
Dataset Balancing in Disease Prediction
Vincenza Carchiolo
a
and Michele Malgeri
b
Dip. Ingegneria Elettrica Elettronica Informatica (DIEEI), Universit
`
a di Catania, Via Santa Sofia 64, Catania, Italy
Keywords:
Machine Learning, Data Analysis, Health Informatics.
Abstract:
The utilization of machine learning in the prevention of serious diseases such as cancer or heart disease is in-
creasingly crucial. Various studies have demonstrated that enhanced forecasting performance can significantly
extend patients’ life expectancy. Naturally, having sufficient datasets is vital for employing techniques to
classify the clinical situation of patients, facilitating predictions regarding disease onset. However, available
datasets often exhibit imbalances, with more records featuring positive metrics than negative ones. Hence,
data preprocessing assumes a pivotal role. In this paper, we aim to assess the impact of machine learning and
SMOTE (Synthetic Minority Over-sampling Technique) methods on prediction performance using a given
set of examples. Furthermore, we will illustrate how the selection of an appropriate SMOTE process signifi-
cantly enhances performance, as evidenced by several metrics. Nonetheless, in certain instances, the effect of
SMOTE is scarcely noticeable, contingent upon the dataset and machine learning methods employed.
1 INTRODUCTION
The importance of machine learning (ML) in health-
care is increasingly evident and significant. ML mod-
els can analyze large amounts of data, such as medi-
cal images, vital signs, and medical histories, to assist
physicians in the early and accurate diagnosis of dis-
eases. This can lead to better outcomes for patients,
as it allows for the timely and precise identification
of conditions. Furthermore, through the analysis of
patient data, advanced customization of treatments is
possible. Indeed, ML can help develop personalized
treatment plans, taking into account individual varia-
tions in biological data, test results, and treatment re-
sponses, thereby improving the effectiveness of care.
Finally, machine learning plays a central role in dis-
ease prevention, since ML models can identify risk
factors for specific medical conditions and help pre-
vent diseases through the early detection of predictive
signs and the implementation of preventive interven-
tions.
Medical datasets often suffer from imbalance, a
critical issue for predictive modelling. When applied
to imbalanced datasets, models may exhibit a bias to-
ward predicting the majority class, resulting in Clas-
sification Bias. This bias can lead to Inaccurate Per-
formance, particularly for underrepresented classes,
a
https://orcid.org/0000-0002-1671-840X
b
https://orcid.org/0000-0002-9279-3129
where the model fails to learn effectively from those
examples or may suffer from overfitting. Balancing
the dataset is thus paramount for building accurate
disease prediction models. It directly impacts the
model’s ability to generalize correctly and make ac-
curate predictions across all disease classes. With-
out proper balancing, models may struggle to gener-
alize from the training data to new instances, impair-
ing their predictive performance. In medical applica-
tions, dataset balancing is one of the most significant
problem for several critical reasons, with the primary
concern being patient safety. There are many charac-
teristics regarding dataset balancing that become sig-
nificant, such as disease prevalence, particularly when
studying the class of rare diseases which might be un-
derrepresented.
Existing literature is considered in Section 2,
while the datasets are introduced in Section 3. In Sec-
tion 4, the methods and results are discussed in detail.
Moreover, a comparative study is presented to point
out the appropriateness of the results with respect to
several metrics. Finally, we consider further works
and concluding remarks in Section 5.
2 DATA SET BALANCING
As commonly acknowledged, there are numerous
methods for balancing a dataset. In this section, we
Carchiolo, V. and Malgeri, M.
Dataset Balancing in Disease Prediction.
DOI: 10.5220/0012755700003756
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Data Science, Technology and Applications (DATA 2024), pages 293-300
ISBN: 978-989-758-707-8; ISSN: 2184-285X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
293

discuss balancing methods for classification and pro-
vide an overview of related work in the literature on
methods to balance datasets, particularly focusing on
health-related research. In many scenarios, the classes
of interest, such as those related to rare diseases or
clinically significant events, can be significantly un-
derrepresented compared to control or normal classes.
This is challenging specially during the training of
machine learning models, as models tend to be influ-
enced more by the majority class, thereby overlooking
the minority class. Consequently, the model’s ability
to generalize to new data and correctly identify posi-
tive cases in the minority class may be compromised.
Therefore, it is crucial to carefully address the issue
of data imbalance in health-related datasets to ensure
the construction of accurate and reliable models.
