
Towards FAIR Data Workflows for Multidisciplinary Science:
Ongoing Endeavors and Future Perspectives in Plasma Technology
Robert Wagner
1a
, Dagmar Waltemath
2b
, Kristina Yordanova
3c
and Markus M. Becker
1d
1
Leibniz Institute for Plasma Science and Technology (INP), Felix-Hausdorff-Str. 2, Greifswald, Germany
2
Medical Informatics Laboratory, University Medicine Greifswald, Felix-Hausdorff-Str. 8, Greifswald, Germany
3
Institute for Data Science, University of Greifswald, Felix-Hausdorff-Str. 18, Greifswald, Germany
Keywords: Data Management, FAIR Data Principles, Graph Database, Plasma Science.
Abstract: This paper focuses on the ongoing process of establishing a FAIR (Findable, Accessible, Interoperable and
Reusable) data workflow for multidisciplinary research and development in applied plasma science. The
presented workflow aims to support researchers in handling their project data while also fulfilling the
requirements of modern digital research data management. The centerpiece of the workflow is a graph
database (utilizing Neo4J) that connects structured data and metadata from multiple sources across the
involved disciplines. The resulting workflow intents to enhance the FAIR compliance of the data, thereby
supporting data integration and automated processing as well as providing new possibilities for user friendly
data exploration and reuse.
1 INTRODUCTION
In times of advancing digitization and global
connectivity, the FAIR (Findable, Accessible,
Interoperable, and Reusable) data principles
(Wilkinson et al., 2016) have become increasingly
crucial for effective data management. Enhancing the
overall FAIR compliance of data can aid in
addressing emerging scientific inquiries. The
growing complexity of these research questions
necessitates a multidisciplinary research approach
(Hadorn et al., 2008). However, multidisciplinary
science poses its own challenges, such as the
variability in data structure, formats, and quality, as
well as the lack of consistent and structured metadata
for data description. One example of these challenges
is the varying structure of generated data and the
frequent lack of measurement-relevant metadata due
to fluctuations of the researchers involved in the
individual projects. The usage of these workflows to
structure the collected metadata and reusing them in
the graph database shall help to present easy to access
a
https://orcid.org/0000-0002-2762-293X
b
https://orcid.org/0000-0002-5886-5563
c
https://orcid.org/0000-0002-6428-1062
d
https://orcid.org/0000-0001-9324-3236
examples for researchers in the coming projects to
lessen these challenges.
Additionally, there is a need for more consistent
implementation of research data management (RDM)
practices (Birkbeck et al., 2022). Improving the
overall FAIR compliance of data can help mitigate
these challenges and lay the foundation for future data
reuse.
In this work the problem of applying the FAIR
data principles to multidisciplinary laboratory
experiments in applied plasma science and plasma
technology is addressed. The spectrum of disciplines
involved in research and development (R&D) in this
field includes engineering sciences (such as
mechanical and electrical engineering), life sciences
(for example environmental sciences, microbiology
and food sciences), medicine and physics. Engineers
and physicists are needed to design and construct the
plasma sources (Schmidt et al., 2019). On the other
hand, researchers from the life sciences and
biomedical research use the designed plasma sources,
e.g. for decontamination of food (Wagner, Weihe, et
Wagner, R., Waltemath, D., Yordanova, K. and Becker, M.
Towards FAIR Data Workflows for Multidisciplinary Science: Ongoing Endeavors and Future Perspectives in Plasma Technology.
DOI: 10.5220/0012808000003756
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 13th International Conference on Data Science, Technology and Applications (DATA 2024), pages 471-477
ISBN: 978-989-758-707-8; ISSN: 2184-285X
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
471

al., 2023) or wound treatment (Emmert et al., 2020).
Their domain knowledge needs to be integrated with
the work of engineers and physicists, forming a
complex scientific network with heterogenous and
domain-specific data sets.
