatenated back to the input image
for further processing.
3.2.2 Convolutional Long Short-Term Memory
The 4D CT captures the movement of the organs and
tumor over time. It consists of a sequence of 3D CTs.
In order to take advantage of the temporal dependency
between images, we can use long short-term memory
that is created to predict time series. The main draw-
back of long short-term memory is not paying atten-
tion to the spatial characteristics of the data because
it loses the spatial properties by vectorizing the data.
This problem can be overcome by using convolutional
long short-term memory, which is designed for spa-
tiotemporal sequence forecasting problems (Shi et al.,
2015).
Chaotic Convolutional Long Short-Term Memory Network for Respiratory Motion Prediction
101

Figure 1: The architecture of the Chaotic Convolutional LSTM Network (CCLSTMNet).
3.2.3 Chaotic Activation Function
In recent studies, oscillatory activation functions have
shown promise in improving the gradient process
and minimizing the network size (Noel et al., 2021).
Traditional activation functions such as the sigmoid,
rectified linear unit (ReLU), and hyperbolic tangent
(Tanh) come with their respective benefits and draw-
backs. The vanishing gradient problem is a challenge
for sigmoid and Tanh functions, slowing the learn-
ing process, whereas ReLU can result in dead neurons
when many neurons’ activation values are zero.
A chaos-based activation function was proposed
to better mirror the neuronal structure of the brain,
incorporating the sigmoid function with the logistic
map to induce chaos (Reid and Ferens, 2021). The lo-
gistic map is utilized for its capacity to create chaotic
behavior.
The sigmoid function, bounded between zero and
one, ensures the logistic map’s input remains within
the required range, defined as follows:
Sigmoid(x) =
1
1 + e
−x
(3)
Subsequently, the output of the sigmoid function
serves as the input to the logistic map:
LogisticMap(x) = r · x · (1 − x) (4)
where r is the excitatory rate of a neuron. This com-
posite function constitutes the chaos-based activation
function, instilling a chaotic dynamic in the network’s
function.
3.3 Implementation Details
The code implementation was performed using the
Keras library in Python, with the computational envi-
ronment provided by Google Colaboratory Pro (Co-
lab Pro), a cloud-based Jupyter Notebook service. A
batch size of one was utilized. We started with a learn-
ing rate of 0.001, implementing a reduction strategy
when the loss ceased improving for a duration of five
epochs. The neural network was trained using the
Mean Squared Error (MSE) loss function to minimize
the difference between the predicted outputs and the
ground truth data. The optimization of the network
weights was conducted using the Adam optimizer.
Considering computational efficiency, the input
images were resized to a resolution of 160× 160. The
’leave one patient out’ cross-validation method was
employed for model evaluation. This involved using
data from one patient as the test set and the rest for
training, iterating through each patient. The initial
neural activity for the GLS neurons was set at 0.7,
with the parameter b in the GLS map from Equation
2 set to 0.467.
4 EXPERIMENTAL RESULTS
In this section, we present the experimental setup used
to evaluate the respiratory motion prediction frame-
work, with comparisons to state-of-the-art methods.
To examine how each component contributes to the
framework’s overall performance, a set of experi-
ments is presented. We achieve this by conducting an
ablation study and experiments focusing on individ-
ual components such as chaotic feature extraction and
chaotic activation function. Additionally, we compare
our method with other approaches that generate im-
ages.
4.1 Experimental Designs
In our study, we employed the CREATIS dataset,
which consists of ten 3D volumes per patient, rep-
resenting distinct phases of the respiratory cycle for
six patients. The imaging was conducted with a 16
Slice Brilliance CT Big Bore Oncology™ configura-
tion from Philips (Vandemeulebroucke et al., 2007).
Axially sliced volumes were saved as reference im-
ages. The number of images in axial view for each
patient is outlined in Table 1. In total, 10090 CT slices
were compiled for the six patients. To manage com-
putation costs, these slices were resized to 160 × 160
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
102

in the preprocessing phase.
Table 1: Number of slices per volume.
Patient Number of slices per volume
Patient 1 141
Patient 2 169
Patient 3 170
Patient 4 187
Patient 5 181
Patient 6 161
The leave-one-patient-out (LOPO) cross-
validation method was applied to evaluate the
model’s robustness, with a training set composed of
the data from five patients and the sixth patient’s data
serving as the test set. This procedure was cycled
through each patient to complete the validation
process.