There are numerous techniques available for bal-
ancing datasets, but in this article, our focus will be
on SMOTE (Synthetic Minority Over-sampling Tech-
nique) (Pradipta et al., 2021). SMOTE is one of
the most widely used methods for addressing the is-
sue of class imbalance in datasets, especially when
there is a significant under-representation of minor-
ity classes compared to others. This technique is
commonly applied in machine learning contexts, in-
cluding classification models used to predict diseases,
frauds, or other rare events. SMOTE operates based
on three main components: 1. Minority Definition:
this component identifies the minority class in the
dataset, which is characterized by having fewer ex-
amples compared to the other classes; 2. Generation
of Synthetic Examples: this step involves generating
synthetic examples of the minority class. These ex-
amples are created by linearly combining nearby sam-
ples in the feature space; 3. SMOTE Procedure: for
each example in the minority class, this procedure se-
lects some of its nearest neighbors and creates new
synthetic examples through a linear combination of
the feature values.
Finally, by adding these synthetic examples to the
dataset, SMOTE increases the amount of data avail-
able for the minority class, thus helping to bal-
ance the dataset. In addition to SMOTE, several
other methods address the issue of class imbalance
in datasets. Among them, we mention the following
methods. The table below (Table 1) reports a com-
parison among them. Each technique has its advan-
tages and disadvantages, and the choice depends on
the specific characteristics of the dataset and the prob-
lem being addressed. In some cases, experimenting
with different approaches may be effective in deter-
mining which one works best for the specific case.
Several authors propose various approaches to
address class imbalance and feature selection prob-
lems in Clinical Decision Support Systems (CDSS).
In (Sreejith et al., 2020) the authors introduce a frame-
work that balances the dataset at the data level and
employs a wrapper approach for feature selection,
utilizing Chaotic Multi-Verse Optimization (CMVO)
for subset selection. Performance evaluation using
the arithmetic mean of Matthews correlation coef-
ficient (MCC) and F-score (F1) indicates compet-
itiveness of the proposed framework. Paper (Xu
et al., 2021) presents a cluster-based oversampling
algorithm (KNSMOTE), which combines Synthetic
Minority Oversampling Technique (SMOTE) and k-
means clustering. This algorithm identifies ”safe
samples” from clustered classes and synthesizes new
samples through linear interpolation, effectively ad-
dressing class imbalance. In a different study (Li
et al., 2021; Xu et al., 2020) SMOTE is highlighted
as a successful method with practical applications,
alongside the introduction of a novel oversampling
approach called SMOTE-NaN-DE, which improves
class-imbalance data by generating synthetic samples.
Additionally, a hybrid sampling algorithm named
RFMSE, combining M-SMOTE and Edited Nearest
Neighbor based on Random Forest, is proposed to
enhance sampling effectiveness. Jakhmola and Prad-
han in (Jakhmola and Pradhan, 2015) propose an in-
teractive algorithm allowing users to customize pre-
processing requirements, yielding higher quality data
suitable for correlation and multiple regression anal-
ysis, as demonstrated on a diabetes dataset. Finally,
in (Khushi et al., 2021) are introduced research in-
vestigates class imbalance techniques for lung can-
cer prediction, employing various methods includ-
ing under-sampling, over-sampling, and hybrid tech-
niques. Evaluation metrics, such as AUC, reveal
the superiority of over-sampling methods, particularly
random forest with random over-sampling, in predict-
ing lung cancer presence.
3 DATASET DESCRIPTION
To analyze the balance issue in the healthcare domain,
we will leverage five diverse datasets. These datasets
differ significantly in terms of the number of features,
observations, and imbalance ratio. Despite these dif-
ferences, they all revolve around predicting medical
situations through binary classification tasks. Given
the inherent imbalance in these datasets, our objec-
tive is twofold: Evaluate the performance of predic-
tions when using the imbalanced dataset and assess
the impact of preprocessing techniques on prelimi-
nary dataset balancing to enhance prediction perfor-
mance.
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
294

Table 1: Comparison of some Imbalanced Data Handling Methods.
Method Advantages Disadvantages
SMOTE Preserves information from the minority class, reducing the risk
of data loss. Can improve the generalization of the model.
May introduce noise in the synthetic data, especially if the data
distribution is complex. Could require more computational time
compared to other methods.
Random Undersampling Simple and fast to implement. Can reduce the training time on
very large datasets.
May lead to loss of important information in the majority class,
increasing the risk of under-representation.
Random Oversampling Simple to implement. Can improve the accuracy of models on
imbalanced datasets.
May lead to overfitting if not used cautiously, especially with ex-
cessive replication.
Cluster Based Oversam-
pling
Effective when minority class examples form distinct clusters. Re-
duces the risk of generating synthetic data in inconsistent regions.
Requires careful parameter tuning and can be computationally ex-
pensive.
Tomek Links Enhances class separation without adding noise. May not be effective in complex class distributions.
ENN Can improve model performance by reducing misclassification. May excessively reduce dataset size, potentially losing important
information.
SMOTE-ENN - Combines benefits of both techniques, enhancing class separa-
tion and mitigating overfitting risks.
Computationally intensive, particularly on large datasets.
ADASYN More effective in complex and non-uniform data distributions. Requires more computational resources compared to SMOTE.
