
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine
Radiology Reports
Klaudia Szab
´
o Ledenyi
1 a
, Andr
´
as Kicsi
1 b
and L
´
aszl
´
o Vid
´
acs
1,2 c
1
Department of Software Engineering, University of Szeged, Szeged, Hungary
2
HUN-REN-SZTE Research Group on Artificial Intelligence, Szeged, Hungary
Keywords:
Radiology, Clinical Reports, NLP, LLM, GPT, Prompt Engineering.
Abstract:
The significant growth of large language models revolutionized the field of natural language processing. Re-
cent advancements in large language models, particularly generative pretrained transformer (GPT) models,
have shown advanced capabilities in natural language understanding and reasoning. These models typically
interact with users through prompts rather than providing training data or fine-tuning, which can save a sig-
nificant amount of time and resources. This paper presents a study evaluating GPT-4’s performance in data
mining from free-text spine radiology reports using a single prompt. The evaluation includes sentence clas-
sification, sentence-level sentiment analysis and two representative biomedical information extraction tasks:
named entity recognition and relation extraction. Our research findings indicate that GPT-4 performs effec-
tively in few-shot information extraction from radiology text, even without specific training for the clinical
domain. This approach shows potential for more effective information extraction from free-text radiology
reports compared to manual annotation.
1 INTRODUCTION
In recent years, there has been a significant growth in
the field of natural language processing (NLP), one
of the most researched fields of artificial intelligence.
The application of NLP in biomedical research has
significantly expanded due to the rapid development
of NLP models. This is particularly evident in the
field of radiology, where a large amount of radio-
logic reports are generated daily. Typically, these are
free-form text reports, a format that includes a large
amount of raw data. These data can be effectively
extracted using various NLP techniques. Biomedical
text mining encompasses various tasks on biomedical
text data, including sentence classification, sentiment
analysis, information extraction, text summarization,
question answering, etc. Sentence classification in-
volves classifying sentences into predefined groups.
Sentence classification plays a role in organizing and
understanding textual data. Sentiment analysis deter-
mines the sentiment of the text data, typically catego-
rized as positive, negative, or neutral. Information ex-
a
https://orcid.org/0009-0001-4478-632X
b
https://orcid.org/0000-0002-3144-9041
c
https://orcid.org/0000-0002-0319-3915
traction is the automated process of extracting struc-
tured information from unstructured data and trans-
forming it into a more usable format. In radiology, the
two main components of information extraction are
the extraction of clinical entities and relations from
radiology reports. These tasks are known as named
entity recognition (NER) and relation extraction (RE).
NER is the most important step in extracting relevant
data, which aims to identify specific terms. RE fo-
cuses on identifying relations between the detected
entities.
1.1 Technical Background
The transformer architecture, a type of neural net-
work architecture that was introduced by (Vaswani
et al., 2017), has revolutionized NLP. It uses attention
mechanisms to identify relationships between words,
effectively capturing long-range dependencies in in-
put sequences. The architecture consist of an encoder-
decoder structure, multiple layers of self-attention
mechanisms, and feedforward networks. This formed
the foundation for both pre-trained language models
(PLMs) and large-sized PLMs, known as large lan-
guage models. PLMs use the transformer architecture
to train on a vast corpus of text data before fine-tuning
82
Szabó Ledenyi, K., Kicsi, A. and Vidács, L.
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine Radiology Reports.
DOI: 10.5220/0012765100003756
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Data Science, Technology and Applications (DATA 2024), pages 82-92
ISBN: 978-989-758-707-8; ISSN: 2184-285X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

for specific downstream tasks. The currently popu-
lar large language models (LLMs) are transformer-
based deep learning models too, that integrate the
concept of pretrained models with an emphasis on
billions of parameters. These models capture con-
textual information from a wide range of texts, en-
abling them to understand and generate human-like
language. LLMs have demonstrated state-of-the-art
(SOTA) results across a wide range of NLP tasks. At
the end of 2022, OpenAI developed ChatGPT, a free-
to-use AI system. ChatGPT has garnered consider-
able public attention because it does not require do-
main expertise for usage, unlike for instance Bidirec-
tional Encoder Representations from Transformers-
based (BERT) (Devlin et al., 2019) models which
are currently still achieving SOTA results in several
domains. The latest version of GPT (GPT-4) (Ope-
nAI, 2023) has 100 trillion parameters, 570 times
more than its predecessor, GPT-3. Because of the
number of parameters, fine-tuning LLMs for specific
tasks is impractical. The primary way to interact
with AI systems for different tasks is prompting (Liu
et al., 2021). Prompt engineering is a novel paradigm
in NLP, which involves designing prompts to guide
LLMs, such as GPT-4, to generate specific outputs for
downstream tasks. A prompt is a text input that guides
the model to generate the desired output. The design
of the prompt significantly influences the model’s per-
formance on specific tasks. Few-shot and zero-shot
prompting are specific techniques within prompt en-
gineering. A well-designed prompt should provide
clear guidance to the model, aiding in accomplish-
ing tasks efficiently. Few-shot prompting involves
providing demonstrations in the prompt to guide the
model’s behaviour to solve specific tasks. This tech-
nique can serve clear and explicit prompt inputs, guid-
ing the model towards desired outputs. Zero-shot
prompting means that the prompt used to interact with
the model does not contain any examples or demon-
strations. In-context learning (ICL) is a method used
to solve complex tasks with LLMs (Brown et al.,
2020). An ICL prompt is a task description presented
in natural language text with a small number of task
examples with the desired input-output pairs. These
examples are also known as few-shot demonstrations.
Prompt-based learning allows GPT to solve various
NLP problems without updating its parameters, re-
sulting in significant time and cost savings.
1.2 Research Objective
Our research objective is to evaluate the capabilities
of GPT-4 in various biomedical text-mining tasks, fo-
cusing on spine radiology reports. We evaluate GPT-
4’s performance in sentence classification, sentence-
level sentiment analysis, NER and RE tasks. We
provide an optimized prompt that enables GPT-4 to
perform these diverse tasks in a single step. Our fi-
nal prompt, as well as the .csv format of one of the
datasets (MTSamples) is available in our online ap-
pendix
1
. We note that while we strived to provide
reproducible results, GPT-4’s results are still not en-
tirely deterministic. We also provided the resulting
outputs produced by GPT-4 for this dataset. The other
evaluated dataset’s text cannot be published as per its
terms of use, thus we omitted this dataset from the on-
line appendix. Our experiments used version 0613 of
GPT-4. Our evaluation result demonstrates that when
given instructions and examples as a prompt, GPT-4
is capable of handling the examined tasks reasonably
well.