The field-specific knowledge of scientists in the
life sciences includes, for example, the diverse targets
of physical plasma in the field of microbiology and
the resulting physico-chemical reactions. The plasma
source-related knowledge of engineers and the
specialist knowledge of physicists in the field of
plasma physics support the optimal application of the
required plasma. Both sides are necessary to include,
since their domain knowledge are polar opposites that
complement each other.
The most important benefit of the proposed
workflows for all researchers involved is easier
access to the information, which are important to
answer the multidisciplinary scientific questions in
the field of plasma science by utilizing the FAIR
principles. The presented paper reports on recent
endeavors and technical solutions that contribute to
the challenge of handling heterogenous, complex
research data in plasma science. The work contributes
to the implementation of the FAIR data principles in
workflows for R&D, that also take the individual
needs of each discipline involved into account. This
includes a comprehensive strategy for data
management, providing detailed insights into the
potential for interlinking metadata about research
studies. This metadata can encompass information
about the devices used, their properties, samples,
preparation and treatment procedures, and more. The
proposed approach enables the connection between
metadata contained in an electronic lab notebook
(ELN) and the corresponding raw data, typically
stored in a repository, through the utilization of a
graph database such as Neo4J (Webber, 2012).
Furthermore, the context of the interlinked data can
be further enriched by incorporating additional
information and by leveraging the categorization of
entities from the ELN. This integrated approach aims
to enhance the overall management and
interconnectivity of research-related data, facilitating
a more comprehensive and contextual understanding
of the research process and its outcomes.
2 WORKFLOW DESCRIPTION
The developed workflow consists of collecting
metadata and linking it to additional information from
a variety of sources such as local data management
platforms or ELN. Given the varying progress in data
management across different disciplines, the
metadata collection was separated into two distinct
workflows. This division takes the individual levels
of development in structured RDM of each discipline
into account, while simultaneously enhancing the
overall FAIR compliance of the collected metadata.
The metadata linking section of the general workflow
embeds the collected metadata of each discipline into
a graph database.
Nevertheless, the general concept of this work is
no novelty, as other institutes are dealing with similar
issues due to the relevance of the topic. As an
example, (de Oliveira, 2022) and (Crystal-Ornelas,
2022), have already shown similar concepts. In (de
Oliveira, 2022), the metadata collected is also stored
in RDF format and in (Crystal-Ornelas, 2022) the
interaction of data from several disciplines was
addressed.
2.1 Research Without
Discipline-Specific Metadata
Collection Standards
The first step in research without any discipline-
specific metadata collection standards is the
unstructured or generic structured (by generic
metadata schema like DataCite (Group, 2021))
collection of experimental results and experimental
metadata. The consisting elements of collected
experiments are used to design the first draft of a
template, which will be used for the following
collection of similar experiments. For this purpose of
collecting metadata and designing a template the ELN
“eLabFTW” (Carpi et al., 2017) is used. The raw and
processed data from the experiments, that are part of
a publication (or a dataset publication itself) can be
published in the interdisciplinary plasma technology
data platform “INPTDAT” (Becker et al., 2019). The
datasets in INPTDAT or from a location in the central
data storage can be linked to the experiments in the
ELN. The different experiments and used resources
(such as devices and consumables) can be interlinked
and categorized to enable better findability of the
provided information. Furthermore, the laboratory
management can be enhanced by proper organization
of resources in the ELN, including the possibility to
setup the booking of resources in eLabFTW.
