
Towards Accurate Cervical Cancer Detection: Leveraging Two-Stage
CNNs for Pap Smear Analysis
Franklin Steven De la Cruz Paucar
1 a
, Carlos Julio Macancela Bojorque
1 b
, Iv
´
an Reyes-Chac
´
on
2 c
,
Paulina Vizcaino-Imaca
˜
na
2 d
and Manuel Eugenio Morocho-Cayamcela
1,2 e
1
Yachay Tech University, School of Mathematical and Computational Sciences, DeepARC Research Group,
Hda. San Jos
´
e s/n y Proyecto Yachay, Urcuqu
´
ı, 100119, Ecuador
2
Universidad Internacional del Ecuador, Faculty of Technical Sciences, School of Computer Science,
Quito, 170411, Ecuador
Keywords:
Convolutional Neuronal Network, Cervical Cancer, Pap Smear Test, Dataset, Computer Vision.
Abstract:
Cervical cancer is a type of cancer that occurs in the cervix. It is caused by the abnormal growth of cells in the
cervix and is often caused by the human papillomavirus. Symptoms can include abnormal vaginal bleeding,
and pelvic pain, among others. It is usually diagnosed with a pelvic exam, biopsy, and Papanicolaou or pap test.
Generally, during the test, a small sample of cells is taken from the cervix and examined under a microscope to
look for any abnormal cells. The test is usually done during a pelvic exam and can be done in a doctor’s office
or clinic, which can cause human errors to exist and lead to a deficit of service or misdiagnosis for patients.
Especially, in Ecuador, cervix cancer is the second with the most prominent incidence and mortality. One of
the obstacles in Latin America to improving the number of cervix cancer screens is the amount of time needed
to give results. This paper proposes a pre-trained artificial neural network and a much larger database than
its paper base, this will allow us to obtain better results and a network with more accurate predictions when
throwing where malignant cells could be located that could lead to cervical cancer. The process to carry it
out is similar to its original process, where the analysis of the Papanicolaou tests is carried out in two stages.
The first focused on finding the coordinates of the anomalous cells observed within each of the images of our
dataset and the second, specializing in being able to obtain an image with a much higher resolution for each of
these coordinates, thus obtaining an improvement and being able to give a much more reliable diagnosis for
each of the patients.
1 INTRODUCTION
Detecting cancer is the process of identifying the
presence of cancer cells in a person’s body. This can
be done through various methods including physical
exams, imaging tests, biopsy, and blood tests. Each
method will depend on the type of cancer that needs
to be recognized and the area in the body where it is
suspected to be (Hasenleithner and Speicher, 2022).
Since the different ways to detection of cancer, the in-
clusion of modern technology such as computer vi-
sion algorithms can quickly and accurately analyze
large amounts of medical images, reducing the need
a
https://orcid.org/0000-0002-0429-5219
b
https://orcid.org/0000-0002-8265-3261
c
https://orcid.org/0009-0002-2731-5531
d
https://orcid.org/0000-0001-9575-3539
e
https://orcid.org/0000-0002-4705-7923
for human interpretation and increasing the speed of
giving a diagnosis (Koh et al., 2022). Object detec-
tion and image classification are two common tech-
niques used in computer vision for detecting cancer
cells in medical images such as microscopy images
and medical scans (Rom
´
an et al., 2023). Object de-
tection algorithms aim to locate instances of objects
within an image and draw bounding boxes around
them. In medical imaging, object detection can be
used to identify and localize cancer cells within a tis-
sue sample (Elyan et al., 2022). Image classification,
on the other hand, aims to assign an input image to a
specific class or category, such as “normal” or “can-
cerous”. In medical imaging, image classification can
be used to classify a patch of an image as cancerous
or non-cancerous (Elyan et al., 2022). In this case,
we will focus on cervical cancer by using two steps, a
low-resolution scanning for quick cell detection and
Paucar, F., Bojorque, C., Reyes-Chacón, I., Vizcaino-Imacaña, P. and Morocho-Cayamcela, M.
Towards Accurate Cervical Cancer Detection: Leveraging Two-Stage CNNs for Pap Smear Analysis.
DOI: 10.5220/0012735800003753
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Conference on Software Technologies (ICSOFT 2024), pages 219-227
ISBN: 978-989-758-706-1; ISSN: 2184-2833
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
219

location and a second high-resolution one, for de-
tailed classification. Cervical cancer is challenging
to detect for several reasons. First, the human papil-
lomavirus (HPV) is the main cause of cervical cancer
and is highly contagious and spreads easily (Franco
et al., 2001). Also, many people infected with HPV
do not have symptoms, making early detection of can-
cer difficult. In addition, HPV can cause different
types of cancer, which can present differently and re-
quire different approaches to detection and treatment.
Finally, current screening methods have limitations in
terms of accuracy and availability, making early and
effective detection of cervical cancer difficult.
On the other hand, given that it is a very challeng-
ing and critical issue in the health field, the World
Health Organization (WHO) in 2020 proposed a sig-
nificant initiative to eradicate cervical cancer globally.
This is based on the three key pillars of HPV vac-
cination, cervical screening, and treatment, and also
has associated intervention targets for the year 2030
(Davies-Oliveira et al., 2021). This initiative is based
on a goal called “90-70-90”, which refers to the fact
that 90% of adolescent girls should receive the HPV
vaccine, 70% of adult women should have at least two
cervical HPV tests in their lifetime, and 90% of them
should receive appropriate treatment if they have an
HPV-related disease, either pre-invasive or invasive.