2 RELATED WORK
The transformer architecture has reformed machine
learning models for NLP. The two foundations of this
architecture are the previously mentioned BERT and
GPT models. Initially, these models were trained
on general texts but later appeared domain-specific
models, too. The objective of these new models
was to outperform the general models in various
domain-specific tasks. BioBERT (Lee et al., 2020)
and BioGPT (Luo et al., 2022) models were trained
on biomedical literature. MedBERT (Rasmy et al.,
2021), a German medical natural language process-
ing model, was fine-tuned using medical texts, clini-
cal notes, research papers, and healthcare-related doc-
uments. RAD-BERT (Bressem et al., 2020) and Ra-
diologyGPT (Susnjak, 2024) models, trained on radi-
ology reports, outperformed previous, more general
biomedical domain models, demonstrating a better
understanding of radiology language.
These models have transformed the field of infor-
mation extraction from unstructured text data. Cur-
rent SOTA results were achieved by encoder-only
models like BERT. These models are typically fine-
tuned on annotated data before being used in NER
and RE tasks. With the introduction of LLMs, like
GPT-3, ChatGPT, and GPT-4, researchers started ex-
perimenting with these models to solve biomedical
NER and RE tasks. Researchers (Agrawal et al.,
2022) found that even without explicit training for
the clinical domain, recent language models like In-
structGPT and GPT-3 can effectively extract clini-
cal information in a few-shot setting. Research has
1
https://github.com/anonymusradiology/Data2024-sub
mission-dataset
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine Radiology Reports
83

Figure 1: Structure of a fictitious report with unique identifier, finding and impression sections.
been made to compare the few-shot performance of
GPT-3’s in-context learning with the fine-tuning of
smaller, BERT-sized PLMs on NER and RE tasks
(Jimenez Gutierrez et al., 2022). They used vari-
ous benchmark datasets and found that GPT-3 could
not outperform fine-tuned BERT-sized PLMs. Chen
et al. (Chen et al., 2023b) employed prompt en-
gineering to evaluate ChatGPT’s performance on
biomedical NER and RE tasks within the BLURB
(Biomedical Language Understanding and Reason-
ing Benchmark) datasets. They used both zero-shot
and few-shot approaches. Experiments have been
(Chen et al., 2023a) established benchmarks for GPT-
3.5 and GPT-4 in biomedical NER and RE at zero-
shot and one-shot settings. They selected examples
from BC5CDRchemical, NCBI-disease, ChemProt,
and DDI datasets, using consistent prompts for eval-
uation. Their experiments showed that LLMs like
ChatGPT, GPT-3.5, and GPT-4 performed less effec-
tively than fine-tuned pretrained models on common
NER and RE datasets. Researchers have also ana-
lyzed free-text CT reports on lung cancer (Fink et al.,
2023). They compared ChatGPT with GPT-4 on the
task of labelling oncologic phenotypes. They found
that GPT-4 outperformed ChatGPT in extracting le-
sion parameters, identifying metastatic disease and
generating correct labels for oncologic progression.
GPT models has greatly benefited sentiment anal-
ysis. Studies observed that the zero-shot performance
of LLMs achieves comparable performance to the
fine-tuned BERT model (Qihuang et al., 2023). There
was a study using ChatGPT for sentiment analysis
task, specifically examining its capacity to manage
polarity shifts, open-domain scenarios, and sentiment
inference issues (Wang et al., 2023). Researchers in-
vestigated the GPT’s sentiment analysis capabilities
in a prompt-based GPT, a finetuned GPT model, and
GPT embedding classification (Kheiri and Karimi,
2023). They demonstrated a significant performance
improvement compared to the SOTA models.
Medical professionals and researchers have uti-
lized LLM technologies in a variety of text-generation
tasks, leading to significant advancements. These
tasks include question answering, automatic impres-
sion generation, summarization of medical docu-
ments, medical education, etc. Studies presented a
comparative evaluation of GPT-4, GPT-3.5 and Flan-
PaLM 540B (Nori et al., 2023). Their findings re-
vealed that GPT-4 significantly outperforms GPT-
3.5’s and Flan-PaLM 540B’s performance in gener-
ating answers for the US Medical Licensing Exam
(USMLE) and on the MultiMedQA dataset. GPT-4’s
performance was evaluated in generating evidence-
based impressions from radiology reports (Sun et al.,
2023), they compared the results to human data.
Impressions composed by radiologists outperformed
GPT-4-generated ones in coherence, comprehensive-
ness, and factual consistency.
In addition, numerous articles deal with explor-
ing LLMs’ opportunities and pitfalls in revolution-
izing radiology (Thapa and Adhikari, 2023; Ak-
inci D’Antonoli et al., 2023; Liu et al., 2023)
We dive deep into the analysis of spine reports,
examine them at several levels, which is not typical
in the radiology research. While multiple research
articles deal with tasks such as sentence classifica-
tion, sentiment analysis, information extraction and
summarisation, to our knowledge, none of the above
studies has combined all these problems into a single
process. While we deal with a thin slice of medical
text processing, our analysis and evaluations require
a high granularity of data on anatomy levels and spe-
cific nomenclature of spine conditions which are, to
the best of our knowledge, not included in any bench-
mark that is currently available. Thus, our results
were evaluated by a human linguist annotator.
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
84

3 METHOD
3.1 Data
This study utilized two databases, the publicly avail-
able MIMIC-III
2
(Johnson et al., 2016) database
(which is accessible through a course) and the MT-
Samples
3
corpus.