In this paper, we employ three evaluation met-
rics to assess the performance of our proposed model:
Root Mean Squared Error (RMSE), Structural Simi-
larity Index (SSIM), and Peak Signal-to-Noise Ratio
(PSNR).
This metric measures the difference between pre-
dicted and actual values. When applied to image pro-
cessing, it measures how closely a predicted image
matches the original image. A lower RMSE indicates
less error between the predicted and original images,
which means the predicted image is more accurate.
RMSE is calculated using Equation 5:
RMSE(x, y) =
s
∑
M
i=1
∑
N
j=1
(x(i, j)− y(i, j))
2
MN
(5)
where x and y represent the ground truth and the pre-
dicted images, respectively, with M representing the
number of rows and N representing the number of
columns in each image.
SSIM measures the similarity between two images
by comparing their structural information, luminance,
and contrast. It provides a score ranging from -1 to 1,
with a higher score indicating greater similarity be-
tween the predicted and original images. SSIM is de-
fined as per Equation 6:
SSIM(x, y) =
(2µ
x
µ
y
+ c
1
)(2σ
xy
+ c
2
)
(µ
2
x
+ µ
2
y
+ c
1
)(σ
2
x
+ σ
2
y
+ c
2
)
(6)
where µ
x
and µ
y
are the average pixel values of images
x and y, σ
2
x
and σ
2
y
are the variances of x and y, and σ
xy
is the covariance of the images. The constants c
1
and
c
2
are small stabilizers to prevent division by zero.
PSNR is another measure of image quality evalu-
ation, which quantifies the quality of a reconstructed
image or video by comparing it to the original, undis-
torted version. A higher PSNR value suggests less
distortion. PSNR is obtained from Equation 7:
PSNR(x, y) = 10log
10
MAX
2
I
MSE(x, y)
(7)
where MAX
I
is the maximum possible pixel value of
the image, and MSE is the mean squared error as cal-
culated in Equation 8.
MSE(x, y) =
1
MN
M
∑
i=1
N
∑
j=1
(x(i, j)− y(i, j))
2
(8)
4.2 Results
In Table 2, we present the quantitative results obtained
from the Chaotic ConvLSTM network for each pa-
tient in terms of RMSE, SSIM, and PSNR. Also, the
weighted average of these results is presented. Fig-
ure 2 shows the RMSE and SSIM between ground
truth and predicted slices across the different patients.
Figure 3 shows some examples of the model out-
put, ground truth, and their difference during the ten
phases.
Table 3 shows the comparison results of the res-
piratory motion prediction algorithms using ConvL-
STM and CGAN on the CREATIS dataset. Compared
to other algorithms, the proposed strategy accuracy
has been improved, and the training time has been re-
duced.
4.3 Ablation Study
In order to better understand the role and contribution
of each module in our framework, an ablation study
was performed. Different configurations of the pro-
posed model have been deployed. The baseline ver-
sion of the model includes a sequence of ConvLSTM
with a chaotic activation function and a CNN module.
This means that the model attempts to learn how to
generate volumes directly by generating slices with-
out considering chaotic features. Table 4 shows the
results of the ablation study, evaluating each configu-
ration based on the quantitative metrics.