The metadata of the collected experiments can be
extracted from the unstructured (before template
design) and (semi-)structured (after usage of
templates) ELN entries. The extracted metadata are
stored as machine-readable JavaScript object notion
(JSON) files. Automatic data processing of the
machine-readable metadata files and the linked raw
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
472
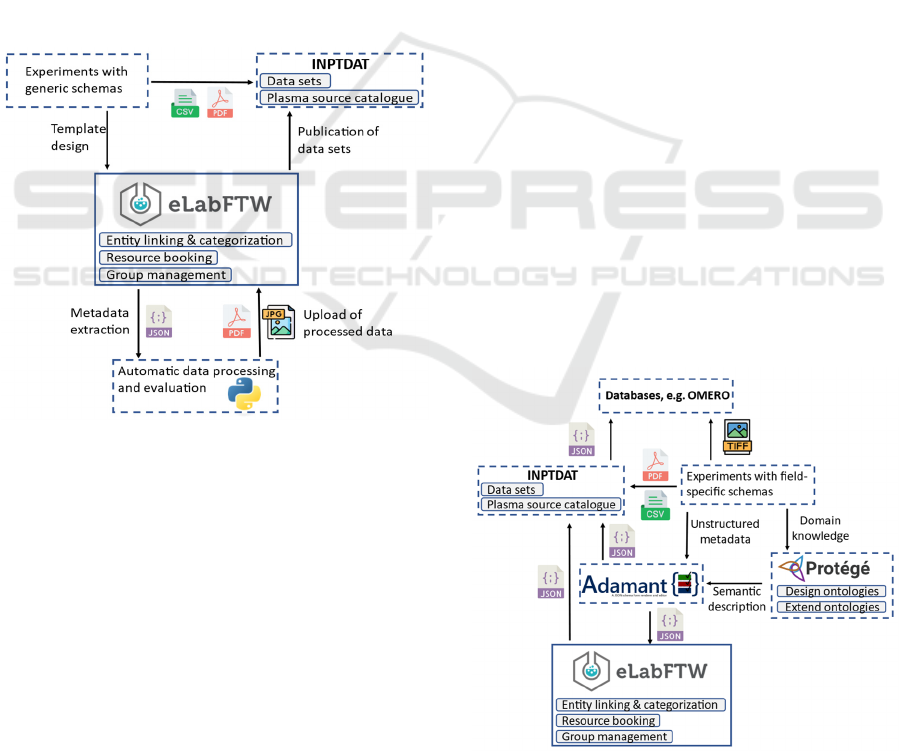
data is part of the workflow (Figure 1). The processed
data in form of tables or graphs can later be linked to
the corresponding ELN entries or the related datasets
published in INPTDAT.
One example of the application of this workflow
in plasma science is the extraction of industrially
relevant ingredients from microalgae (Sommer et al.,
2021). Due to the new developments of systems for
these tasks, the experiments are still subject to
constant changes, so that no schemes have yet been
established. Therefore, this workflow shall assist the
researchers during the metadata collection and shall
also enable the possibility to process and evaluate
their scientific data while utilizing the machine-
readable metadata, which can be extracted from their
experiments. However, it must be noted that the
extracted metadata is strongly influenced by the still
changing structure of the experiments and the lack of
structured metadata schemata and is therefore a
temporary solution.
Figure 1: Workflow schema for R&D without discipline-
specific metadata collection standards.
2.2 Research Using Discipline-Specific
Metadata Collection Standards
The second case (Figure 2) covers research in
disciplines, which can utilize existing discipline-
specific metadata collection standards or RDM
standards. These standards/schemas are used to
structure the collected metadata. The structured
collection of metadata based on metadata schemas is
realized by the RDM tool “Adamant” (Chaerony Siffa
et al., 2022). Adamant can also directly push the
collected metadata into the ELN.
One example for such schemas is based on the
community standard REMBI (Sarkans et al., 2021).
REMBI (REcommended Metadata for Biological
Images) defines the structure for community accepted
descriptions of imaging metadata. The images from
related experiments can be stored along with their
metadata according to REMBI in databases like
“OMERO” (Allan et al., 2012). OMERO is a database
for images that also allows the annotation of images
and datasets with their corresponding metadata via
scripts. The REMBI-structured metadata in OMERO
are further supplemented by the addition of
discipline-specific elements (e.g. via Plasma-MDS
(Franke et al., 2020)) and the OME schema (Goldberg
et al., 2005).