MIMIC-III is a large, de-identified, and publicly
available collection of medical reports. The dataset
is freely accessible, making it a valuable resource for
researchers. The dataset contains approximately 60K
medical reports from ICUs. It contains detailed in-
formation on medications, laboratory measurements,
procedure codes, diagnostic codes, imaging reports,
hospital length of stay, and more. After complet-
ing a web course offered by the National Institutes
of Health (NIH), we gained access to the MIMIC-III
database. The database consists of 26 tables. In this
article, we work with radiology reports, so we utilize
only the NOTEEVENTS table. This table contains
unstructured text data, including nursing and physi-
cian notes, ECG reports, radiology reports, and dis-
charge summaries. These free-text radiology reports
contain the finding, the impression, the date of the
study, and additional information depending on the
study type. We filtered the spine radiology reports
based on the technical area of the study, which is
also provided. After this selection process, we struc-
tured these reports and kept only the findings and the
impressions sections. We note that the MIMIC-III
dataset’s website expressly forbids the dataset’s use
with online GPT services, with the caveat of using
them with one of two services with the appropriate
settings. We used the Azure OpenAI service for our
experiments, as suggested by the website.
The MTSamples database, a publicly accessible
resource of transcribed medical reports, comprises
transcribed medical transcription sample reports and
examples across 40 medical specialities, such as ra-
diology, neurology, and surgery. These are free text
reports with headings, which change according to the
speciality. For the purpose of our research, we created
a small corpus from the MTSamples website focus-
ing on spinal reports. Our final collection includes 53
spine reports’s findings and impression sections.
Figure 1 illustrates the structure of a report after
its extraction from the table and preprocessing. This
structure includes three main sections: a unique iden-
tifier generated by us, the findings section, and the im-
pression section. The report in Figure 1 is an entirely
fictitious example.
2
https://physionet.org/
3
https://mtsamples.com/
3.2 Tasks and Methods
Our project encompassed five tasks: sentence classi-
fication, sentence-level sentiment analysis, NER, RE
and anatomy level determination.
The sentence classification task involved two la-
bels, ”spinal” and ”extraspinal”. During the in-
terpretation of radiologic studies, radiologists often
identify abnormalities outside the region of interest.
For instance, a lumbar spinal MRI can detect many
extraspinal abnormalities, which may carry signifi-
cant clinical implications and are crucial to recognize
(Dilli et al., 2014). Common extraspinal regions in-
clude the renal area, uterus, kidney, prostate, and in-
frarenal aorta. This classification was essential as our
focus was only on sentences related to spine anoma-
lies. In the following steps, we focused on sentences
labelled ”spinal”.
We determined the sentiment, categorizing them
as either positive or negative. A sentence was la-
belled positive if the radiologist found no concerning
issues and reported a normal or unremarkable state.
Such sentences typically report normal results or reas-
suring information about a patient’s imaging studies.
Negative sentiment indicates concerns or abnormali-
ties in a patient’s imaging results, providing crucial
insights for professionals into potential issues or ar-
eas requiring further investigation. It’s common to
encounter sentences describing both normal and ab-
normal conditions in different anatomical areas. We
labelled these sentences with negative sentiment be-
cause, from our point of view, it is better to draw at-
tention to a normal condition than to ignore an abnor-
mal state. Please note that our current nomenclature
can potentially be misleading for medical profession-
als, as they usually use the ”positive” term to indicate
the presence of an anomaly (such as positive for her-
nia), and ”negative” for its absence. We used these
terms in the positive and negative sentiment sense,
not in the medical sense. In the information extrac-
tion task, we worked only with the negative sentiment
sentences.
In the named entity recognition task, we deter-
mined two entity types: anatomy and disorder. A term
was considered anatomy, if it described a specific part
of the human body in the spine area, like disc, neuro-
foramen. Disorders are various pathologies observed
by the radiologist like hernia, and lysthesis. If there
are both normal and abnormal conditions in a sen-
tence, only negative conditions were labelled with the
disorder label.
In our relation extraction task, we instructed the
model to determine the relations between anatomy
and disorder entities that were extracted by the pre-
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine Radiology Reports
85

Figure 2: Results of a finding processed with the proposed method, categorized into three sections: ”extraspinal” sentences,
positive sentiment sentences, and negative sentiment sentences with various extracted information.
vious step. The anatomy-disorder relation connects
the disorder with its anatomic locations.
In the case of anatomies, we determined the sec-
tions and the levels within them. We also determined
the intervals (”L3-L5”) and various other referring
phrases (”at this level” and ”the two lower discs at the
cervical spine”). When there was no level or section
in a sentence, we instructed the model to determine
it considering the previous sentences, even if it had
a positive sentiment. If the report did not contain any
level, then we determined just the section of the spine.
Figure 2 illustrates the results of the fictitious find-
ing seen in Figure 1 processed using the proposed
method and our final prompt. The results can be cat-
egorized into three sections. The first section con-
tains the output of the sentence classifier. This section
includes sentences labelled as ”extraspinal”, which
were omitted during the further processing. The mid-
dle section lists sentences expressing positive senti-
ments. Lastly, the lower box contains sentences de-
scribing negative sentiments, from which various in-
formation was extracted.
3.3 Prompt
Initially, we performed experiments to find the most
effective prompt for use in subsequent analyses. The
development of the final prompt was a multi-step pro-
cess, with the help of a development set (5 new re-
ports along the lines of the ones in the MTSamples
dataset), which was separated from the available re-
ports. We tested the different prompts, each incorpo-
rating all five radiology reports from the development
set. Upon identifying errors, we tried to correct them
by adjusting the prompt. We have repeatedly tested
and modified the prompt to optimize output quality.
During our experiments to find the optimal prompt,
it was revealed that each stage of the prompt signifi-
cantly influenced the performance and the efficiency
of the entire process. The process of creating the final
prompt is shown in Figure 3. Initially, we evaluated a
starting prompt. Following this, we improved it cycli-
cally, produced the output using GPT-4, and evaluated
the outcome manually to measure the effectiveness of
prompt variations. Meanwhile, we attempted to refine
the prompt along the evaluation output.