5 DISCUSSION
In this paper, we introduced a Chaotic ConvLSTM
network designed for predicting 3D CT scan slices
from preceding scans. The innovation of this model
lies in harnessing chaotic features inherent in respi-
ratory signals, leading to notable enhancements in
learning speed and prediction accuracy compared to
existing methods. Such improvements underscore the
Chaotic Convolutional Long Short-Term Memory Network for Respiratory Motion Prediction
103
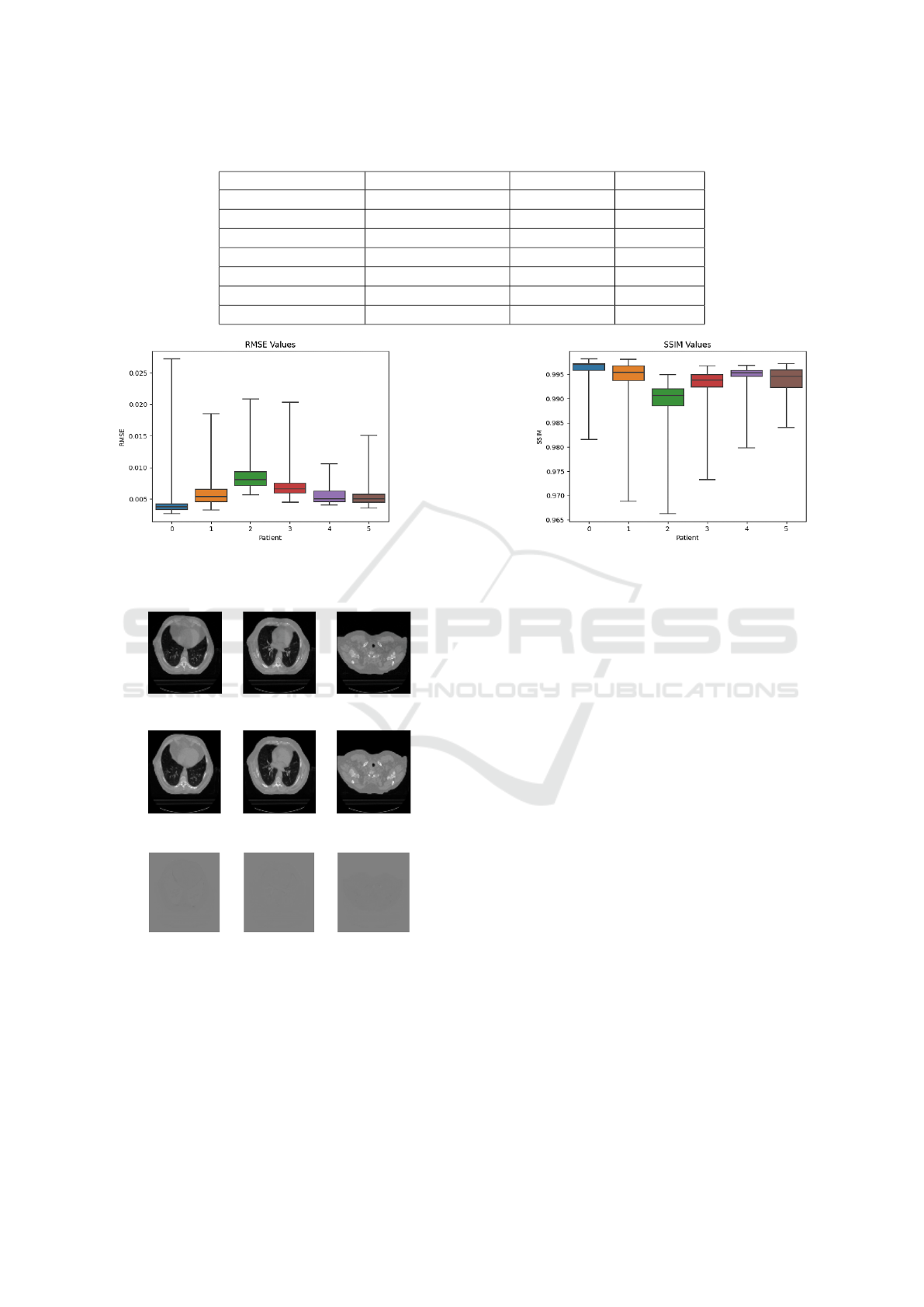
Table 2: Quantitative metrics result.
Patient RMSE SSIM PSNR
Patient 1 experiment 4 × 10
−3
0.995 48.09
Patient 2 experiment 5 × 10
−3
0.994 44.93
Patient 3 experiment 8 × 10
−3
0.988 41.49
Patient 4 experiment 7 × 10
−3
0.992 43.13
Patient 5 experiment 5 × 10
−3
0.993 45.17
Patient 6 experiment 5 × 10
−3
0.993 45.53
Weighted average 6 × 10
−3
± 1 × 10
−3
0.993 ± 0.002 44.60 ± 1.6
(a) (b)
Figure 2: (a) RMSE values obtained for 6 patients. (b) SSIM values obtained for 6 patients.
(a) Reference slices.
(b) Predicted slices.
(c) Difference between reference and predicted
slices.
Figure 3: The predicted pulmonary motion. The first row
represents the reference slices; the second row represents
the predicted slices, and the third row represents the differ-
ence between the reference image and the predicted image.
model’s potential for clinical application, particularly
in optimizing radiation therapy through respiratory
motion prediction.