Random Oversampling with
replacement
Simple to implement. Can enhance model performance on imbal-
anced datasets.
Risk of overfitting if replication is excessive, especially on small
datasets.
Cost-Sensitive Learning Improves model performance on imbalanced datasets without syn-
thetic data addition.
Requires careful weight selection and may not be universally ef-
fective.
We will define the Imbalance Ratio as the propor-
tion between the number of examples in the minor-
ity class and the number of examples in the majority
class. This ratio provides a quantitative measure of
the degree of class imbalance within each dataset. For
example, If there are 100 negative examples (major-
ity class) and 20 positive examples (minority class)
the imbalance ratio will be 20/100 = 0.2. Obviously,
the more imbalanced the dataset, the closer this value
to zero.
The first dataset we discuss, Wisconsin Diagnos-
tic Breast Cancer (WDBC) (Repository, ), is the well-
known dataset that collects data for breast cancer pre-
diction. Since breast cancer is the most common
cause of cancer deaths in women and is a type of
cancer that can be treated when diagnosed early, pre-
diction is a very important aspect. This dataset has
been extensively studied in the literature (Elter et al.,
), which is why it is utilized in this paper. The dataset
is from the University Hospital of California and can
be downloaded from both the UCI Machine Learning
Repository and Kaggle. It consists of 569 samples
and 33 features, computed from a digitized image of
a fine needle aspiration (FNA) of a breast mass and re-
lated to some characteristics of each cell nucleus (e.g.,
radius, texture, perimeter, area, etc.). Some of these
features are more selective and decisive than others,
and the determination of these features significantly
increases the success of the models, which is why
Feature Selection is applied to select them.
The second dataset, also widely referenced in
literature, is the Heart Failure Clinical Records
dataset (Chicco and Jurman, 2020a). Cardiovascular
diseases (CVDs) are the leading cause of death glob-
ally, claiming approximately 17.9 million lives each
year, representing 31% of all deaths worldwide. Heart
failure, a common occurrence resulting from CVDs,
is the focus of this dataset, which comprises 12 fea-
tures aimed at predicting mortality associated with
heart failure. Many CVDs are preventable through ad-
dressing behavioral risk factors such as tobacco use,
poor diet, obesity, physical inactivity, and excessive
alcohol consumption via population-wide interven-
tions. Individuals with existing CVD or those at high
cardiovascular risk, often due to hypertension, dia-
betes, hyperlipidemia, or other established diseases,
require early detection and management, where ma-
chine learning models can offer significant assistance.
This dataset includes medical records from 299 heart
failure patients, gathered at the Faisalabad Institute of
Cardiology and Allied Hospital in Faisalabad, Punjab,
Pakistan, between April and December 2015. It en-
compasses 13 features encompassing clinical, physi-
ological, and lifestyle-related information.
The third dataset used is Pima Indians Diabetes
Database (Sigillito, ). The Pima Indians Diabetes
Database is a well-known dataset in the field of ma-
chine learning and healthcare research. It contains
medical data from the Pima Indian population, specif-
ically focused on women aged 21 and above from the
Gila River Indian Community near Phoenix, Arizona.
The dataset includes various health-related attributes
such as glucose level, insulin level, BMI (Body Mass
Index), age, and the presence or absence of diabetes
within a five-year period following the initial exami-
nation. This dataset is widely used for developing pre-
dictive models to identify individuals at risk of devel-
oping diabetes. Due to its large sample size and com-
prehensive health information, the Pima Indians Di-
abetes Database has been instrumental in advancing
research in diabetes prediction and management. De-
spite its significance, the dataset also poses challenges
due to its inherent class imbalance and missing data,
necessitating careful preprocessing and model evalu-
ation techniques. Its availability in the public domain
has facilitated numerous studies aimed at improving
diabetes diagnosis and treatment strategies, contribut-
ing significantly to the broader efforts in public health
and medical informatics.
The fourth dataset is a more recent data set. The
Dataset Balancing in Disease Prediction
295

Differentiated Thyroid Cancer Recurrence dataset
(Borzooei et al., 2023) is a valuable resource in the
domain of thyroid cancer research. It comprises
clinical data from patients diagnosed with differenti-
ated thyroid cancer (DTC) who underwent thyroidec-
tomy and subsequent treatment. The dataset includes
various demographic and clinical variables such as
age, sex, tumor size, histopathological characteristics,
treatment modalities, and follow-up information. A
key focus of the dataset is to predict the recurrence
of thyroid cancer following initial treatment based on
these factors. Researchers utilize machine learning
and statistical methods to develop predictive mod-
els that can identify patients at higher risk of recur-
rence, thereby aiding in personalized treatment strate-
gies and follow-up care. Due to its specialized nature
and importance in thyroid cancer management, the
Differentiated Thyroid Cancer Recurrence dataset has
garnered attention from researchers worldwide. How-
ever, challenges such as limited sample size and data
heterogeneity need to be addressed to enhance the ro-
bustness and generalizability of predictive models de-
rived from this dataset. Overall, it serves as a valuable
tool in advancing our understanding of thyroid cancer
recurrence and improving patient outcomes through
tailored interventions.