During the development of the optimal prompt,
we utilized a prompt engineering method, in-context
learning. Each prompt included a task description and
the expected output for two reports. In the initial at-
tempts, we did the whole process in a single step, pro-
viding a brief task description and the expected output
for two example reports. In the initial prompt the task
description consisted of a few sentences: ”Your task
is to analyze radiological reports about the spine. Fo-
cus solely on statements made about the spine, extract
sentences indicating a negative status, and from these
sentences, extract information on disorder, anatomy,
and the level of the anatomy.” Upon evaluating the
development reports with this prompt, we identified
several issues: The results lacked sentences indicat-
ing negative status on the spine, included sentences
with negated negative statuses such as ”no herniation
is seen”, incorporated anatomy and descriptive fea-
tures within the extracted disorder, contained anatom-
ical terms of location within the extracted anatomy, as
well as other, less general mistakes.
The initial approach to fixing the prompt involved
formulating a set of rules to avoid the identified errors.
We created a list containing ten specific rules, and di-
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
86

rected the model to adhere to these during the pro-
cessing of the reports. Throughout the rule develop-
ment, we performed multiple tests on the incorrectly
processed sentences identified in the prior evaluation.
We observed that too many rules adversely impacted
the output, leading to new errors. Despite our efforts,
the list of rules did not bring the expected improve-
ments; the output still included the previously noted
errors.
Figure 3: Creation of the final prompt involving iterative
output generation using GPT-4, evaluation and improve-
ment.
Our subsequent strategy for enhancing the prompt
involved dividing the task into several subtasks, con-
sidering the errors we had identified. We experi-
mented with various task divisions until we found the
one that appeared optimal based on the development
reports. This final prompt comprises five subtasks
and a final step, which instructs the model to pro-
duce the final output, without displaying the outputs
of the intermediate steps. In the first step, we instruct
the model to generate a list of sentences from the in-
put report. This step was crucial because it missed
the sentence segmentation task, which forms the ba-
sis of the subsequent steps. The following step in-
volves sentence classification and sentence-level sen-
timent analysis. Here, we guided the model to an-
notate each sentence with an ”extraspinal” label if it
contained information only about a non-spine region,
like the lung, aorta, and head, otherwise the sentence
was labelled as ”spinal”. The sentiment analysis task
then assigns a positive or negative sentiment to the
remaining sentences. The third step works with sen-
tences labelled as negative, performing NER and RE
tasks. Initially, it identifies anatomies and disorders
then establishes relations between these detected enti-
ties. The next step deals with these anatomy-disorder
pairs and assigns the level of the anatomy to each one.
If there is no level in the current sentence, the model
attempts to determine it, considering the report’s pre-
vious sentences. The fifth step comprises rules that
verify the absence of irrelevant information in the ex-
tracted anatomy or disorder entity. Almost each step
includes specified rules for the current task and a tem-
plate for the output. In the prompt we provided illus-
trative examples of complex rules. We developed two
versions of the final prompt. In the first version, the
prompt contained the expected output to each step.
The second version of the final prompt excluded the
output of the inner phases, only featuring the expected
output of the entire process. After testing both ver-
sions on the development dataset, we found no sig-
nificant difference between the two prompts. Con-
sequently, we decided to proceed with the prompt,
which only included the final output in the example.
We utilized GPT-4 for the project’s development.
We created a Python script to repeatedly evaluate the
prompt on different development examples. Within
this script, we adjusted the temperature and top p val-
ues to zero, aiming for the most probable word to
achieve consistent and deterministic output. We also
set the seed value to zero, further enhancing the de-
terministic response. This adjustment ensures nearly
identical output for a given input and seed combina-
tion. Despite our attempts to make the model deter-
ministic, some outputs remained unpredictable in cer-
tain situations.
4 RESULTS
The evaluation process involved 100 reports from the
MIMIC-III database and 53 reports from the MTSam-
ples collection. We selected these reports from the
previously structured ones (structured reports have
unique identifier, finding and impression sections),
focusing solely on the findings section. We measured
the response time for each report and analyzed the
output. The response time mostly depends on the
number of output characters generated by the model.
In our case, the number of output characters was influ-
enced by several factors, such as the length of the in-
put report and the presence of extraspinal and positive
sentences, which required no further processing. The
output length was the longest when the majority of
the findings included degenerative statements about
the spine, leading to an increase in processing time.