Given the chaotic nature of respiratory signals, our
approach employs chaotic feature extraction, mim-
icking the complex firing patterns observed in neural
structures. This strategy not only refines the predic-
tion process but also contributes to the precision of
radiation therapy, potentially increasing its efficacy.
The process of generating images, a core compo-
nent of our method, is inherently non-invasive. By
predicting 4D CT scans within a respiratory cycle, our
approach offers detailed insights into organ motion,
encompassing anatomical details and tissue charac-
teristics beyond mere motion tracking. This wealth
of information holds significant promise for enhanc-
ing treatment planning and diagnostic accuracy.
Recent advances in medical imaging technolo-
gies have significantly broadened our capabilities in
diagnosing and treating complex health conditions.
Among these, 4D CT imaging stands out for its com-
prehensive visualization of organ and tissue dynam-
ics. Nonetheless, the application of 4D CT imaging is
not without its drawbacks, notably the high radiation
exposure required and the potential for image distor-
tion due to the compilation of multiple CT scans over
time.
Our proposed Chaotic ConvLSTM network
presents a viable alternative, especially in settings
lacking advanced 4D CT imaging capabilities. Its
ability to simulate 4D imaging from a single 3D
CT scan could substantially reduce radiation expo-
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
104
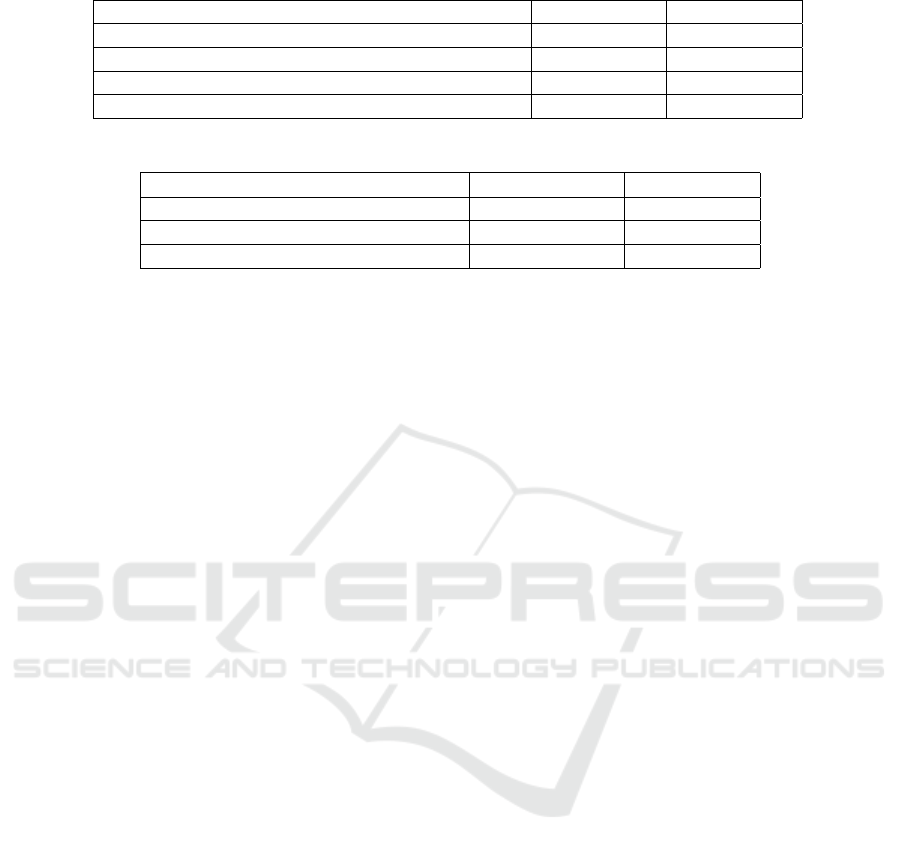
Table 3: Performance comparison of video next frame prediction models on a 4D MRI video.
Model RMSE SSIM
cGAN(Isola et al., 2017) 0.015 0.937
ConvLSTM(Nabavi et al., 2020) 0.009 0.943
ConvLSTM(Ghasemi and Samadi Miandoab, 2022) 0.012 0.988
Proposed Method 0.006 ± 0.001 0.993 ± 0.002
Table 4: Quantitative metrics comparison between different configurations.