In 2009, a survey of 81 women in both urban
and rural areas of Ecuador revealed dissatisfaction
with the lengthy wait for test results. The delays
were caused by insufficient equipment and the lim-
ited capacity of trained professionals, especially in
rural areas (Strasser-Weippl et al., 2015). Addition-
ally, nearly half of women in Ecuador over the age of
18 have never undergone a cervical cancer screening
(ins, 2018). Hence, the analysis of cervical screening
can only be performed by three specialists: anatomic
pathologists, cytologists, and clinical pathologists.
This limited number of specialties for evaluating pap
smear tests increases the difficulty of evaluating more
tests. According to a 2021 WHO report, in Ecuador
showed that the number of medical personnel per
10,000 cancer patients in Ecuador was as follows in
2019: 0 radiation oncologists, 3 medical physicists,
154 radiologists, and 4 nuclear medicine physicians
(wor, 2021). The lack of equipment, trained profes-
sionals, and the large amount of tests that are per-
formed and have to be analyzed are problems that do
not allow for achieving the goals proposed by WHO.
In this paper, we propose a pre-trained artificial
neural network and a much larger database than its
paper base, this will allow us to obtain better results
and a network with more accurate predictions when
throwing where malignant cells could be located that
could lead to cervical cancer. The process to carry it
out is similar to its original process, where the anal-
ysis of the Papanicolaou tests is carried out in two
stages. The first focused on finding the coordinates
of the anomalous cells observed within each of the
images of our dataset and the second, specializing in
being able to obtain an image with a much higher res-
olution for each of these coordinates, thus getting an
improvement and being able to give a much more re-
liable diagnosis for each of the patients.
2 RELATED WORKS
Because of the improvement in the area for taking cy-
tological images. It is possible to develop computer-
aided cancer diagnosis methods. One of the first ones
is from Yamal et al. started the work related to the de-
tection of cervical cancer by using hierarchical data.
The logistic regression algorithm is used to distin-
guish cervical cancer at the cellular level and use an
add Hoc approach to classify it at the patient level.
The dataset was about 1728 patients obtaining a 61%
and specificity of 89% on the independent dataset
(Yamal et al., 2015).
Having this first algorithm used, there is also an-
other from Su et al. that proposes a two-level cascade
integration system to classify the cervical cells into
normal and abnormal, and logistic regression (LR)
was used individually as the work presented before.
However, by the integration of a two-level cascade,
the false positives were lower than in the traditional
pap smear review. Obtaining the recognition rate of
95.6 % (Su et al., 2016).
Other traditional machine learning methods were
applied. In this case, Kurniawati et al. propose differ-
ent methods for cervical cancer prediction using a pap
smear, such as Na
¨
ıve Bayes, support vector machines,
and random forest. The data used were obtained from
the medical records of the pap smear test results. The
performance metric used is accuracy. The best accu-
racy of 80.18% was obtained by random forest (Kur-
niawati et al., 2016).
Alsalatie et al. focus on cervical cancer screening
and the challenges that traditional screening methods
approach face. Alsalatie mentions the importance of
computer-assisted diagnosis to improve the accuracy
of cervical cancer screening. According to the paper,
conventional screening methods take into account the
knowledge of a pathologist which can result in mis-
diagnosis and low diagnostic effectiveness. In addi-
tion, the article proposes a deep learning model de-
signed for the automatic diagnosis of whole-slide im-
ages (WSI) in cervical cancer samples. The proposed
ICSOFT 2024 - 19th International Conference on Software Technologies
220

network has a high accuracy rate of up to 99.6%, and
considers the entire staining slice image, not just a
single cell. The deep learning architecture considers
overlapping and non-overlapping cervical cells in the
WSI. Finally, they mention that the work is distinct
from existing research in terms of simplicity, accu-
racy, and speed (Alsalatie et al., 2022).
Hussain et al. discuss the importance of using
clinical data in AI systems for automated disease di-
agnosis, prediction, or classification. The article re-
marks the importance of a publicly available bench-
mark dataset, and that the hospital data collected from
a clinical setup is also important. The data must
be frequently updated to guarantee that developed
AI systems are as accurate as possible. Thus, Hus-
sain provides a liquid-based cytology (LBC) reposi-
tory with images collected from 460 patients visiting
a public hospital’s O&G department. The repository
consists of 963 LBC images split into four classes,
high squamous intra-epithelial lesion, low squamous
intra-epithelial lesion, negative for intra-epithelial
malignancy, and squamous cell carcinoma represent-
ing pre-cancerous and cancerous lesions. The images
were taken with a Leica ICC50 HD microscope and
categorized by experts from the pathology department
(Hussain et al., 2020).
Rezende et al. treat the difficulties faced in ac-
curately detecting cervical cancer through the con-
ventional pap smear test. Furthermore, the authors
mentioned the importance of computational tools to
support screening efficiently, especially during the
current health crisis. The article said that machine
learning can reduce the test limitations, but the lack
of high-quality datasets has break the development
of strategies to improve cervical cancer screening.
The Center for Recognition and Inspection of Cells
(CRIC) platform has created the CRIC Cervix col-
lection, which currently contains 400 images of con-
ventional pap smears with manual classification of
11,534 cells. The dataset is a good beginning for
improving machine learning algorithms to automate
tasks in cytopathological analysis (Rezende et al.,
2021).
Zhang et al. explore the use of deep convolutional
neural networks for cervical cell classification. The
authors describe how they used a deep convolutional
neural network to analyze cervical cell images and
classify them into relevant categories for cervical can-
cer detection. They propose a method to directly clas-
sify cervical cells based on deep features using con-
volutional neural networks (ConvNets), without prior
segmentation. Unlike previous methods that relied on
hand-crafted features and cytoplasm/nucleus segmen-
tation, this method automatically extracts deep fea-
tures embedded in the cell patch for classification. It
involves extracting image patches centered roughly
on the nucleus as input to the network, transferring
features from a pre-trained model to a new ConvNet
to fine-tune the cell image patches, and aggregating
multiple predictions to form the final network output.