The average processing time for the 100 MIMIC-III
reports was 35 seconds, the average input contained
approximately 685 characters per report and the aver-
age output contained approximately 1200 characters
per report. Figure 4 illustrates the correlation between
processing time and the total number of input charac-
ters and generated output characters.
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine Radiology Reports
87
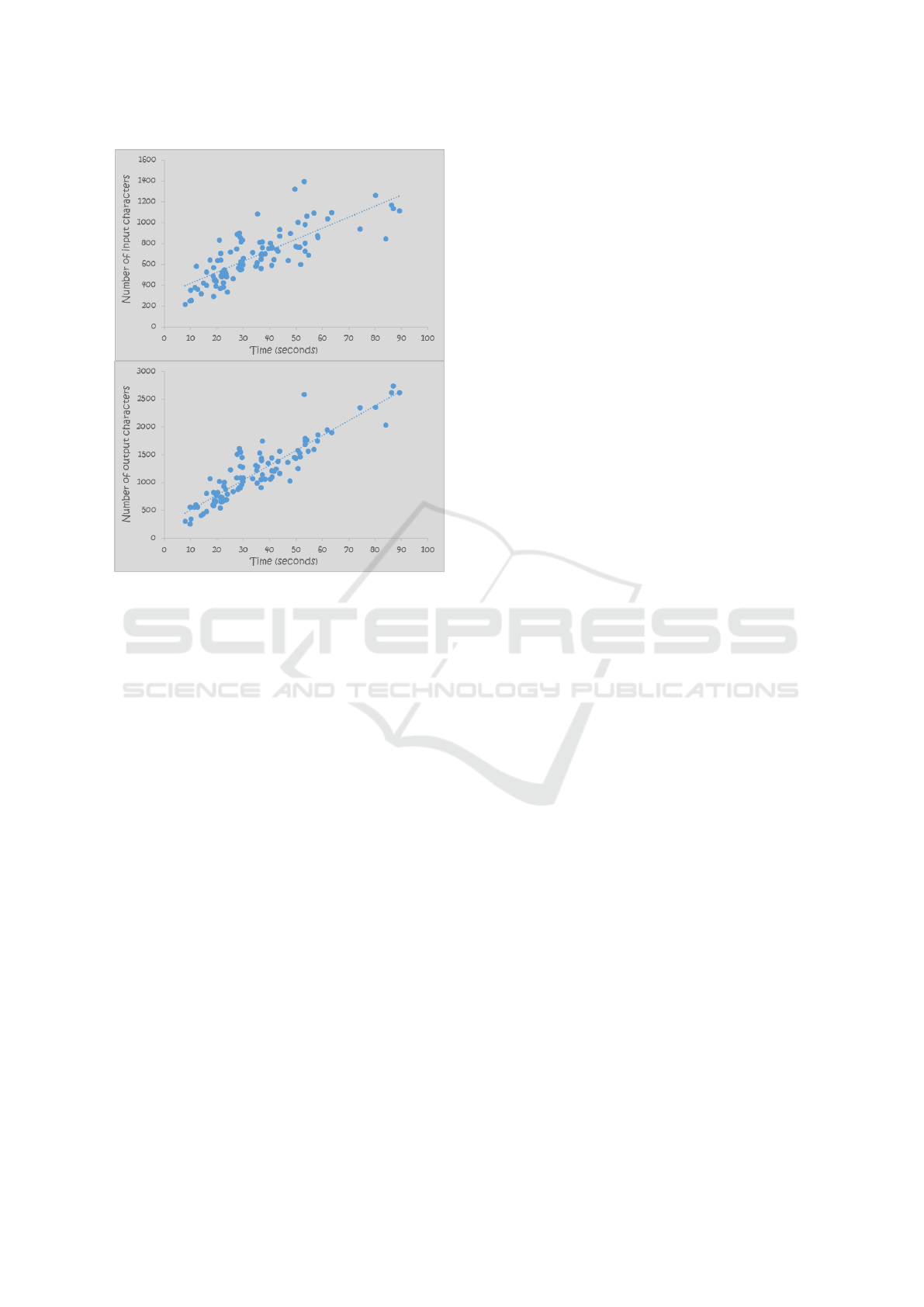
Figure 4: Correlation between processing time and the
number of input (top diagram) and output (bottom diagram)
characters.
We evaluated the final prompt using a complex
evaluation process. We generated the output for each
report, then a linguist annotator who had previously
worked on tasks related to radiological spinal reports
performed the manual evaluation. Finally, one of the
authors with years of experience in processing spine
radiology reports reviewed the evaluation results once
more. The model’s performance was evaluated us-
ing accuracy, precision, recall, and F1-value, which
are based on the concepts of True Positive (TP), True
Negative (TN), False Positive (FP), and False Nega-
tive (FN) values. Accuracy measures the proportion
of correctly classified instances among all instances.
Precision describes the proportion of TP predictions
made by the model. Recall indicates the number of
positives the model rightly predicted. The F1-value is
the harmonic mean of precision and recall. Table 1
provides a comprehensive overview with these met-
rics for the evaluated tasks in the study. The values of
accuracy, precision, recall, and F1-value are presented
for each task separately.
For the sentence classification (SC), sentence
analysis (SA), and anatomy level determination tasks,
the manual annotator had to provide a number rep-
resenting the accurately labelled sentences and cor-
rectly determined levels. In the case of anatomy, de-
generation, and relation testing, FP and FN values
were determined on entity level. The annotator in-
creased the FP value if the model labelled a non-
disorder phrase as a disorder. Similarly, if the model
did not identify a negative sentiment disorder in a
sentence, the FN value was increased. To better dis-
tinguish the different types of errors, we divided the
report-processing workflow into four categories. The
first level of Figure 5 represents sentence segmenta-
tion. However, we did not evaluate this capability
of GPT-4; it is included only to increase the figure’s
comprehensibility. Each category had specific rules
for calculating different measures. As can be seen in
Figure 5, each category is built upon the outcomes of
the previous one, treating errors from earlier stages as
valid inputs for the subsequent layer. The categories
included:
• Sentence classification: We analyzed a list of
sentences derived from the original report. We
checked whether the sentences were correctly cat-
egorized into the ”spinal” and ”extraspine” cate-
gories.
• Sentiment analysis: This step involved a detailed
examination of every sentence labelled as ”spinal”
in the previous step. We checked if each sentence
was correctly labelled as ’positive’ or ’negative’
based on the presence of disorder within the sen-
tence.
• Information extraction: This step involved ana-
lyzing every sentence labelled as ’negative’ in the
sentiment analysis step. We evaluated the labels
for anatomy and disorder entities and the rela-
tionships between the identified entities. Note
that this is the point where entities are considered
rather than sentences.
• Level extraction: This step involved analyzing the
anatomy-disorder pairs generated in the informa-
tion extraction step. Not all sentences included
the level, so the annotator and the model were re-
quired to infer this information from the context.
In the context of anatomy detection, there were 7
instances where the sentence did not contain any
anatomical reference. However, the model inferred
an anatomical location based on previous sentences
and basic knowledge. We classified these instances as
FP, given that the objective of the named entity recog-
nition task does not include inferring missing enti-
ties, and the model was also not instructed to provide
these.
In the case of detected anatomies, there were
46 occurrences in the MTSamples and 10 occur-
rences in the MIMIC-III reports where the entity
included supplementary information beyond the ex-
tracted anatomy. This additional information often
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
88

Figure 5: The report-processing workflow is hierarchically
divided into five categories to distinguish various types of
errors. Each category builds on the results of the previous
one.
specified anatomies, using terms like ”posterior”, ”su-
perior”, and ”distal”. In the disorder detection, there
were 73 identified entities within MTSamples and 62
instances in the MIMIC-III reports that contained ad-
ditional information. These terms were used to spec-
ify the location of the disorder, using words such as
”anterior”, ”posterior”, and ”congenital”. The extra
information also described the characteristics of dis-
orders, like ”small”, ”‘comminuted”, ”partially”, and
”non-displaced”. During the evaluation, we did not
classify these instances as errors. This is because,
in each case, the model accurately determined the
anatomy and disorder terms, along.
5 DISCUSSION
In our study, we evaluated 100 reports from the pub-
licly available MIMIC-III database and 53 reports
from our MTSamples set. We aimed to select diverse
and representative samples that provide a solid basis
for our analyses.