Ontologies can also be integrated into the
workflow to increase the FAIR compliance of the
collected metadata. Semantic annotations contribute
to the machine-readable description of the metadata,
metadata schema and collected data. It is preferable
to reuse existing domain ontologies. However, the
extension or the design of new ontologies is also part
of the later workflow, if no fitting ontology or
ontology terms exist or necessary entities are not well
enough described. The software “Protégé” (Musen &
Protege, 2015) is used to build and maintain the
ontologies in this project. One example of the
application of this workflow in plasma science is the
analysis of plasma-treated liquids in ion
chromatography. Ion chromatography does not yet
have a common metadata schema, but as part of
related research, a metadata schema based on ASTM
1151 ("ASTM E1151:1993 Standard Practice for Ion
Chromatography Terms and Relationships," 1993)
has been designed. This metadata schema has since
been used in this field to collect structured metadata.
Figure 2: Workflow for research, that utilizes discipline-
specific metadata collection standards.
Towards FAIR Data Workflows for Multidisciplinary Science: Ongoing Endeavors and Future Perspectives in Plasma Technology
473
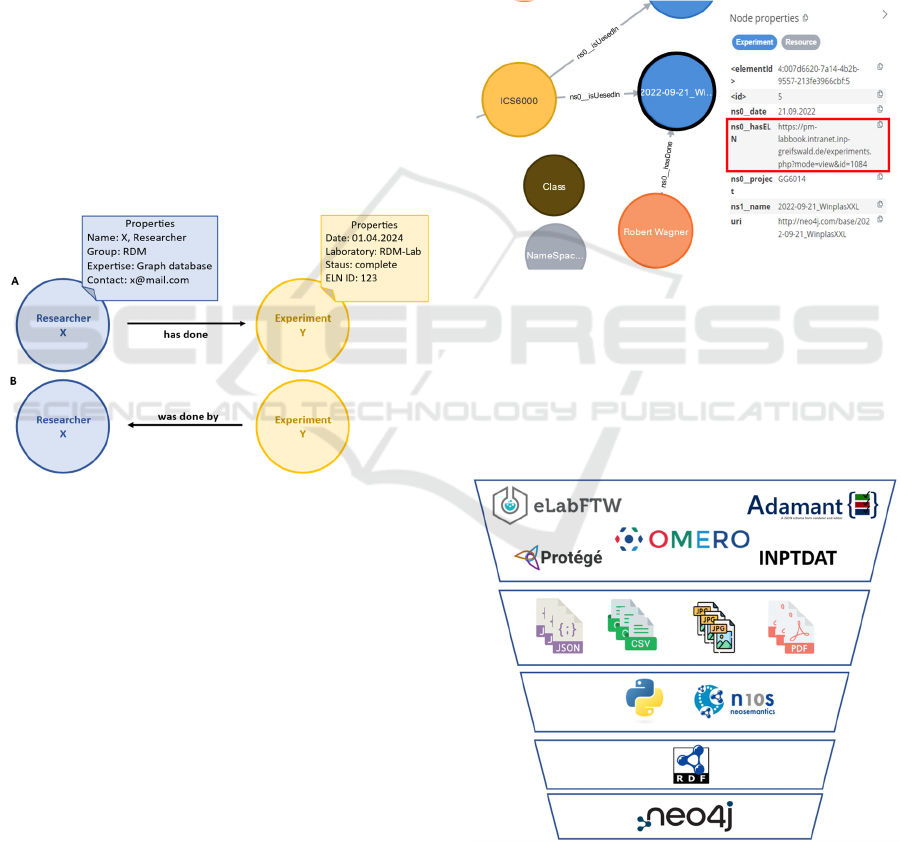
2.3 Graph Database for
Multidisciplinary Data
The structured metadata and experimental data
resulting from steps (2.1) or (2.2) need to be
contextualized and interlinked in a flexible and
extensible manner. One approach is the design of an
overarching graph database, see for example (Mazein
et al., 2024). In our design, the entities (so-called
nodes) represent the experiments, researchers,
devices and projects involved in the multidisciplinary
research. The context that describes the relationships
between each entity are represented as directed
arrows, i.e. the edges of the graph. However, a simple
graph consisting of edges and nodes is not sufficient
to cover the complexity of multidisciplinary scientific
questions and to provide a good readability for the
end users. To meet both needs, a property graph (used
in most graph databases) is used. A property graph as
shown in Figure 3 allows to add information to nodes
and edges to describe the context.