Model RMSE (×10
−3
) SSIM
ConvLSTM 744 ± 163 0.959
Baseline (ConvLSTM + CAF) 653 ± 152 0.992 ± 0.002
Proposed (ConvLSTM + CAF + CFE) 626 ± 141 0.993 ± 0.002
sure and eliminate the distortions and artifacts typical
of traditional 4D CT reconstructions. These advan-
tages are particularly appealing for radiotherapy cen-
ters with limited access to cutting-edge imaging tech-
nology, offering a cost-effective yet efficient solution.
Furthermore, the comparative analysis of error
rates and the fidelity of generated images to their
ground truth counterparts confirm the superiority of
our model over traditional ConvLSTM-based ap-
proaches. This efficacy stems from our model’s
unique integration of chaotic dynamics, enhancing its
ability to predict pulmonary motion with remarkable
accuracy.
In light of these findings, our work contributes to
the ongoing evolution of medical imaging and pre-
dictive modeling. By leveraging the chaotic pat-
terns inherent in biological systems, we offer a novel
perspective on motion prediction that could signifi-
cantly impact the field of radiation therapy and be-
yond. Future work will aim to expand the application
of chaotic feature extraction and explore its potential
in other areas of medical imaging and treatment plan-
ning, paving the way for broader clinical adoption.
6 CONCLUSIONS
In this study, we developed and presented a novel
Chaotic Convolutional LSTM Network aimed at en-
hancing the accuracy of respiratory motion prediction
for radiation therapy planning. This method predicts
each subsequent CT slice based on the preceding one.
This approach, tested on the CREATIS dataset, un-
derscores the method’s superiority over traditional 4D
CT-generating techniques by offering significant im-
provements in prediction accuracy.
The promising outcomes of this research highlight
not just the methodological advancements but also
the potential for substantial clinical impact, particu-
larly in the field of radiation therapy where precise
tumor targeting is critical. The reduced error rates and
the method’s non-invasive nature represent significant
steps forward in the pursuit of safer, more effective
treatment strategies.
Looking ahead, our research opens several av-
enues for further investigation and application. A
key direction for future work involves validating the
proposed Chaotic ConvLSTM network against dy-
namic breathing lung phantoms, a crucial step to-
wards confirming its applicability and effectiveness in
real-world clinical settings. Additionally, exploring
the integration of this network into existing radiation
therapy planning systems could provide actionable in-
sights into its operational benefits and challenges.
The successful application of chaotic character-
istics in respiratory motion prediction underscores a
promising path for future investigations. We plan
to explore the potential of the Chaotic ConvLSTM
network in modeling physiological processes with
similar chaotic dynamics. This exploration aims to
broaden the utility of our approach, offering a com-
prehensive tool for analyzing and predicting complex
physiological behaviors that exhibit chaotic proper-
ties. By focusing on these areas, we intend to deepen
our understanding of chaotic phenomena in biology
and their implications for medical imaging and diag-
nostics.
In conclusion, the Chaotic ConvLSTM network
represents an advancement in the predictive model-
ing of respiratory motion. Its development not only
contributes to the body of knowledge in medical
imaging and computational biology but also sets the
stage for groundbreaking applications in personalized
medicine and advanced healthcare delivery.
ACKNOWLEDGEMENTS
The authors are grateful to L
´
eon B
´
erard Cancer Cen-
ter and CREATIS Laboratory, Lyon, for sharing their
Chaotic Convolutional Long Short-Term Memory Network for Respiratory Motion Prediction
105

imaging data.
REFERENCES
Balakrishnan, H. N., Kathpalia, A., Saha, S., and Nagaraj,
N. (2019). Chaosnet: A chaos based artificial neural
network architecture for classification. Chaos: An In-
terdisciplinary Journal of Nonlinear Science, 29(11).
Bukhari, W. and Hong, S. (2014). Real-time prediction
and gating of respiratory motion using an extended
kalman filter and gaussian process regression. Physics
in Medicine & Biology, 60(1):233.
Ghasemi, Z. and Samadi Miandoab, P. (2022). Feasibility
study of convolutional long short-term memory net-
work for pulmonary movement prediction in ct im-
ages. Journal of Biomedical Physics and Engineering.
Gottwald, G. A. and Melbourne, I. (2004). A new test
for chaos in deterministic systems. Proceedings of
the Royal Society of London. Series A: Mathematical,
Physical and Engineering Sciences, 460(2042):603–
611.