As can be seen in the results section, based on
the evaluated reports, the total F1-value of our ap-
proach is 98.18% and 98.34% on the two different
databases. This F1-value includes the sentence clas-
sification, sentence analysis, anatomy and disorder
detection, relation extraction and the anatomy level
determination tasks. As the complexity of the task
increases, there is a slight decrease in result qual-
ity. The task of annotating spinal and extraspinal
sentences is relatively simple, requiring only a de-
cision on whether the anatomy includes the spine.
Sentences referring to extraspinal only mention the
body part, such as the lung, without going into its
detailed anatomy. The next task, sentiment analysis,
remains relatively straightforward. The process takes
into account two things. The sentiment is positive if
the sentence mentions only positive aspects related to
the given anatomy, such as ”normal” or ”patent”, and
the mention of disorder in negative sentences, for in-
stance, ”no herniation”. Otherwise, the sentiment is
negative. Thus, this task achieved a high level of ac-
curacy. The task of detecting anatomies and disorders
is more difficult than the tasks of sentence classifica-
tion, as evidenced by the results tables. The F1-value
of anatomy detection is somewhat higher than the de-
tection of disorders. This can be because anatomical
terminologies are more standardized and well-defined
across different sources. This consistency enhances
the performance of recognition algorithms. Anatomi-
cal terms may occur more frequently in text data com-
pared to disorder terms. Disorders can vary widely
in representation of their complexity and linguistic
expressions, making it harder for models to identify
them accurately. The task of relation extraction in
our methodology can be relatively simple, so it also
reached a high F1-value. The model is designed to
link the identified anatomical and degenerative enti-
ties in a sentence. In most cases, this is not complex
because, the findings’ sentences have various state-
ments about a particular anatomy. Therefore, a clause
or sentence typically includes one anatomical and one
degenerative entity, making the link between them
straightforward.
Each MTSamples report had an average of 11 sen-
tences, with around 5.5% discussing an extraspinal re-
gion. About 61% of these spinal sentences were neg-
ative, talking about concerns or abnormalities in a pa-
tient’s imaging result. The rest of the sentences were
positive, reporting a normal or unremarkable state. In
comparison, the MIMIC-III reports averaged 8.5 sen-
tences per report in the evaluated 100 reports. Ap-
proximately 20% of the sentences talked about an ex-
traspinal region. We also checked the sentiment of
these spinal sentences. Surprisingly, just more than
half of them, about 51%, were negative, talking about
concerns or abnormalities in a patient’s imaging re-
sult. The rest of the sentences were positive. A sum-
mary of these analyses can be seen in Figure 6.
The other aspect we analysed in the reports is
the contained number of anatomy and disorder en-
tities, as well as the relations between them. Our
approach found about 515 and 430 mentions related
to various parts of the spine and 640 and 550 in-
stances describing different disorders on the spine in
the 53 MTSamples and 100 MIMIC-III reports, re-
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine Radiology Reports
89
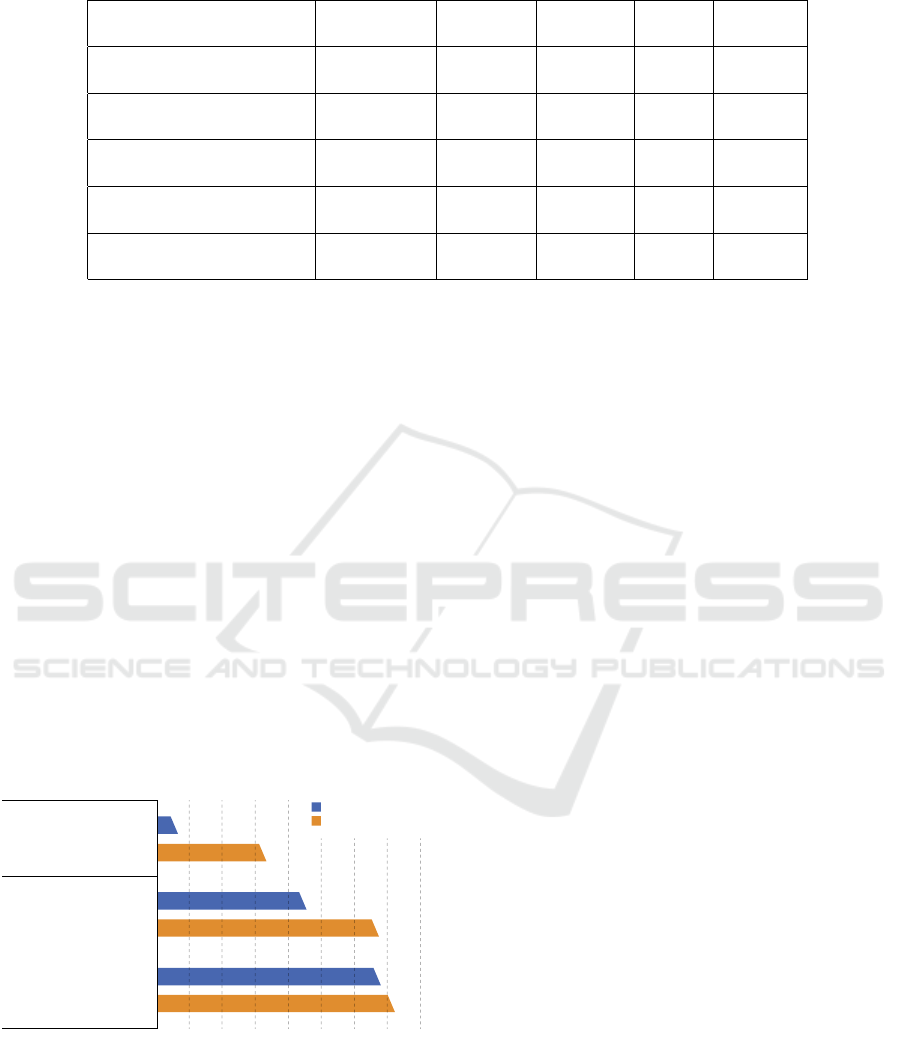
Table 1: Evaluation results on the various examined subtask of the extraction.