Finally, the last dataset used is the Sepsis Sur-
vival Minimal Clinical Records (Chicco and Jur-
man, 2020b). The Sepsis Survival Minimal Clini-
cal Records dataset is an essential and widely used
dataset in sepsis study and research, a severe medi-
cal condition caused by a systemic inflammatory re-
sponse to an infection. This dataset contains clinically
relevant and minimal information about patients with
sepsis, including demographic data, vital signs, labo-
ratory test results, and treatment information. Its sim-
plified structure makes it particularly suitable for de-
veloping predictive models of sepsis survival and for
evaluating clinical management strategies. Thanks to
its availability and focused nature, the Sepsis Sur-
vival Minimal Clinical Records dataset has signifi-
cantly contributed to the understanding of sepsis and
the research of effective clinical interventions to im-
prove outcomes for patients with this severe medical
condition. However, it is important to consider limita-
tions and potential biases in the data to obtain accurate
and generalizable results.
The common feature shared by the aforemen-
tioned datasets is the presence of only two classes.
The main data of the five datasets are summarized in
Table 2, demonstrating varying numbers of features,
observations, and Imbalance Ratios.
4 EXPERIMENT AND
DISCUSSION
We implemented the following 10 supervised algo-
rithms. 1. Logistic Regression (LG) is a machine
learning method used for binary classification prob-
lems. Its principle of operation is based on estimat-
ing the conditional probabilities that an instance be-
longs to one of the two classes. It uses the logistic
function (or sigmoid function) to transform a linear
combination of features into a value between 0 and
1, representing the estimated probability. This value
is then used as a threshold to assign the instance to
one of the two classes. 2. Support Vector Machine
(SVM) operates by seeking to find the optimal hy-
perplane of separation between classes in the case of
binary classification. The separation hyperplane is
defined as the hyperplane that maximizes the margin
between the nearest class instances, which are called
support vectors. SVM can effectively handle datasets
with many features, and it tends to generalize well to
test data, reducing the risk of overfitting. 3. Gaus-
sian Naive Bayes (GNB) is based on Bayes’ theo-
rem and assumes that features are independent and
follow a Gaussian distribution. 4. Decision Tree DT
recursively splits the dataset into subsets based on the
value of features, aiming to maximize the purity of
each subset in terms of class labels. 5. Random For-
est RF is an ensemble learning method that builds
multiple decision trees and combines their predictions
through voting or averaging. 6. Extra Tree (ET) is
similar to RF but introduces additional randomness
in the feature selection process. 7. K-Nearest Neigh-
bors (KNN) operates by classifying an instance based
on the majority class among its k nearest neighbors
in the feature space. The distance metric (e.g., Eu-
clidean distance) is used to measure the similarity be-
tween instances. 8. Hist Gradient Boosting (HGB is
a boosting algorithm that builds a series of decision
trees sequentially, each one correcting the errors of its
predecessors. It uses histogram-based techniques to
speed up training. 9. Bagging Classifier (BC) is an
ensemble learning method that trains multiple mod-
els on bootstrap samples of the dataset and combines
their predictions. It reduces variance and improves
stability. 10. Finally, Multilayer Perceptron (MLP) is
a type of artificial neural network consisting of mul-
tiple layers of interconnected neurons. The selection
of these methods is intended to facilitate experiments
showcasing the diverse impacts of various smoothing
techniques. Through these experiments, we aim to
ascertain the complexity of asserting a universally su-
perior smoothing method. As we will observe, the ef-
ficacy of a particular smoothing method, which may
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
296

Table 2: Dataset Information.
Dataset # Instances # Features Imbalance Ratio
Feature Types
# Numeric # Symbolic
Breast Cancer Wisconsin (Diagnostic) 699 9 0.59 9 0
Hearth failure 299 12 0.94 12 0
Pima Indians Diabetes Database 768 8 0.54 8 0
Differentiated Thyroid Cancer Recurrence dataset 383 16 0.39 6 10
Sepsis Survival Minimal Clinical Records 137 3 0.21 3 0
excel in certain scenarios, could result in inferior out-
comes compared to the unbalanced dataset in other
cases.
Furthermore, we will employ four distinct over-
sampling techniques, commonly utilized in address-
ing imbalanced datasets, which will be referred to
throughout the remainder of the paper as Smote1,
Smote2, Smote3, and Smote4.
BorderlineSMOTE with 20 neighbors=20
(Smote1) is a variant of the SMOTE algorithm that
generates synthetic samples only for those minority
class instances that are misclassified or lie near the
decision boundary (i.e., borderline instances). It
generates synthetic samples by selecting a minority
class instance and finding its k nearest neighbors.
