
A Lightweight, Computation-Efficient CNN Framework for an
Optimization-Driven Detection of Maize Crop Disease
Shahinza Manzoor
1 a
, Muhammad Rizwan Mughal
2 b
, Syed Ali Irtaza
1 c
, Saif ul Islam
3 d
and Jalil Boudjadar
4 e
1
Department of Computer Sciences, Institute of Space Technology, Pakistan
2
Department of Electrical and Computer Engineering, Sultan Qaboos University, Oman
3
WMG, The University of Warwick, U.K.
4
Department of Electrical and Computer Engineering, Aarhus University, Denmark
Keywords:
Convolutional Neural Networks, Computation Efficiency, Optimization, Crop Disease Detection.
Abstract:
Detecting and mitigating crop diseases can prevent significant yield losses and economic damage. How-
ever, most state-of-the-art solutions can be expensive computation-wise. This paper presents an efficient
Lightweight multi-layer convolutional neural network (ML-CNN) to identify maize crop diseases. The pro-
posed model aims to improve disease identification accuracy and reduce computational costs. The model was
optimized by adjusting parameters, setting convolutional layers, changing the combinations of the pooling
layer, and adding dropout layers. By optimizing the model architecture, we create a software tool that can
be deployed in resource-limited environments, an ideal choice for deployment on embedded platforms. The
PlantVillage dataset was used to train and test the model implementation, including images of healthy and
two disease-affected leaves. The performance of the proposed model was compared with pre-trained models
such as InceptionV3, VGG16, VGG19, and ResNet50. The analysis results show that the proposed model im-
proved identification accuracy by 16.32%, 1.48%, 1.28%, and 2.26%, respectively. Additionally, the proposed
model achieved identification accuracy of 99.60% on the training data and 98.16% on the testing data and also
reduced iteration convergences and computational costs.
1 INTRODUCTION
Most farmers in the farming industry have small to
medium land for crops. Therefore, they rely heavily
on the quality of crop yield (FAO, 2020). However,
regional environmental conditions, crop diseases and
other factors influence crop yield (Zimmermann et al.,
2017). Due to a lack of resources, small-to-medium
farmers cannot use high-quality fertilizers to increase
crop yield. Farmers frequently use non-technological
methods to detect disease, making it challenging to
assess its severity (Al Bashish et al., 2010). As a re-
sult, a solution that can provide a farmer with authen-
tic, accurate, and timely agro advice at a low cost is of
capital interest. A farmer typically seeks agronomic
advice on disease or pests by calling an expert and
describing visible symptoms. Maize is a member of
a
https://orcid.org/0009-0001-1432-7675
b
https://orcid.org/0000-0002-0660-2761
c
https://orcid.org/0000-0001-5979-4448
d
https://orcid.org/0000-0002-9546-4195
e
https://orcid.org/0000-0003-1442-4907
the Gramineae family, which ranks third in terms of
overall yield and cultivated area after wheat and rice
(Kaur et al., 2020); with a productivity of 5.82t/ha, the
growing global yield was approximately 1.17 billion
MT in 2020 (FAO, 2020).
The literature has extensively utilized deep learn-
ing (DL), or machine learning, as a successful means
of identifying plant diseases (Mohanty et al., 2016;
Olawuyi and Viriri, 2022; Divyanth et al., 2023;
Tirkey et al., 2023; Jasrotia et al., 2023; Chauhan
et al., 2022; Haque et al., 2023; Karlekar and Seal,
2020; Vallabhajosyula et al., 2022; Ji et al., 2020;
Manzoor et al., 2023).
However, due to data disparities, identifying an
efficient DL architecture with optimal parameters
and classification functions is always a difficult task
(Tirkey et al., 2023; Uchida et al., 2016). Moreover,
most of the proposed studies in the literature, to pro-
vide accurate estimation and classification of the crop
health state, rely on the processing of massive data
(graphical images) using deep neural networks and
machine learning models (Yang et al., 2023; Esgario
et al., 2020; Demilie, 2024) to achieve high accuracy,
Manzoor, S., Mughal, M., Irtaza, S., Islam, S. and Boudjadar, J.