Figure 3: Property graph example of the two nodes
“Researcher X” and “Experiment Y” linked by a directed
edge and their assigned properties. Graph A and B show the
difference in labelling an edge considering the origin of the
edge.
In the current approach, these properties are
extracted via python scripts from the structured
metadata provided by (2.1) and (2.2). The edge
properties can include semantic descriptions from
either ontologies or open vocabularies, like
schema.org for persons (Schema.org, 2024). The
properties of the nodes also provide the opportunity
to cater the provenance of the specific nodes. For
example, the raw data location in a research data
repository (e.g. INPTDAT), the specific stack of
images in an image database (e.g. OMERO), the
original experiment in an ELN (e.g. eLabFTW), the
description of the used plasma source from a plasma
device catalogue (e.g. the plasma source catalogue
that is also part of INPTDAT) or the link to the formal
description of the involved laboratory devices in a
device database can be automatically attached to
nodes via scripts.
Figure 4 depicts a small example of a graph in
Neo4J, that contains nodes for experiments from
different researchers using the same device. The
graph also contains a node with the configurations of
the graph and a node with the namespaces used for
the semantic description. Each class of nodes can
easily be distinguished by their color and label. The
properties of each node can be accessed by clicking
on the specific node in Neo4J as shown in Figure 4.
Figure 4: Example of a property graph and the node
property utilization in Neo4J. The red box shows the
incorporation of the ELN into the graph database.
The process of building a graph from these
different sources is automatized by the formulation of
a triple-based Resource Description Framework
(RDF) file, as illustrated by Figure 5.
Figure 5: Generation of the graph based on a RDF file,
containing the extracted information from different sources
for graph design via python scripts.
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
474

Here, the RDF file is translated into the final graph
by a python script in combination with the Neo4J
plugin “neosemantics”, which is also known as
“n10s” (Barrasa & Cowley). The usage of basic
constraints such as node and relationship property
uniqueness shall prevent the accidental duplication of
already ingested data. For example, if two different
experiments done by “Researcher X” are ingested,
only three nodes are created. During the ingestion of
the second experiment, only the experiment node is
created, since the defined node uniqueness prevents
the creation of the already existing “Researcher X”.
The second experiment is then linked to the existing
node of “Researcher X”. Note that the creation of the
RDF as an intermediate step instead of the direct
graph translation of the extracted information via
Cypher (Francis et al., 2018) enables the easy storage
of RDF files, sharing with the community or use by
other graph database management systems (Das et al.,
2020).
3 CONCLUSIONS AND
OUTLOOK
In this paper an approach of applying the FAIR data
principles to a metadata collection and interlinking
workflow for multidisciplinary experiments in
applied plasma science and plasma technology is
described. Two workflows for the collection of
metadata, one for research with discipline-specific
metadata collection standards and another one for
research without such standards, were presented. The
metadata interlinking is achieved by the generation of
a graph database. The foundation for the metadata
collection is set and is also demonstrated in (Wagner,
Chaerony Siffa, et al., 2023) and (Ahmadi et al.,
2023).
The FAIR principles are implemented as follows.
The findability of the relevant metadata is ensured by
the graph database used. The graph database links all
metadata collected by the workflows with the
scientists and devices involved in order to place them
in a scientific context. The clear and user-friendly
structure of a graph also makes it possible to access
the data. The interoperability of the metadata linked
in the graph is achieved by integrating the
information from various relevant media such as the
ELN, INPTDAT and the device database and can be
accessed by the user by clicking on the respective
properties. The connection and clarity achieved in
this way should also facilitate the reuse of the data.
To evaluate the implementation of the FAIR
principles, a structured FAIR assessment for the
proposed workflows is planned and the results of this
assessment will be used to optimize the workflows.