It then selects one of these neighbors randomly
and generates a synthetic sample along the line
segment joining the original instance and the selected
neighbor. By setting m neighbors=20, it specifies
the number of nearest neighbors to consider when
generating synthetic samples.
BorderlineSMOTE with neighbors = 10 and
sampling strategy =
′
minority
′
(Smote2) is a variant
of BorderlineSMOTE also generates synthetic sam-
ples near the decision boundary between the minor-
ity and majority classes. Additionally, it adjusts
the sampling strategy to focus on the minority class
by specifying sampling strategy=’minority’. By set-
ting m neighbors = 10, it specifies a different num-
ber of nearest neighbors to consider when generat-
ing synthetic samples compared to the previous vari-
ant. SMOTE with 10 neighbors (Smote3) is a pop-
ular oversampling technique that generates synthetic
samples by interpolating between existing minority
class instances. It selects a minority class instance
and finds its k nearest neighbors. It then selects one
of these neighbors randomly and generates a syn-
thetic sample along the line segment joining the orig-
inal instance and the selected neighbor. By setting
k neighbors = 10, it specifies the number of nearest
neighbors to consider when generating synthetic sam-
ples. SMOTE with sampling strategy =
′
minority
′
and neighbors = 10(Smote4), similar to Smote3, gen-
erates synthetic samples by interpolating between ex-
isting minority class instances. It further adjusts the
sampling strategy to focus on the minority class by
specifying sampling strategy =
′
minority
′
. By set-
ting k neighbors = 10, it specifies a different number
of nearest neighbors to consider when generating syn-
thetic samples compared to the previous variant. Both
BorderlineSMOTE and SMOTE aim to address class
imbalance by oversampling the minority class.
To assess the impact of balancing, we will utilize
Accuracy and AUC. Accuracy is a general measure of
the model’s precision and represents the percentage of
instances classified correctly out of the total instances,
it’s calculated as the ratio of the number of correct
predictions to the total number of predictions made,
and it is particularly useful when classes in the dataset
are balanced but can be misleading in presence of un-
balancing. AUC measures the model’s discriminative
ability, i.e., its ability to correctly classify positive ex-
amples as positive and negative examples as negative.
All the figures are on a logarithmic scale and the blue
bar refers to the analysis of the imbalanced dataset,
while the others refer to datasets obtained with the
four balancing methods
The result of Breast Cancer accuracy and Auc
score are shown in figure 1, let us note that while
some datasets yield consistent accuracy values across
methods, others exhibit significant variability depend-
ing on the method used. For the Breast Cancer
dataset, the highest accuracy (see figure 1a) values
are achieved with the ET and HGB methods, whether
smoothing is applied or not. The maximum score of
0.973684 is obtained for ET when no balancing is per-
formed and for HGB when Smote1 is applied. The
maximun Auc score, 0.974206, (see figure 1b) it ob-
tained for HGB with Smote1. This, considering that
AUC is less prone to overfitting, allows us to affirm
that smoothing allows us to gain an advantage, albeit
small
The result of Heart
f ailure accuracy and AUC
score are shown in fig 2. The accuracy values reach 1
(see figure 2a) using various methods, but almost al-
ways with smoothing. Note that this data set consis-
tently performs quite well in terms of accuracy (the
worst value being 0.829268). Analyzing the results
for AUC (see figure 2b) allows us to make the same
considerations and therefore its study is not particu-
larly significant for our purposes.