Accuracy
(%)
Precision
(%)
Recall
(%)
F1-value
(%)
Sentence classification
MTSamples
MIMIC-III
98.65
99.42
99.11
99.43
99.46
99.86
99.28
99.64
Sentiment analysis
MTSamples
MIMIC-III
98.09
98.00
97.43
96.48
99.42
99.72
98.41
98.07
Named entity recognition
MTSamples
MIMIC-III
89.89
91.67
97.78
97.50
91.77
93.91
94.68
95.66
Relation extraction
MTSamples
MIMIC-III
98.22
98.24
99.67
99.11
98.54
99.11
99.10
99.11
Level of anatomy
MTSamples
MIMIC-III
98.88
98.42
98.88
98.42
100.00
100.00
99.44
99.20
spectively. Additionally, our analysis uncovered 615
and 560 connections between the mentioned anatomy
and disorder entities in these reports. This suggests
that, on average, each MTSamples report contains ap-
proximately 12 negative sentiment disorders and 10
anatomies with disorder and each MIMIC-III report
contains approximately 5.5 negative sentiment disor-
ders and 4.3 anatomies with disorder. The method’s
output helps to measure how often these anatomy and
disorder entities appear and how they are linked, lead-
ing to a better understanding of the medical informa-
tion in the reports.
From these analyses, it can be seen that there is a
significant difference between the two used databases.
Comparing the databases, it is clear that the MTSam-
ples reports contain more sentences, with a majority
referring to the spine region. However, only 40% of
these sentences do not contain degeneration. In con-
trast, the MIMIC-III reports have 2.5 fewer sentences
per report, and half of the sentences describe normal
conditions with positive sentiments.
NON-DEGENERATIVE
SPINAL EXTRASPINAL
DEGENERATIVE
MTSamples (53 reports)
MIMIC-III (100 reports)
0 50 100 150 200 250 300 350 400
34
167
228
338
341
362
Figure 6: Number of spinal (degenerative, non-degeneraive)
and extraspinal sentences in the 53 MTSamples and 100
MIMIC-III reports.
In processing radiology reports, the application
of PLMs and LLMs can yield both positive and
negative effects. The problem of processing spine
radiology reports can also be solved without rely-
ing on GPT-4. With PLMs, it could be solved by
employing, for instance, four separate BERT-based
models, which is a more task-specific approach. A
BERT model fine-tuned for sentence classification
tasks would be employed to categorize sentences into
predefined classes. Another BERT model, fine-tuned
for sentiment analysis, would be utilized to determine
positive or negative sentiment of sentences. A dedi-
cated BERT model fine-tuned for NER tasks would be
applied to recognize and classify anatomy and disor-
der entities within the text. The last fine-tuned model
would be applied to detect relations between the de-
tected entities. The fine-tuning process relies on an-
notated task-specific data. In the medical field, find-
ing freely available, large and representative anno-
tated datasets can be challenging, and the annotation
of these reports is time-consuming and expensive.
In addition fine-tuning models requires substantial
computational resources, including high-performance
GPUs and significant memory. Despite the disad-
vantages task-specific BERT models might provide
clearer interpretability for individual tasks, so it could
be easier to further develop the models by analyzing
the errors in their outputs. Advantages include that,
unlike LLMs, working with such on-premise, fine-
tuned models typically involves less privacy concerns
when dealing with sensitive information.
Compared to PLMs, GPT’s few-shot capability
can effectively guide these models to generate the
desired outputs, eliminating the need for extensive
fine-tuning. This approach can save computational
resources. Moreover, GPT models typically require
less preprocessing of the input text. This can sim-
plify the data preparation process, especially for tasks
involving free-form text like radiology reports. The
method’s disadvantages include that utilizing cloud-
based GPT models may raise privacy concerns, espe-
cially when dealing with sensitive medical data such
as radiology reports. The models can also have a lack
of interpretability and results are harder to reproduce,
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
90

making it challenging to understand and improve the
model’s decision-making process. The outputs are
also less controllable compared to models like BERT,
which is a drawback, particularly in situations where
precise output is essential. Similarly to BERT, it is
possible to fine-tune a GPT-based model for specific
tasks, but this can also require extensive computa-
tional resources and data.
Our study has limitations. First, the evaluation
was implemented on a relatively small sample size of
just 153 spine reports. This may not accurately reflect
the processing capabilities of GPT-4 for spine radi-
ology reports in real-world clinical settings. Second,
using only the MIMIC-III and MTSamples database
restricts the diversity of data. Different databases
may include different language uses and terminolo-
gies, which might affect the model’s performance.
Third, each report was evaluated once, yet the output
of the model is not deterministic. Repeated evalua-
tions could provide a more accurate assessment.
6 CONCLUSION
In our study, we utilized GPT-4 for processing radi-
ology reports, completing the entire task with a sin-
gle prompt. We classified the sentences, determined
the sentiment of each spine-related sentence and ex-
tracted the level of anatomy, anatomy and disorder
triplets. Finally, we evaluated the method on two
different databases, 100 radiology spine reports from
the MIMIC-III database and 53 radiology spine re-
ports from the MTSamples collection. These results
highlight how prompt-learning large language models
can find information from free-text radiology reports
without needing expert knowledge or task-specific
fine-tuning. According to our findings, the GPT-4
model performed with over 91% accuracy and F-score
values in each of our five subtasks of information ex-
traction of the reports. Our MTSamples input and out-
put data, as well as our final prompt are available in
our online appendix.
ACKNOWLEDGEMENTS
The research presented in this paper was supported
in part by the European Union project RRF-2.3.1-21-
2022-00004 within the framework of the Artificial In-
telligence National Laboratory. The national project
TKP2021-NVA-09 also supported this work. Project
no TKP2021-NVA-09 has been implemented with the
support provided by the Ministry of Culture and Inno-
vation of Hungary from the National Research, De-
velopment and Innovation Fund, financed under the
TKP2021-NVA funding scheme.
REFERENCES
Agrawal, M., Hegselmann, S., Lang, H., Kim, Y., and Son-
tag, D. (2022). Large language models are few-shot
clinical information extractors. In Proceedings of the
2022 Conference on Empirical Methods in Natural
Language Processing, pages 1998–2022.