The results show that the DeepPap neural network is
effective in the task of cervical cell classification and
this proposed method was evaluated on Papanicolaou
and LBC smear datasets. The results show that their
method outperforms previous algorithms in classifi-
cation accuracy (98.3%) (Zhang et al., 2017).
Sompawong et al. This study investigates the
use of the neural network known as Mask Region-
Based convolutional neural network (R-CNN) for de-
tecting cervical cancer from Papanicolaou tests. The
researchers claim that this is the first time that this
technology is used to identify and examine the nu-
cleus of cervical cells. Tissue samples obtained from
a hospital were used to train the algorithm. The aver-
age accuracy of the model was 57.8%, with an accu-
racy of 91.7%, sensitivity of 91.7%, and specificity of
91.7% per image. Additionally, this model was com-
pared to another implemented in (Zhang et al., 2017),
using the proposed data set, achieving an accuracy of
89.8%, sensitivity of 72.5%, and specificity of 94.3%.
Also, the authors discovered that the accuracy in im-
age classification is high (91.7%). This is because if
an abnormal cell was found, the entire image was con-
sidered to be abnormal. The idea behind this was to
reduce the workload of the histopathologist and de-
crease the time needed for cell analysis. However,
when evaluating each cell with its corresponding nu-
cleus, the experimental results showed low accuracy
due to the dataset containing both typical and atypi-
cal cells, different from other studies reviewed in the
literature (Sompawong et al., 2019).
Ghoneim et al. suggest a system for detecting
and categorizing cervical cancer cells using convolu-
tional neural networks (CNNs) and Extreme Learn-
ing Machines (ELMs). They first input cell images
into the CNNs model to extract important features and
then use the ELM-based classifier to categorize the
images. The authors also examine alternative clas-
sifiers using Multi-layer Perceptron (MLP) and Au-
toencoder (AE). The study was conducted using the
Herlev database, and the proposed system showed a
99.5% accuracy rate in detecting cancer (2-class) and
91.2% accuracy in classifying it (7-class) (Ghoneim
et al., 2020).
Other authors have propose a cervical cancer cell
detection and classification system based on deep
convolutional neural networks (CNNs) (Macancela
et al., 2023). The cell images are fed into a CNN
Towards Accurate Cervical Cancer Detection: Leveraging Two-Stage CNNs for Pap Smear Analysis
221
model to extract deeply learned features. Then, an ex-
treme learning machine (ELM) based classifier clas-
sifies the input images. The CNNs model is used via
transfer learning and fine-tuning. Alternatives to the
ELM, multi-layer perceptron (MLP), and autoencoder
(AE) based classifiers are also investigated. Experi-
ments were performed using the Herlev database. The
proposed CNN-ELM-based system achieved 99.5%
accuracy in the detection problem (2 classes) and
91.2% in the classification problem (7 classes) (Xia
et al., 2020).
3 SYSTEM MODEL AND
METHODOLOGY
3.1 Cell Recognition Model
3.1.1 Data Preparation
It is a common practice to split data into three sepa-
rate folders (train with 80% of data, test, and valida-
tion with the rest) when training a convolutional neu-
ral network. This helps prevent overfitting, which is
when the model performs well on training data but not
on new data. Overfitting is undesirable as it results in
a less accurate model. The dataset contains 2000 im-
ages, which have a size of 150 × 150 pixels, and they
are split in cells and no cells images, see Figure 1 and
Figure 2.
Figure 1: Examples of cell’s images.
Figure 2: Examples of no cell’s images.
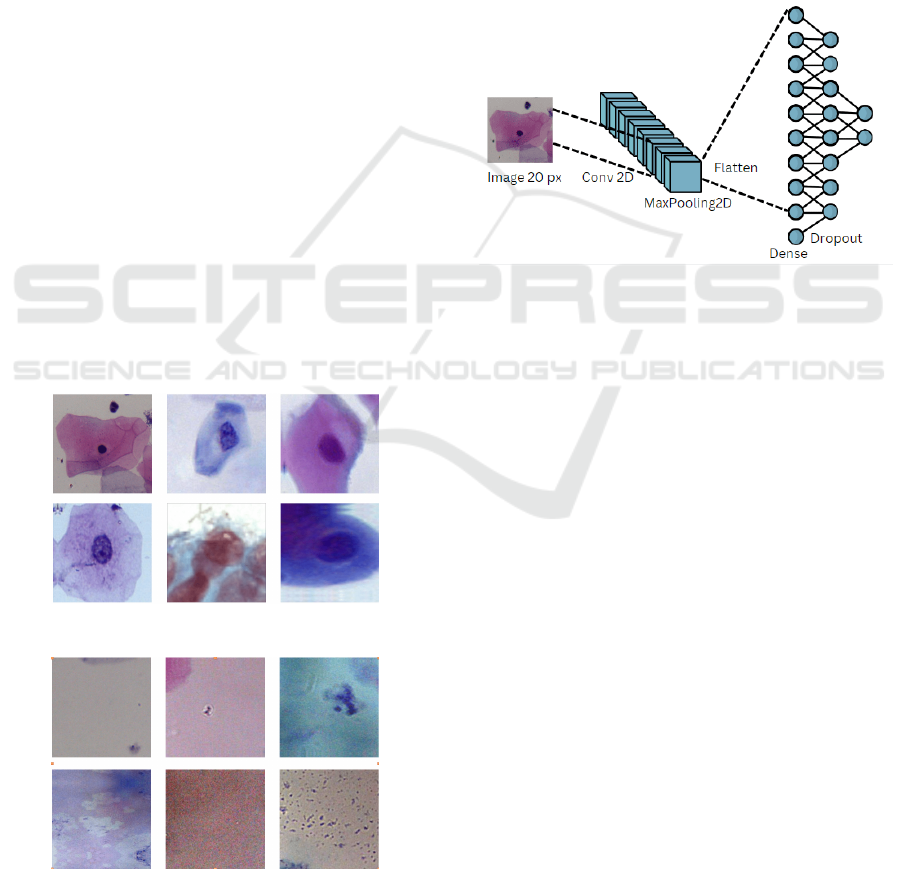
3.1.2 Architecture
The first stage of the fast scanning process uses a sim-
ple Convolutional Neural Network (CNN) to iden-
tify cell shapes and reduce the hardware resources
required for predictions. After experimentation, the
chosen architecture has four layers. The first layer is
a 10x10 convolutional layer with 10 filters, receiving
an input of 20x20 pixels. The second layer is a 2x2
max-pooling layer, followed by a dense layer of 16
neurons. The first and third layers use a ReLu acti-
vation function, while the output layer uses a softmax
activation function. We will refer to this architecture
as CNN 1 see Figure3.