For the third dataset (Pima), all methods perform
Dataset Balancing in Disease Prediction
297
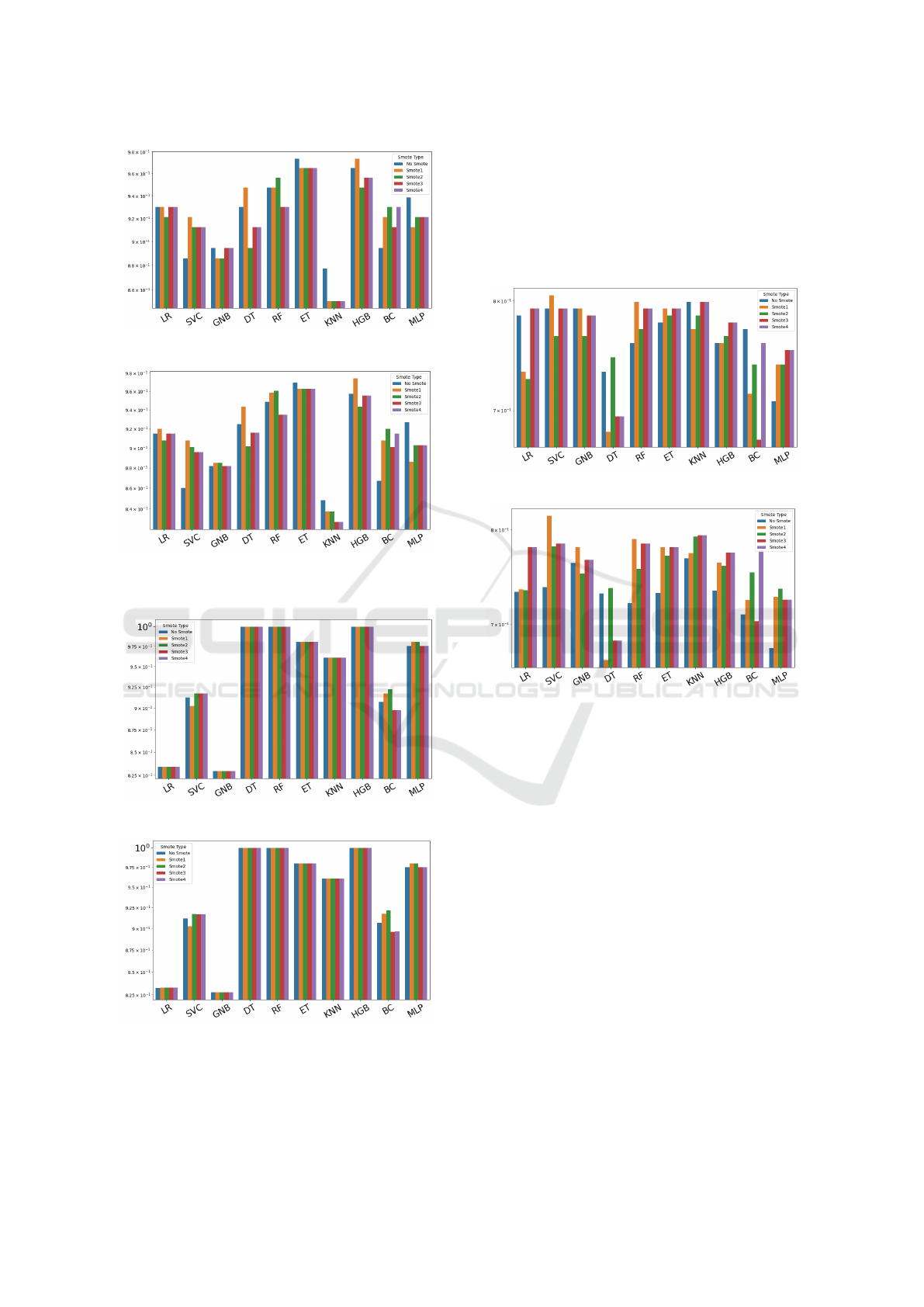
(a) Accuracy.
(b) AUC.
Figure 1: Breast cancer scores.
(a) Accuracy.
(b) AUC.
Figure 2: Heart failure scores.
better with appropriate balancing, both in terms of
accuracy and AUC score 3. In this case, several
methods with SMOTE allow achieving an accuracy
of 0.892857 (3a). The figure 3b clearly shows that
the best AUC (DT or ET method) is consistently ob-
tained with datasets that have had SMOTE2 applied
(0.856522). This result allows us to conclude that for
this dataset, characterized by an imbalance ratio of
0.54, balancing is often beneficial.
(a) Accuracy.
(b) AUC.
Figure 3: Pima scores.
For the fourth dataset, the Differentiated Thyroid
Cancer Recurrence dataset (see figure 4), the high-
est accuracy value (0.961039) was achieved using the
DT method with Smote1 (refer to figure 4a). Inter-
estingly, all methods improved with balancing, which
is significant considering this dataset has a higher
imbalance ratio than the previous three, making the
impact of balancing generally beneficial. The AUC
analysis further distinguishes the methods, confirm-
ing the most effective solutions (refer to figure 4b).
The top AUC score is 0.945455, obtained using the
DT method with Smote1.