Also, the feedback of all involved researchers has to
be taken into account.
The next steps intend to expand the linking of the
collected metadata. The usage of the generated graph
database by the intended end-users is conceptualized
by the integration of graph exploration and
visualization tools (Jong, 2021). Another aspect that
is planned in the future is the involvement of the end-
users as a fast survey of needed improvements. One
possible approach for the user integration is to enable
the access to the graph database in Adamant via React
hooks as described in (Cowley, 2020). The
combination of both tools is intended to unify the
structured metadata collection and metadata
representation on the one hand and reduce the amount
of software that users need to familiarize themselves
with on the other.
Another but more specific challenge for plasma
science is the absence of a plasma science ontology
and therefore the design and implementation of such
an ontology is crucial to solidify the collected
metadata by the addition of a proper semantic
description. A correct semantic description can avoid
possible misunderstandings between scientists in
different fields of plasma science. An example for this
is the term "matrix", which can describe a carrier
material of analytical samples in the field of
chemistry, the inner fluid of cell organelles in biology
or a mathematical order of numbers. Thus, the usage
of ontologies can explain the context and then clearly
explain the analysis of the matrix for the scientists
involved.
ACKNOWLEDGEMENTS
The work is funded by the Deutsche
Forschungsgemeinschaft (DFG, German Research
Foundation) under the National Research Data
Infrastructure – [NFDI46/1] - 501864659.
REFERENCES
Ahmadi, M., Wagner, R., Plathe, N., Mattern, P.,
Bekeschus, S., Becker, M. M., Stöter, T., &
Weidtkamp-Peters, S. (2023). Data stewardship and
research data management tools for multimodal linking
of imaging data in plasma medicine [Poster].
https://doi.org/10.5281/zenodo.10069368
Towards FAIR Data Workflows for Multidisciplinary Science: Ongoing Endeavors and Future Perspectives in Plasma Technology
475

Allan, C., Burel, J.-M., Moore, J., Blackburn, C., Linkert,
M., Loynton, S., MacDonald, D., Moore, W. J., Neves,
C., Patterson, A., Porter, M., Tarkowska, A., Loranger,
B., Avondo, J., Lagerstedt, I., Lianas, L., Leo, S.,
Hands, K., Hay, R. T., Patwardhan, A., Best, C.,
Kleywegt, G. J., Zanetti, G., Swedlow, J. R. (2012).
OMERO: flexible, model-driven data management for
experimental biology. Nature Methods, 9(3), 245-253.
https://doi.org/10.1038/nmeth.1896
ASTM E1151:1993 Standard Practice for Ion
Chromatography Terms and Relationships. (1993).
https://dx.doi.org/10.1520/E1151-93R19
Barrasa, J., & Cowley, A. neosemantics (n10s): Neo4j RDF
& Semantics toolkit. Retrieved 11.03.2024 from
https://neo4j.com/labs/neosemantics/
Becker, M. M., Paulet, L., Franke, S., & O'Connell, D.
(2019). INPTDAT – a new data platform for plasma
technology 72nd Annual Gaseous Electronics
Conference (GEC), College Station, Texas, USA.
https://doi.org/10.5281/zenodo.3500283
Birkbeck, G., Nagle, T., & Sammon, D. (2022). Challenges
in research data management practices: a literature
analysis. Journal of Decision Systems, 31(sup1), 153-
167. https://doi.org/10.1080/12460125.2022.2074653
Carpi, N., Minges, A., & Piel, M. (2017). eLabFTW: An
open source laboratory notebook for research labs. The
Journal of Open Source Software.
https://doi.org/10.21105/joss.00146
Chaerony Siffa, I., Schäfer, J., & Becker, M. M. (2022).