Figure 3: Cell recognition model architecture.
3.2 Cell Classification
For the second step, the cell classification stage, a pre-
trained CNN was used. A Residual Network (ResNet)
capable of categorizing cells into the following cate-
gories is implemented:
• Negative for intraepithelial lesion (NIL).
• Low-grade squamous intraepithelial lesion (LSIL).
• High-grade squamous intraepithelial lesion (HSIL).
• Squamous cell carcinoma (SCC).
In a total dataset of 4800 images. For the dataset of
this step, not all the images were centered. All images
were standardized to a dimension of 250 x 250 pixels.
Residual Network (ResNet) is a deep convolu-
tional neural network architecture introduced in 2015
by Kaiming He et al. in the paper “Deep Residual
Learning for Image Recognition”, mentions that the
main idea behind ResNet is to address the issue of
vanishing gradients in deep neural networks. Deep
networks with many layers can suffer from vanish-
ing gradients, where the magnitude of the gradients
used to update the weights during training becomes
very small, leading to slow convergence or even non-
convergence. ResNet solves this problem by adding
residual connections to the network, which allow the
ICSOFT 2024 - 19th International Conference on Software Technologies
222
gradients to bypass one or more layers and flow more
easily through the network (Targ et al., 2016).
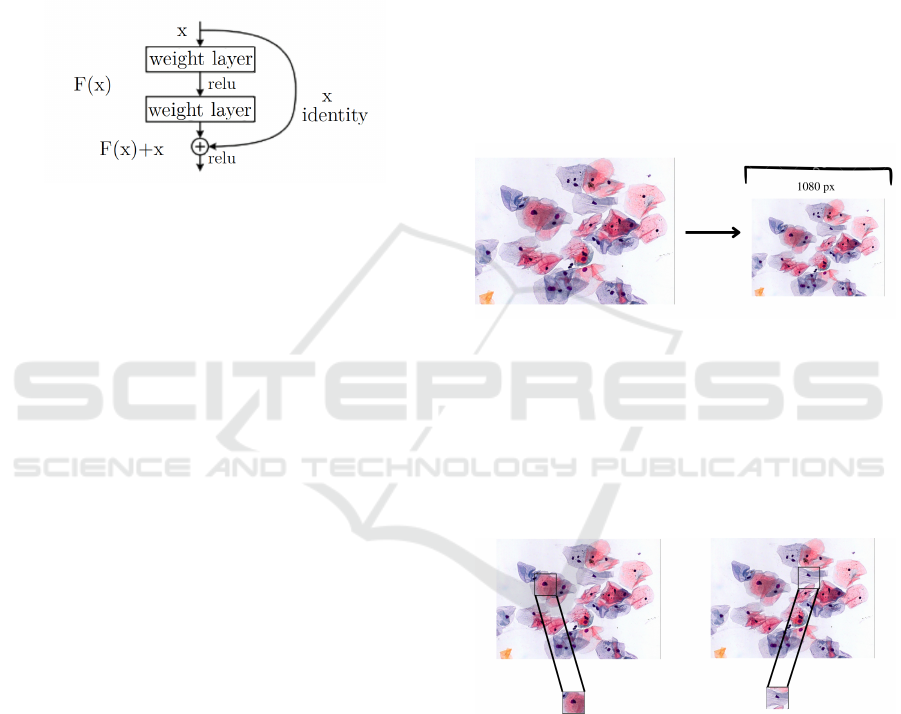
A residual block in ResNet consists of several
convolutional layers, with the output of each layer be-
ing added to the input of the next layer (the “resid-
ual connection”). This allows the network to learn
residual functions, or the difference between the de-
sired output and the input, instead of trying to learn
the whole mapping from scratch (He et al., 2020) see
Figure. 4.
Figure 4: Architecture of a ResNet residual block uses a
shortcut connection to bypass one or more layers, allowing
the model to capture and reuse the original image informa-
tion. Retrieved from (He et al., 2020).
ResNet also introduced the concept of deep super-
vision, where multiple loss functions are used for dif-
ferent parts of the network to help improve conver-
gence.
The ResNet architecture achieved state-of-the-art
results on several computer vision benchmarks and
has been widely used in many subsequent deep learn-
ing models and systems. It is also one of the most
popular models for transfer learning, where a pre-
trained ResNet model is fine-tuned for a new task (He
et al., 2020).