Finally, for the fifth dataset (the most imbalanced),
the behavior is nearly equivalent for any method, as
adopting the appropriate smoothing method (not al-
ways the same) 5. The best choice allows achieving
a value of 0.892857. A particular situation occurs for
the MLP method, which performs poorly using any of
the four balancing methods. In terms of AUC score,
the results are still extremely diverse depending on
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
298
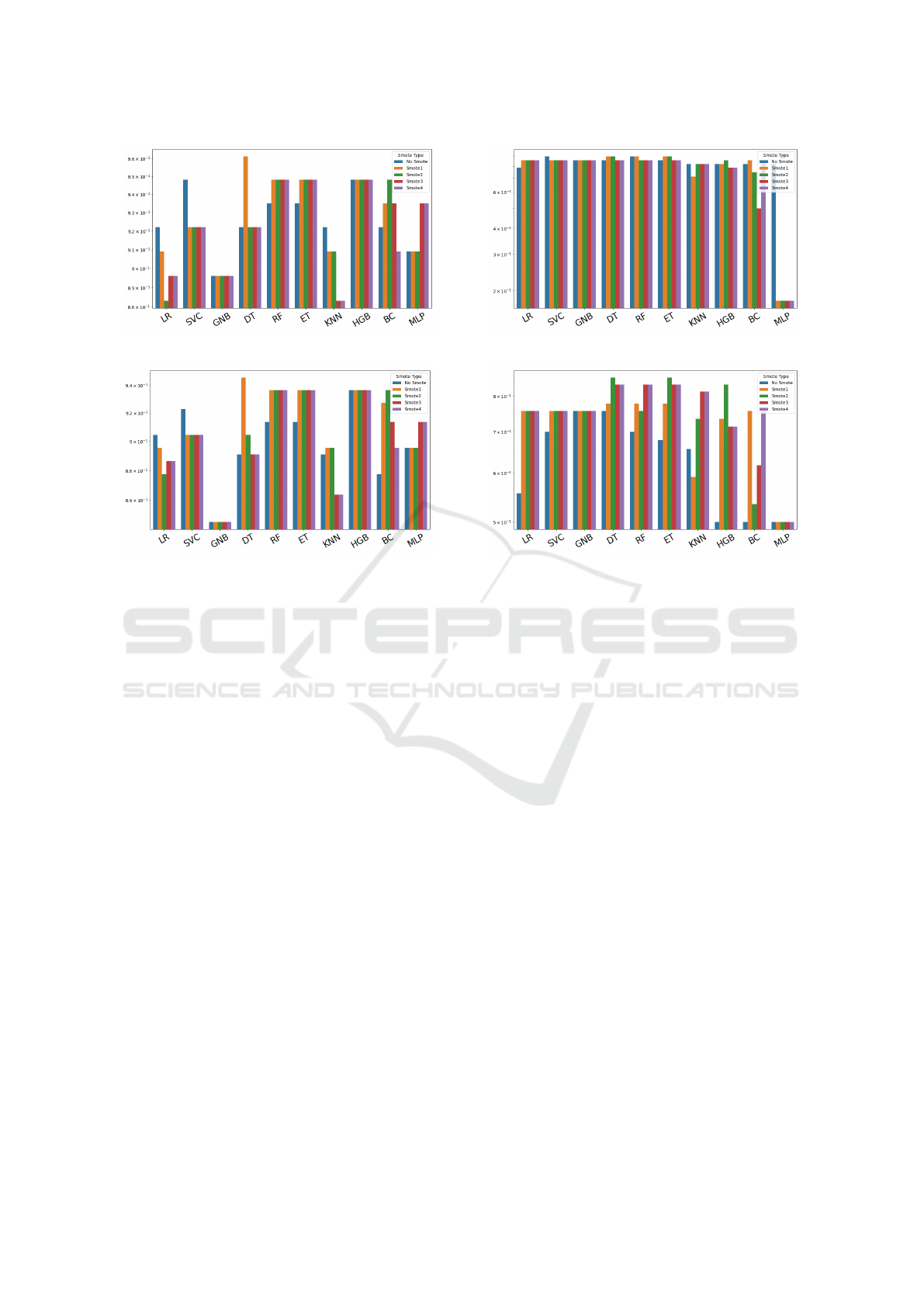
(a) Accuracy.
(b) AUC.
Figure 4: Thyroid score.
the method and smoothing used, much like with accu-
racy. However, it should be noted that for this dataset,
which is the most imbalanced one, balancing can be
crucial as it allows us to achieve the best result. At the
same time, if used inadequately, it can even worsen
the results. In terms of ACU score, the best score is
obtained with DT and ET using Smote2.
The positive effect of smoothing is better appre-
ciated by analyzing AUC, which is less influenced
by overfitting compared to accuracy. For datasets
where the choice of method substantially alters ac-
curacy values, the impact of data balancing can be
significant. The figures from 1b to 5b summarize the
variation of accuracy and AUC values for each dataset
and method. Therefore, it can be stated that there
is no single most effective Smote method, but rather,
the (method, Smote) pair yielding better performance
should be sought.
5 CONCLUSION
A detailed analysis was conducted on five distinct
datasets, utilizing various machine learning tech-
niques to assess the impact of data preprocess-
ing (Carchiolo et al., 2022). For each dataset, we ex-
plored the behavior of ten distinct algorithms, each
with its own characteristics and tuning parameters. In
(a) Accuracy.
(b) AUC.