Adamant: a JSON schema-based metadata editor for
research data management workflows [version 2; peer
review: 3 approved]. F1000Research, 11(475).
https://doi.org/10.12688/f1000research.110875.2
Cowley, A. (2020). Connecting your React app to Neo4j
with React Hooks. Retrieved 11.03.2024 from
https://medium.com/neo4j/connecting-to-react-app-to-
neo4j-148881d838b8
Crystal-Ornelas, R., Varadharajan, C., O’Ryan, D.,
Beilsmith, K., Bond-Lamberty, B., Boye, K., Burrus,
M., Cholia, S., Christianson, D. S., Crow, M.,
Damerow, J., Ely, K. S., Goldman, A. E., Heinz, S. L.,
Hendrix, V. C., Kakalia, Z., Mathes, K., O’Brien, F.,
Pennington, S. C., Robles, E., Rogers, A., Simmonds,
M., Velliquette, T., Weisenhorn, P., Welch, J. N.,
Whitenack, K., Agarwal, D. A. (2022). Enabling FAIR
data in Earth and environmental science with
community-centric (meta)data reporting formats.
Scientific Data, 9(1), 700.
https://doi.org/10.1038/s41597-022-01606-w
Das, A., Mitra, A., Bhagat, S. N., & Paul, S. (2020). Issues
and Concepts of Graph Database and a Comparative
Analysis on list of Graph Database tools. 2020
International Conference on Computer Communication
and Informatics (ICCCI), 1-6. https://doi.org/10.
1109/ICCCI48352.2020.9104202
de Oliveira, N. Q., Borges, V., Rodrigues, H. F., Campos,
M. L. M., & Lopes, G. R. (2022). A Practical Approach
of Actions for FAIRification Workflows.
https://doi.org/10.48550/arXiv.2201.07866
Emmert, S., van Welzen, A., Masur, K., Gerling, T.,
Bekeschus, S., Eschenburg, C., Wahl, P., Bernhardt, T.,
Schäfer, M., Semmler, M. L., Grabow, N., Fischer, T.,
Thiem, A., Jung, O., & Boeckmann, L. (2020). Cold
atmospheric pressure plasma for the treatment of acute
and chronic wounds. Hautarzt, 71(11), 855-862.
https://doi.org/10.1007/s00105-020-04696-y
Francis, N., Green, A., Guagliardo, P., Libkin, L.,
Lindaaker, T., Marsault, V., Plantikow, S., Rydberg,
M., Selmer, P., & Taylor, A. (2018). Cypher: An
Evolving Query Language for Property Graphs
Proceedings of the 2018 International Conference on
Management of Data, Houston, TX, USA.
https://doi.org/10.1145/3183713.3190657
Franke, S., Paulet, L., Schafer, J., O'Connell, D., & Becker,
M. M. (2020). Plasma-MDS, a metadata schema for
plasma science with examples from plasma technology.
Sci Data, 7(1), 439. https://doi.org/10.1038/s41597-
020-00771-0
Goldberg, I. G., Allan, C., Burel, J.-M., Creager, D.,
Falconi, A., Hochheiser, H., Johnston, J., Mellen, J.,
Sorger, P. K., & Swedlow, J. R. (2005). The Open
Microscopy Environment (OME) Data Model and
XML file: open tools for informatics and quantitative
analysis in biological imaging. Genome Biology, 6(5),
R47. https://doi.org/10.1186/gb-2005-6-5-r47
Group, D. M. W. (2021). DataCite Metadata Schema
Documentation for the Publication and Citation of
Research Data and Other Research Outputs. Version
4.4. https://doi.org/10.14454/3w3z-sa82
Hadorn, G. H., Biber-Klemm, S., Grossenbacher-Mansuy,
W., Hoffmann-Riem, H., Joye, D., Pohl, C., Wiesmann,
U., & Zemp, E. (2008). The Emergence of
Transdisciplinarity as a Form of Research. In G. H.
Hadorn, H. Hoffmann-Riem, S. Biber-Klemm, W.
Grossenbacher-Mansuy, D. Joye, C. Pohl, U.