There are many architectures of ResNet. In this
work, we will use ResNet-50. The main difference
between them is the depth of the network which is
determined because of the number of layers. Each
number represents the total number of layers. Never-
theless, it is necessary to emphasize that as the num-
ber of layers increases the computational cost too. All
these architectures can be trained by using pre-default
weights. This is useful for increasing the efficiency
and precision of the model. As a consequence, it
helps the network to converge faster, having an ac-
curate model in less time. ResNet-50 is trained with a
set of data from ImageNet that can be used to initialize
the network before being trained with different data.
This network already knows how to recognize and ex-
tract image features. To avoid the over-fitting prob-
lem presented during the training a dropout will be
added in the last layer of ResNet-50. The dropout is a
hyper-parameter, and regularization technique where
the main idea is forcing some neurons to not activate
during the training (Chen et al., 2020). For this ar-
chitecture, the dropout will be added in the last layer
with a rate of 0.3. It will compare the difference with
ResNet-50 without dropout and with dropout. In this
context, we will refer to ResNet-50 as CNN 2.
3.3 Scanning Process
The prediction process needs 6 steps, explained in de-
tail below:
Firstly both previously trained CNN models are
charged at the beginning of the process. In CNN 1
we use the architecture presented before, and in CNN
2 we use the model ResNet-50. Then, in Figure 5, the
pap smear image is loaded and resized to N pixels to
standardize all images while preserving the width-to-
height ratio.
Figure 5: Pap smear image is loaded and resized.
In Figure 6, it can be seen that the image is made
up of a matrix, which is linked with 70-pixel steps ver-
tically and horizontally, extracting and storing ROI-
sized matrix sections. Once this is done, the extracted
images are stored in List A, and the quadruple coor-
dinates indicating where each of the extracted areas
begins and ends are stored in List B.
Figure 6: Extraction and storing ROI-sized matrix sections
from pap smear image.
Then, two versions of every section taken out are
saved as vectors to satisfy the demands of the previ-
ously loaded TensorFlow CNN. To keep track of them
using their indexes, each version is put into a sepa-
rate list; one is made smaller to 250 x 250 pixels and
placed in list C, while the other is reduced to 20 x 20
pixels and put into list D. This is seen in the Figure 7.
In Figure 8, the process of recognizing cells in a
list D involves feeding each element of the list to a
CNN for cell recognition. If the prediction for “Cell”
Towards Accurate Cervical Cancer Detection: Leveraging Two-Stage CNNs for Pap Smear Analysis
223
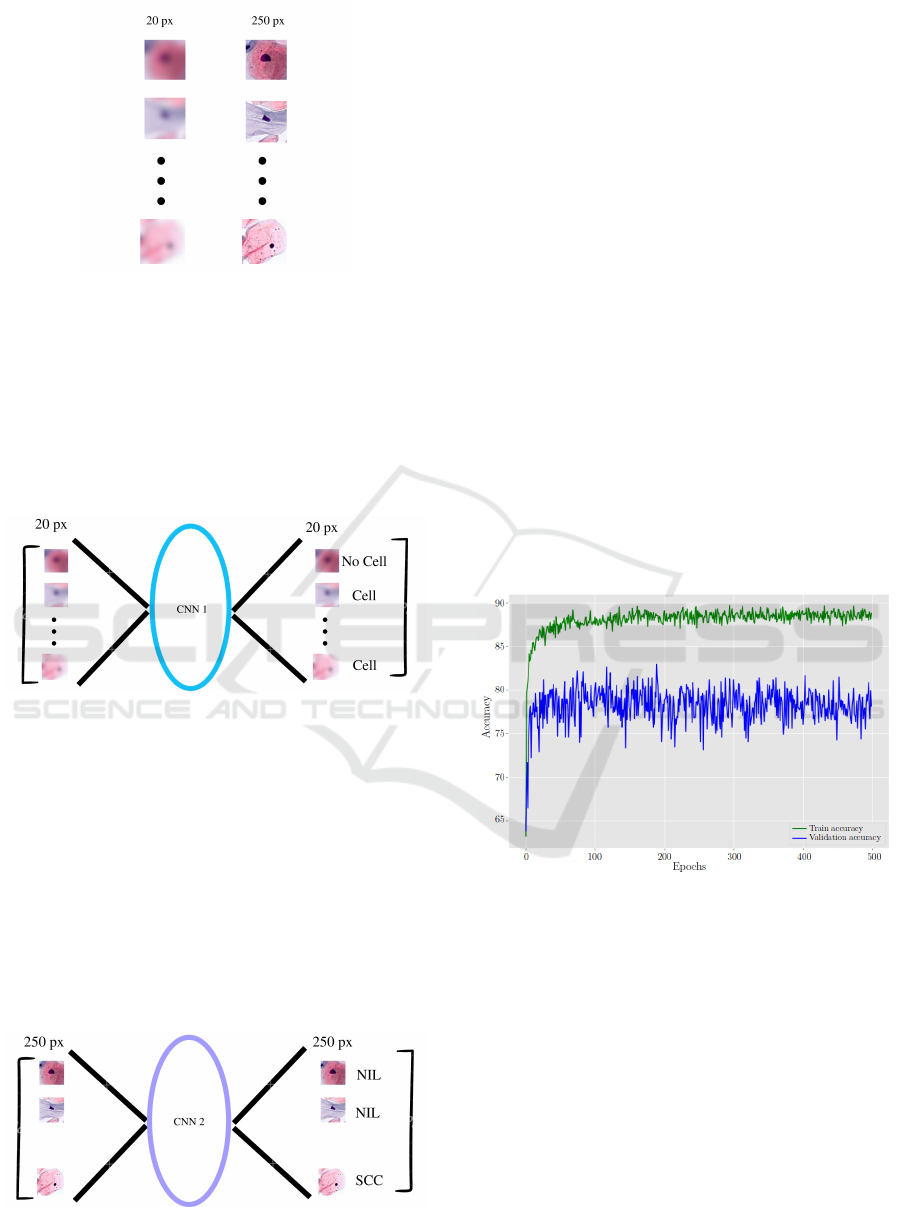
Figure 7: Two versions of every section saved as vectors.
is accurate (above 80%), the corresponding image,
coordinates, and vector are stored in new lists (E, F,
and G) using the current index of list D as a reference.