Figure 5: Sepsis scores.
order to assess the impact of data balancing, we per-
formed the analysis both with and without data bal-
ancing techniques, considering four different smooth-
ing approaches to handle the presence of underrepre-
sented classes. The results obtained highlighted a sig-
nificant variation in model performance based on the
different combinations of machine learning method,
data balancing, and smoothing techniques. It became
clear that the choice of machine learning method and
the application of balancing strategies must be closely
integrated to achieve optimal results. In particular,
we observed that while data balancing can signifi-
cantly improve model performance on heavily imbal-
anced datasets, inadequate implementation could lead
to inferior results. Furthermore, we recognized that
parameter optimization for datasets characterized by
imbalance requires a particularly careful and targeted
approach, as the specific dataset characteristics can
significantly influence the effectiveness of proposed
solutions. While various approaches exist, compar-
ing them can be challenging due to numerous tun-
ing parameters and variations within articles. How-
ever, ongoing research suggests the importance of ex-
ploring diverse classifiers and imbalance techniques,
including deep learning models, to enhance predic-
tion capabilities and address imbalance issues effec-
tively. In conclusion, our analysis underscored the
importance of carefully considering the specific con-
Dataset Balancing in Disease Prediction
299

text of each dataset and adopting a flexible and tar-
geted approach to address the issue of data imbal-
ance in machine learning contexts. From the analy-
sis conducted, it emerged that in the vast majority of
cases, the solutions’ accuracy and AUC with the ap-
plication of balancing are better. Nonetheless, as fu-
ture work, the analysis will be extended to a greater
number of datasets and balancing methods. Another
activity for future work concerns the application to
datasets that involve non-binary classification to an-
alyze whether balancing is advantageous in this case
as well. Finally, precision and recall analysis could be
conducted to add further confidence in the quality of
the results.
ACKNOWLEDGEMENTS
The work is partially supported by UDMA project,
CUP: G69J18001040007.
REFERENCES
Borzooei, S., Briganti, G., and Golparian, M. e. a. (2023).
Machine learning for risk stratification of thyroid
cancer patients: a 15-year cohort study. European
Archives of Oto-Rhino-Laryngology.
Carchiolo, V., Grassia, M., Malgeri, M., and Mangioni, G.
(2022). Co-authorship networks analysis to discover
collaboration patterns among italian researchers. Fu-
ture Internet, 14(6).
Chicco, D. and Jurman, G. (2020a). Machine learning can
predict survival of patients with heart failure from
serum creatinine and ejection fraction alone. BMC
Medical Informatics and Decision Making, 20(16).
Chicco, D. and Jurman, G. (2020b). Survival prediction of
patients with sepsis from age, sex, and septic episode
number alone. Sci Rep, 10:17156.
Elter, M., Schulz-Wendtland, R., and Wittenberg, T. The
prediction of breast cancer biopsy outcomes using two
cad approaches that both emphasize an intelligible de-
cision process.
Jakhmola, S. and Pradhan, T. (2015). A computational ap-
proach of data smoothening and prediction of diabetes
dataset. In Proceedings of the Third International
Symposium on Women in Computing and Informatics,
WCI ’15, page 744–748, New York, NY, USA. Asso-
ciation for Computing Machinery.
Khushi, M., Shaukat, K., Alam, T. M., Hameed, I. A., Ud-
din, S., Luo, S., Yang, X., and Reyes, M. C. (2021). A
comparative performance analysis of data resampling
methods on imbalance medical data. IEEE Access,
9:109960–109975.
Li, J., Zhu, Q., Wu, Q., Zhang, Z., Gong, Y., He, Z., and
Zhu, F. (2021). Smote-nan-de: Addressing the noisy
and borderline examples problem in imbalanced clas-
sification by natural neighbors and differential evolu-
tion. Knowledge-Based Systems, 223:107056.
Pradipta, G. A., Wardoyo, R., Musdholifah, A., Sanjaya,
I. N. H., and Ismail, M. (2021). Smote for handling
imbalanced data problem : A review. In 2021 Sixth
International Conference on Informatics and Comput-
ing (ICIC), pages 1–8.
Repository, U. M. L. UCI Machine Learning Repository:
Mammographic Mass Data Set. http://archive.ics.uci.
edu/ml/datasets/mammographic+mass.
Sigillito, V. National Institute of Diabetes and Digestive and
Kidney Diseases, note = Donor of database: Vincent
Sigillito (vgs@aplcen.apl.jhu.edu) Research Center,
RMI Group Leader Applied Physics Laboratory The
Johns Hopkins University Johns Hopkins Road Lau-
rel, MD 20707 (301) 953-6231 (c) Date received: 9
May 1990.
Sreejith, S., Khanna Nehemiah, H., and Kannan, A. (2020).
Clinical data classification using an enhanced smote
and chaotic evolutionary feature selection. Computers
in Biology and Medicine, 126:103991.
Xu, Z., Shen, D., Nie, T., and Kou, Y. (2020). A hybrid sam-
pling algorithm combining m-smote and enn based on
random forest for medical imbalanced data. Journal
of Biomedical Informatics, 107:103465.
Xu, Z., Shen, D., Nie, T., Kou, Y., Yin, N., and Han, X.
(2021). A cluster-based oversampling algorithm com-
bining smote and k-means for imbalanced medical
data. Information Sciences, 572:574–589.
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
300