Akinci D’Antonoli, T., Stanzione, A., Bl
¨
uthgen, C., Vernuc-
cio, F., Ugga, L., Klontzas, M., Cuocolo, R., Cannella,
R., and Koc¸ak, B. (2023). Large language models in
radiology: fundamentals, applications, ethical consid-
erations, risks, and future directions. Diagnostic and
Interventional Radiology.
Bressem, K. K., Adams, L. C., Gaudin, R. A., Tr
¨
oltzsch, D.,
Hamm, B., Makowski, M. R., Sch
¨
ule, C.-Y., Vahldiek,
J. L., and Niehues, S. M. (2020). Highly accurate clas-
sification of chest radiographic reports using a deep
learning natural language model pretrained on 3.8 mil-
lion text reports. Bioinformatics, 26(21):5255–5261.
Brown, T., Mann, B., Ryder, N., Subbiah, M., Kaplan, J. D.,
Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G.,
Askell, A., Agarwal, S., Herbert-Voss, A., Krueger,
G., Henighan, T., Child, R., Ramesh, A., Ziegler, D.,
Wu, J., Winter, C., Hesse, C., Chen, M., Sigler, E.,
Litwin, M., Gray, S., Chess, B., Clark, J., Berner,
C., McCandlish, S., Radford, A., Sutskever, I., and
Amodei, D. (2020). Language models are few-shot
learners. In Advances in Neural Information Process-
ing Systems, volume 33, pages 1877–1901.
Chen, Q., Du, J., Hu, Y., Keloth, V., Peng, X., Raja, K.,
Zhang, R., Lu, Z., and Qi, W. (2023a). Large lan-
guage models in biomedical natural language process-
ing: benchmarks, baselines, and recommendations.
arXiv preprint arXiv:2305.16326.
Chen, Q., Sun, H., Liu, H., Jiang, Y., Ran, T., Jin, X., Xiao,
X., Lin, Z., Chen, H., and Niu, Z. (2023b). An exten-
sive benchmark study on biomedical text generation
and mining with ChatGPT. Bioinformatics, 39(9).
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Dilli, A., Ayaz, U. Y., Turanlı, S., Saltas, H., Karabacak,
O. R., Damar, C., and Hekimoglu, B. (2014). Clinical
research incidental extraspinal findings on magnetic
resonance imaging of intervertebral discs. Archives of
Medical Science, 10(4):757–763.
Fink, M. A., Bischoff, A., Fink, C. A., Moll, M., Kroschke,
J., Dulz, L., Heussel, C. P., Kauczor, H.-U., and We-
ber, T. F. (2023). Potential of chatgpt and gpt-4 for
data mining of free-text ct reports on lung cancer. Ra-
diology, 308 3.
Jimenez Gutierrez, B., McNeal, N., Washington, C., Chen,
Y., Li, L., Sun, H., and Su, Y. (2022). Thinking about
GPT-3 in-context learning for biomedical IE? think
A Deep Dive into GPT-4’s Data Mining Capabilities for Free-Text Spine Radiology Reports
91

again. In Findings of the Association for Computa-
tional Linguistics: EMNLP 2022, pages 4497–4512.
Johnson, A. E. W., Pollard, T. J., Shen, L., wei H. Lehman,
L., Feng, M., Ghassemi, M. M., Moody, B., Szolovits,
P., Celi, L. A., and Mark, R. G. (2016). Mimic-iii,
a freely accessible critical care database. Scientific
Data, 3.
Kheiri, K. and Karimi, H. (2023). Sentimentgpt: Exploit-
ing gpt for advanced sentiment analysis and its depar-
ture from current machine learning. arXiv preprint
arXiv:2307.10234.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). Biobert: a pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Liu, P., Yuan, W., Fu, J., Jiang, Z., Hayashi, H., and Neubig,
G. (2021). Pre-train, prompt, and predict: A system-
atic survey of prompting methods in natural language
processing. ACM Computing Surveys, 55(9):1–35.
Liu, Z., Zhong, T., Li, Y., Zhang, Y., Pan, Y., Zhao, Z.,
Dong, P., Cao, C., Liu, Y., Shu, P., Wei, Y., Wu, Z.,
Ma, C., Wang, J., Wang, S., Zhou, M., Jiang, Z., Li,
C., Xu, S., and Liu, T. (2023). Evaluating large lan-
guage models for radiology natural language process-
ing. arXiv preprint arXiv:2307.13693.
Luo, R., Sun, L., Xia, Y., Qin, T., Zhang, S., Poon, H.,
and Liu, T.-Y. (2022). Biogpt: generative pre-trained
transformer for biomedical text generation and min-
ing. Briefings in Bioinformatics, 23.
Nori, H., King, N., McKinney, S., Carignan, D.,
and Horvitz, E. (2023). Capabilities of gpt-4
on medical challenge problems. arXiv preprint
arXiv:2303.13375.
OpenAI (2023). Gpt-4 technical report. arXiv preprint
arXiv:2303.08774.
Qihuang, Z., Ding, L., Liu, J., Du, B., and Tao, D.
(2023). Can chatgpt understand too? a comparative
study on chatgpt and fine-tuned bert. arXiv preprint
arXiv:2302.10198.
Rasmy, L., Xiang, Y., Xie, Z., Tao, C., and Zhi, D. (2021).
Med-bert: pretrained contextualized embeddings on
large-scale structured electronic health records for dis-
ease prediction. npj Digital Medicine, 4:86.
Sun, Z., Ong, H., Kennedy, P., Tang, L., Chen, S., Elias, J.,
Lucas, E., Shih, G., and Peng, Y. (2023). Evaluating
gpt-4 on impressions generation in radiology reports.
Radiology, 307(5).
Susnjak, T. (2024). Applying BERT and ChatGPT for Senti-
ment Analysis of Lyme Disease in Scientific Literature,
pages 173–183.
Thapa, S. and Adhikari, S. (2023). Chatgpt, bard, and large
language models for biomedical research: Opportuni-
ties and pitfalls. Annals of Biomedical Engineering,
51.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, L. u., and Polosukhin, I.
(2017). Attention is all you need. In Advances in
Neural Information Processing Systems, volume 30.
Wang, Z., Xie, Q., Ding, Z., Feng, Y., and Xia, R. (2023).
Is chatgpt a good sentiment analyzer? a preliminary
study. arXiv preprint arXiv:2304.04339.
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
92