This enables the visualization of the recognition pro-
cess by drawing lines in the Pap test sample image us-
ing the coordinates from list B. This step is efficient
due to the use of a low-resolution CNN with limited
layers.
Figure 8: Cell recognition using the CNN 1.
Finally, in Figure 9, list C is processed using the
classification cell CNN, similarly to the previous step.
Whenever a prediction matches one of the following
categories with the specified accuracy, the coordinates
from list F are used to highlight their location with a
color code:
• NIL
• LSIL
• HSIL
• SCC
Figure 9: Cell classification using the CNN 2.
As a consequence, cells that are determined to
have no Intraepithelial Lesion are designated as Nor-
mal Cells, while all other categories of cells fall under
the Abnormal Cell Category.
4 RESULTS
The system implemented for the cell classification
process was through the use of the ResNet-50 model,
having better results in comparison to the benchmark
paper, following the dataset that we had previously
proposed.
The proposed model was trained, in the first in-
stance, with 500 epochs, see Figure 10 and Figure
11. The figures help to identify problems during the
training. The first indication of overfitting in our
ResNet-50 implementation was observed through the
disparities between the training and validation accu-
racy curves. The training accuracy continued to in-
crease while the validation accuracy stagnated or de-
creased, creating a significant gap between the two. A
similar pattern emerged in the training and validation
loss curves, further confirming the overfitting issue.
Figure 10: ResNet-50 Behavior in training and validation
accuracy without dropout layer.
To combat the overfitting problem, we turned to
data augmentation techniques. Data augmentation in-
volves introducing variability into the training dataset
by applying transformations to the images. We ap-
plied several augmentation techniques, including ro-
tation, brightness adjustments, contrast alterations,
and zoom operations which are differences that Pap
smear images can contain. These modifications aimed
to diversify the training data, making the model more
robust and adaptable to varying conditions.
In addition to data augmentation, we introduced a
dropout layer with a rate of 0.3 in the flattened layer of
the ResNet-50 architecture. Dropout is a regulariza-
tion technique that temporarily deactivates a fraction
ICSOFT 2024 - 19th International Conference on Software Technologies
224
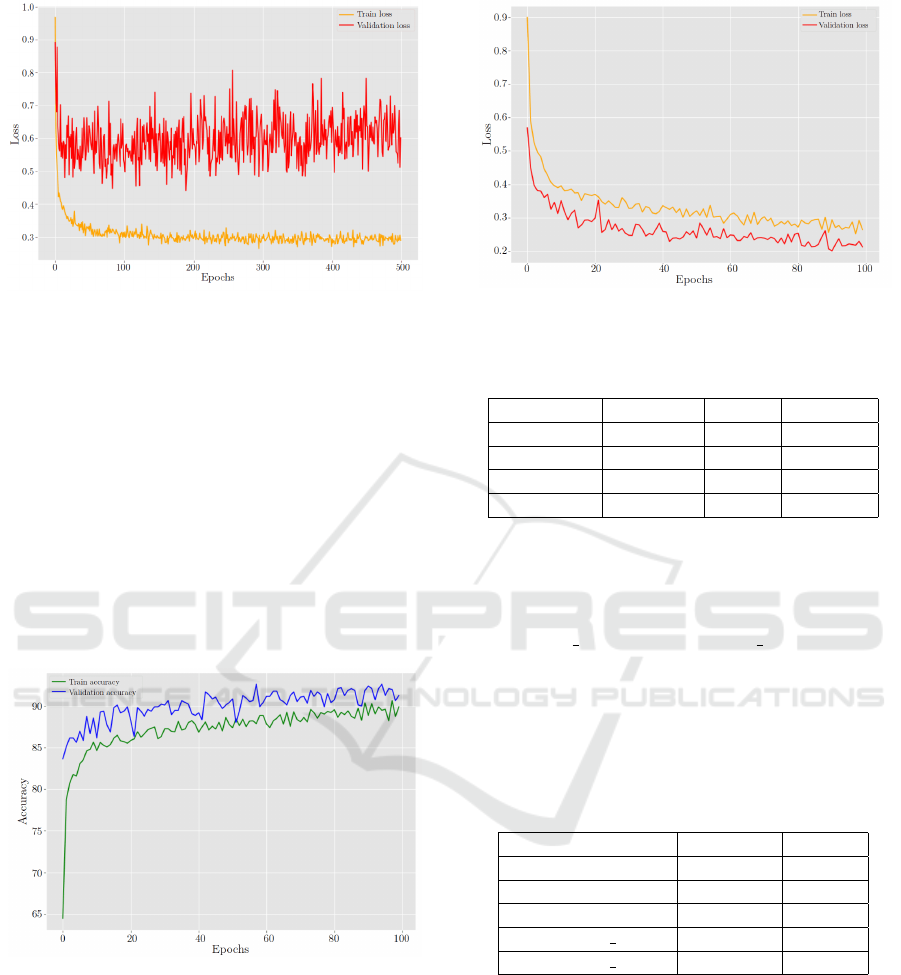
Figure 11: ResNet-50 Behavior in training and validation
loss without dropout layer.
of neurons during training, preventing over-reliance
on particular features. This encourages the network
to learn a more robust representation of the data.