Harikrishnan, N. B. and Nagaraj, N. (2019). A novel chaos
theory inspired neuronal architecture. In 2019 Global
Conference for Advancement in Technology (GCAT),
pages 1–6. IEEE.
Isola, P., Zhu, J.-Y., Zhou, T., and Efros, A. A. (2017).
Image-to-image translation with conditional adversar-
ial networks. In Proceedings of the IEEE conference
on computer vision and pattern recognition, pages
1125–1134.
Kai, J., Fujii, F., and Shiinoki, T. (2018). Prediction of lung
tumor motion based on recurrent neural network. In
2018 IEEE International Conference on Mechatronics
and Automation (ICMA), pages 1093–1099. IEEE.
Lee, S. J., Motai, Y., and Murphy, M. (2011). Respiratory
motion estimation with hybrid implementation of ex-
tended kalman filter. IEEE Transactions on Industrial
Electronics, 59(11):4421–4432.
Lin, H., Shi, C., Wang, B., Chan, M. F., Tang, X., and Ji,
W. (2019). Towards real-time respiratory motion pre-
diction based on long short-term memory neural net-
works. Physics in Medicine & Biology, 64(8):085010.
Lotter, W., Kreiman, G., and Cox, D. (2016). Deep predic-
tive coding networks for video prediction and unsu-
pervised learning. arXiv preprint arXiv:1605.08104.
McCall, K. and Jeraj, R. (2007). Dual-component model of
respiratory motion based on the periodic autoregres-
sive moving average (periodic arma) method. Physics
in Medicine & Biology, 52(12):3455.
Michalski, D., Huq, M., Bednarz, G., Lalonde, R., Yang, Y.,
and Heron, D. (2014). Su-e-j-261: Statistical analysis
and chaotic dynamics of respiratory signal of patients
in bodyfix. Medical Physics, 41(6Part10):218–218.
Nabavi, S., Abdoos, M., Moghaddam, M. E., and Moham-
madi, M. (2020). Respiratory motion prediction using
deep convolutional long short-term memory network.
Journal of Medical Signals & Sensors, 10(2):69–75.
Noel, M. M., Trivedi, A., Dutta, P., et al. (2021). Grow-
ing cosine unit: A novel oscillatory activation func-
tion that can speedup training and reduce parame-
ters in convolutional neural networks. arXiv preprint
arXiv:2108.12943.
Rehailia-Blanchard, A., De Oliveira Duarte, S., Baury, M.,
Auberdiac, P., Diard, A., Brun, C., Talabard, J., Ran-
coule, C., Magn
´
e, N., et al. (2019). Use of 4d-ct: Main
technical aspects and clinical benefits. Cancer Radio-
therapie: Journal de la Societe Francaise de Radio-
therapie Oncologique, 23(4):334–341.
Reid, S. and Ferens, K. (2021). A hybrid chaotic activation
function for artificial neural networks. In Advances in
Artificial Intelligence and Applied Cognitive Comput-
ing: Proceedings from ICAI’20 and ACC’20, pages
1097–1105. Springer.
Rostampour, N., Jabbari, K., Esmaeili, M., Mohammadi,
M., and Nabavi, S. (2018). Markerless respiratory tu-
mor motion prediction using an adaptive neuro-fuzzy
approach. Journal of medical signals and sensors,
8(1):25.
Ruan, D. (2010). Kernel density estimation-based real-time
prediction for respiratory motion. Physics in Medicine
& Biology, 55(5):1311.
Shi, X., Chen, Z., Wang, H., Yeung, D.-Y., Wong, W.-K.,
and Woo, W.-c. (2015). Convolutional lstm network:
A machine learning approach for precipitation now-
casting. Advances in neural information processing
systems, 28.
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A.
(2022). Cancer statistics, 2022. CA: a cancer journal
for clinicians, 72(1):7–33.
Sun, W., Jiang, M., Ren, L., Dang, J., You, T., and Yin, F.
(2017). Respiratory signal prediction based on adap-
tive boosting and multi-layer perceptron neural net-
work. Physics in Medicine & Biology, 62(17):6822.
Vandemeulebroucke, J., Sarrut, D., Clarysse, P., et al.
(2007). The popi-model, a point-validated pixel-based
breathing thorax model. In XVth international con-
ference on the use of computers in radiation therapy
(ICCR), volume 2, pages 195–199.
IMPROVE 2024 - 4th International Conference on Image Processing and Vision Engineering
106