Wiesmann, & E. Zemp (Eds.), Handbook of
Transdisciplinary Research (pp. 19-39). Springer
Netherlands. https://doi.org/10.1007/978-1-4020-6699-
3_2
Jong, N. d. (2021). 15 Tools for Visualizing Your Neo4j
Graph Database. Retrieved 11.03.2024 from
https://neo4j.com/developer-blog/15-tools-for-
visualizing-your-neo4j-graph-database/
Mazein, I., Rougny, A., Mazein, A., Henkel, R., Gütebier,
L., Michaelis, L., Ostaszewski, M., Schneider, R.,
Satagopam, V., Jensen, L., Waltemath, D., Wodke, J.,
& Balaur, I. (2024). The Use of Graph Databases in
Systems Biology: A Systematic Review. Preprints.
https://doi.org/10.20944/preprints202403.1289.v1
Musen, M. A., & Protege, T. (2015). The Protege Project:
A Look Back and a Look Forward. AI Matters, 1(4), 4-
12. https://doi.org/10.1145/2757001.2757003
Sarkans, U., Chiu, W., Collinson, L., Darrow, M. C.,
Ellenberg, J., Grunwald, D., Hériché, J.-K., Iudin, A.,
Martins, G. G., Meehan, T., Narayan, K., Patwardhan,
A., Russell, M. R. G., Saibil, H. R., Strambio-De-
Castillia, C., Swedlow, J. R., Tischer, C., Uhlmann, V.,
Verkade, P., . . . Brazma, A. (2021). REMBI:
Recommended Metadata for Biological Images—
DATA 2024 - 13th International Conference on Data Science, Technology and Applications
476

enabling reuse of microscopy data in biology. Nature
Methods, 18(12), 1418-1422.
https://doi.org/10.1038/s41592-021-01166-8
Schema.org. (2024). Person A Schema.org Type. Retrieved
11.03.2024 from https://schema.org/Person
Schmidt, M., Hahn, V., Altrock, B., Gerling, T., Gerber, I.
C., Weltmann, K.-D., & von Woedtke, T. (2019).
Plasma-Activation of Larger Liquid Volumes by an
Inductively-Limited Discharge for Antimicrobial
Purposes. Applied Sciences, 9(10).
https://doi.org/10.3390/app9102150
Sommer, M. C., Balazinski, M., Rataj, R., Wenske, S.,
Kolb, J. F., & Zocher, K. (2021). Assessment of
Phycocyanin Extraction from Cyanidium caldarium by
Spark Discharges, Compared to Freeze-Thaw Cycles,
Sonication, and Pulsed Electric Fields.
Microorganisms, 9(7). https://doi.org/10.3390/micro
organisms9071452
Wagner, R., Chaerony Siffa, I., Becker, M. M., &
Waltemath, D. (2023). Design of metadata schemas for
ion chromatography in applied plasma sciences
[Conference Presentation]. https://doi.org/10.5281
/zenodo.10216005
Wagner, R., Weihe, T., Winter, H., Weit, C., Ehlbeck, J., &
Schnabel, U. (2023). Reducing Storage Losses of
Organic Apples by Plasma Processed Air (PPA).
Applied Sciences, 13(23). https://doi.org/10.
3390/app132312654
Webber, J. (2012). A programmatic introduction to Neo4j
Proceedings of the 3rd annual conference on Systems,
programming, and applications: software for humanity,
Tucson, Arizona, USA. https://doi.org/10.1145/
2384716.2384777
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J.,
Appleton, G., Axton, M., Baak, A., Blomberg, N.,
Boiten, J.-W., da Silva Santos, L. B., Bourne, P. E.,
Bouwman, J., Brookes, A. J., Clark, T., Crosas, M.,
Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T.,
Finkers, R., Mons, B. (2016). The FAIR Guiding
Principles for scientific data management and
stewardship. Scientific Data, 3(1), 160018.
https://doi.org/10.1038/sdata.2016.18
Towards FAIR Data Workflows for Multidisciplinary Science: Ongoing Endeavors and Future Perspectives in Plasma Technology
477