The impact of these modifications was immedi-
ately evident. We just needed 100 epochs to see that
the disparity between training and validation accuracy
and loss substantially decreased, indicating that over-
fitting had been successfully mitigated, see Figure 12
and Figure 13. The validation accuracy now exhibited
a more consistent growth pattern, and the validation
loss stabilized. These outcomes demonstrated the im-
proved generalization ability of the model, making it
more suitable for real-world applications.
Figure 12: ResNet-50 Behavior in training and validation
accuracy.
Table 1 shows a summary of the final results ob-
tained by applying the adjustments to the CNN after
the training. It is important to notice that the metrics
were assessed with 200 images.
4.1 Comparison with Other Studies
In this comparative analysis, ResNet50V2* and
ResNet101V2* from (Wong et al., 2023) emerge
as primary contenders, achieving accuracy rates
Figure 13: ResNet-50 Behavior in training and validation
loss.
Table 1: Summary of metrics obtained with ResNet50 by
category.
Categories Precision Recall F1-score
NILM 1.00 0.97 0.98
LSIL 0.94 0.92 0.93
HSIL 0.78 0.92 0.84
SCC 0.91 0.86 0.85
of 0.97 and 0.95, respectively, demonstrating their
robust performance. However, our proposed
ResNet50 model maintains a noteworthy accuracy of
0.91, indicating competitive effectiveness. Notably,
ResNetXt29 264d and ResNetXt29 464d from (Zhao
et al., 2022) exhibit similar levels of accuracy. It’s
worth mentioning that (Wong et al., 2023) and (Zhao
et al., 2022), these findings together provide valuable
insights into optimizing convolutional neural network
models.
Table 2: Comparison with other studies ordered by accu-
racy.
AI Methods Accuracy Classes
ResNet50V2* 0.97 4
ResNet101V2* 0.95 4
ResNet50 0.91 4
ResNetXt29 2*64d 0.91 10
ResNetXt29 4*64d 0.91 10
5 CONCLUSIONS
This study addresses a critical challenge in cervical
cancer diagnosis by introducing a rapid and efficient
two-stage CNN approach for the analysis of high-
resolution conventional pap smear images. Tradition-
ally, the process of pap smear analysis has been ham-
pered by the substantial computational resources and
time required, often leading to delays in the diagno-
Towards Accurate Cervical Cancer Detection: Leveraging Two-Stage CNNs for Pap Smear Analysis
225

sis of cervical cancer. Our innovative two-stage CNN
method represents a significant contribution to this
domain. In the first stage, we employ a swift and high-
precision model for cell detection, achieving preci-
sion rates exceeding 90%. This initial stage ensures
prompt identification of cells, effectively reducing the
computational overhead.
In the second stage, we leverage the power of the
ResNet-50 architecture, renowned for its exceptional
top-1 accuracy and efficiency, to perform cell clas-
sification. By employing this pre-trained model, we
not only enhance accuracy but also optimize compu-
tational resources, streamlining the classification pro-
cess, but our journey was not without its challenges.
During our work, we encountered the issue of overfit-
ting in the ResNet-50 model. However, our commit-
ment to excellence and the early recognition of this
challenge allowed us to swiftly address it by introduc-
ing a dropout layer with a rate of 0.3 in the flattened
layer of ResNet-50 architecture. This correction en-
sured that our model not only excelled in accuracy
but also maintained its robustness, further enhancing
its reliability.
This approach aligns with the World Health Orga-
nization’s objectives for cervical cancer screening, as
it expedites the analysis while maintaining high ac-
curacy standards. Our work is not only a technologi-
cal advancement but a potential game-changer in the
field of medical diagnostics, as it holds the promise of
accelerating the detection and, subsequently, the pre-
vention of cervical cancer.
Looking ahead, future research endeavors could
explore further improvements in the scanning pro-
cess, offering even greater efficiency and accuracy.
Additionally, expanding the dataset for training mod-
els may yield enhanced results, reinforcing the ro-
bustness of the method. Despite the limitation of a
small dataset, we can confidently assert that our mod-
els have been successfully trained, marking a pivotal
step toward a future where the early and accurate de-
tection of cervical cancer is not only achievable but
a cornerstone in global healthcare. Our contribution
paves the way for a world where cervical cancer is
no longer an insurmountable threat, but a preventable
and treatable disease.
REFERENCES
(2018). Instituto nacional de estad
´
ıstica y censos.
(2021). Cervical cancer ecuador 2021 country profile.
Alsalatie, M., Alquran, H., Mustafa, W. A., Mohd Yacob,
Y., and Ali Alayed, A. (2022). Analysis of cytology
pap smear images based on ensemble deep learning
approach. Diagnostics, 12(11):2756.
Chen, L., Gautier, P., and Aydore, S. (2020). Dropcluster: A
structured dropout for convolutional networks. arXiv
preprint arXiv:2002.02997.
Davies-Oliveira, J., Smith, M., Grover, S., Canfell, K.,
and Crosbie, E. (2021). Eliminating cervical cancer:
Progress and challenges for high-income countries.
Clinical Oncology, 33(9):550–559. Latest Advances
in Endometrial and Cervical Cancer - A Global View.
Elyan, E., Vuttipittayamongkol, P., Johnston, P., Martin, K.,
McPherson, K., Jayne, C., Sarker, M. K., et al. (2022).
Computer vision and machine learning for medical
image analysis: recent advances, challenges, and way
forward. Artificial Intelligence Surgery, 2.
Franco, E. L., Duarte-Franco, E., and Ferenczy, A. (2001).
Cervical cancer: epidemiology, prevention and the
role of human papillomavirus infection. Cmaj,
164(7):1017–1025.
Ghoneim, A., Muhammad, G., and Hossain, M. S. (2020).
Cervical cancer classification using convolutional
neural networks and extreme learning machines. Fu-
ture Generation Computer Systems, 102:643–649.
Hasenleithner, S. O. and Speicher, M. R. (2022). How to
detect cancer early using cell-free dna. Cancer Cell,
40(12):1464–1466.
He, F., Liu, T., and Tao, D. (2020). Why resnet works?
residuals generalize. IEEE transactions on neural net-
works and learning systems, 31(12):5349–5362.
Hussain, E., Mahanta, L. B., Borah, H., and Das, C. R.
(2020). Liquid based-cytology pap smear dataset for
automated multi-class diagnosis of pre-cancerous and
cervical cancer lesions. Data in Brief, 30:105589.
Koh, D.-M., Papanikolaou, N., Bick, U., Illing, R., Kahn Jr,
C. E., Kalpathi-Cramer, J., Matos, C., Mart
´
ı-Bonmat
´
ı,
L., Miles, A., Mun, S. K., et al. (2022). Artificial
intelligence and machine learning in cancer imaging.
Communications Medicine, 2(1):133.
Kurniawati, Y. E., Permanasari, A. E., and Fauziati, S.
(2016). Comparative study on data mining classifica-
tion methods for cervical cancer prediction using pap
smear results. In 2016 1st International Conference
on Biomedical Engineering (IBIOMED), pages 1–5.
IEEE.
Macancela, C., Morocho-Cayamcela, M. E., and Chang, O.
(2023). Deep reinforcement learning for efficient dig-
ital pap smear analysis. Computation, 11(12).
Rezende, M. T., Silva, R., de O. Bernardo, F., Tobias, A.
H. G., Oliveira, P. H. C., Machado, T. M., Costa, C. S.,
Medeiros, F. N. S., Ushizima, D. M., Carneiro, C. M.,
and Bianchi, A. G. C. (2021). Cric searchable im-
age database as a public platform for conventional pap
smear cytology data. Scientific Data, 8(1).
Rom
´
an, K., Llumiquinga, J., Chancay, S., and Morocho-
Cayamcela, M. E. (2023). Hyperparameter tuning
in a dual channel u-net for medical image segmenta-
tion. In Maldonado-Mahauad, J., Herrera-Tapia, J.,
Zambrano-Mart
´
ınez, J. L., and Berrezueta, S., editors,
Information and Communication Technologies, pages
337–352, Cham. Springer Nature Switzerland.
Sompawong, N., Mopan, J., Pooprasert, P., Himakhun, W.,
Suwannarurk, K., Ngamvirojcharoen, J., Vachiramon,
ICSOFT 2024 - 19th International Conference on Software Technologies
226

T., and Tantibundhit, C. (2019). Automated pap smear
cervical cancer screening using deep learning. In 2019
41st Annual International Conference of the IEEE En-
gineering in Medicine and Biology Society (EMBC),
pages 7044–7048. IEEE.
Strasser-Weippl, K., Chavarri-Guerra, Y., Villarreal-Garza,
C., Bychkovsky, B. L., Debiasi, M., Liedke, P.
E. R., de Celis, E. S.-P., Dizon, D., Cazap, E.,
de Lima Lopes, G., Touya, D., Nunes, J. S., Louis,
J. S., Vail, C., Bukowski, A., Ramos-Elias, P., Unger-
Salda
˜
na, K., Brandao, D. F., Ferreyra, M. E., Luciani,
S., Nogueira-Rodrigues, A., de Carvalho Calabrich,
A. F., Carmen, M. G. D., Rauh-Hain, J. A., Schmeler,
K., Sala, R., and Goss, P. E. (2015). Progress
and remaining challenges for cancer control in latin
america and the caribbean. The Lancet Oncology,
16(14):1405–1438.
Su, J., Xu, X., He, Y., and Song, J. (2016). Automatic de-
tection of cervical cancer cells by a two-level cascade
classification system. Analytical Cellular Pathology,
2016.
Targ, S., Almeida, D., and Lyman, K. (2016). Resnet
in resnet: Generalizing residual architectures. arXiv
preprint arXiv:1603.08029.
Wong, L., Ccopa, A., Diaz, E., Valcarcel, S., Mauricio, D.,
and Villoslada, V. (2023). Deep learning and trans-
fer learning methods to effectively diagnose cervical
cancer from liquid-based cytology pap smear images.
International Journal of Online and Biomedical Engi-
neering (iJOE), 19(04):pp. 77–93.
Xia, M., Zhang, G., Mu, C., Guan, B., and Wang, M.
(2020). Cervical cancer cell detection based on deep
convolutional neural network. In 2020 39th Chinese
Control Conference (CCC), pages 6527–6532. IEEE.
Yamal, J.-M., Guillaud, M., Atkinson, E. N., Follen, M.,
MacAulay, C., Cantor, S. B., and Cox, D. D. (2015).
Prediction using hierarchical data: Applications for
automated detection of cervical cancer. Statistical
Analysis and Data Mining: The ASA Data Science
Journal, 8(2):65–74.
Zhang, L., Lu, L., Nogues, I., Summers, R. M., Liu, S.,
and Yao, J. (2017). Deeppap: deep convolutional net-
works for cervical cell classification. IEEE journal of
biomedical and health informatics, 21(6):1633–1643.
Zhao, S., He, Y., Qin, J., and Wang, Z. (2022). A semi-
supervised deep learning method for cervical cell clas-
sification. Analytical Cellular Pathology, 2022:1–12.
Towards Accurate Cervical Cancer Detection: Leveraging Two-Stage CNNs for Pap Smear Analysis
227