A Lightweight, Computation-Efficient CNN Framework for an Optimization-Driven Detection of Maize Crop Disease.
DOI: 10.5220/0012836900003753
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 19th International Conference on Software Technologies (ICSOFT 2024), pages 271-282
ISBN: 978-989-758-706-1; ISSN: 2184-2833
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
271

making the computation cost one of the barriers to
adopt and deploy such solutions on resource-limited
computation systems such as embedded platforms
(Barbedo, 2016; Demilie, 2024; Jensen et al., 2023)
and consumer devices such mobile phones (Waheed
et al., 2023).
This paper proposes multi-layer convolutional
neural networks (ML-CNN) to identify maize crop
diseases. The proposed ML-CNN is designed to
have fewer parameters, less memory usage, and lower
computational cost than existing models while main-
taining high accuracy. Our model is designed to learn
relevant features directly from the input images with-
out relying on pre-trained architectures. By optimiz-
ing the model architecture, we designed and imple-
mented a software tool to identify and classify maize
crop state of health, deployable in resource-limited
environments.
The main contributions of this paper are as fol-
lows:
• An efficient multilayer convolutional neural
network-based model is proposed to identify
maize crop disease. This model can accurately
identify maize crop diseases and reduce iteration
convergences in a complex environment.
• The proposed model achieves a high identifica-
tion accuracy compared to pre-trained CNN mod-
els and architectures.
• The number of parameters in the proposed model
is optimal compared to state-of-the-art models.
• The implementation of the proposed ML-CNN
significantly reduces the computational and mem-
ory resources required for crop disease identifica-
tion, making it practical for use in real-world em-
bedded systems.
• Thorough experimental analysis and comparison
to the state of the art for validation.
The rest of the paper is structured as follows: Sec-
tion 2 is a backrgound. Section 3 cites relevant related
work. Section 4 depicts the architecture and modules
of the proposed ML-CNN model. Section 5 describes
the dataset and processing methods we utilized in this
paper. Section 6 discusses the results and comparison,
whereas Section 7 concludes the paper.
2 BACKGROUND
Convolutional neural network (CNN) (Saleem et al.,
2022) is a powerful deep learning model for computer
vision applications including image classification, ob-
ject detection, and recognition. In general, the main
layers of CNN are convolutional, pooling, ReLu, and
fully connected layers. Some variants of CNN, such
as InceptionV3, VGG19, VGG16, and ResNet50, are
given below, along with the proposed model for dis-
ease identification in maize crops.
InceptionV3 Model. Inception models are the
types of CNN designed mainly for image classifica-
tion. Google develops these models through differ-
ent versions (V1, V2, V3), where each model opti-
mizes on the previous architecture. It comprises in-
ception blocks, and InceptionV3 is a pre-trained 48-
layer model trained on millions of ImageNet dataset
images with a 224x224 input size. The model extracts
general features and classifies images with new fully
connected layers of 256 and 128 units size and the
softmax activation function in the output with three
classes for classification.
VGG-16 Model. VGG16 is a 16-layer pre-trained
CNN-based model used for image classification.
There are 13 convolutional layers and three fully con-
nected layers. It features a unique architecture with
a small 3x3 filter size and stride of 1 in the convolu-
tional layers and a 2x2 max-pooling layer with stride
2. The model uses 64, 128, 256, and 512 filters in the
first to fifth convolutional layers, followed by three
fully Connected (FC) layers with 4096 neurons each.
This model represents a baseline for our paper, where
the first two FC layers are to contain 256 and 128 units
of neurons. In contrast, the output layer has a soft-
max activation function for classifying into a speci-
fied number of classes.
VGG-19 Model. VGG19 model is a 19-layer vari-
ation of the VGG model, including 16 convolutional
layers, 5 Maxpooling layers, 3 FC layers, and an out-
put layer. The model uses a ReLu activation func-
tion and a 3x3 kernel with a 1-pixel stride to classify
images effectively. The first two layers comprise 64
filters with a 3x3 kernel and stride 1. Max-pooling
layers with a 2x2 window size and stride two are used
to reduce the image dimensions. Additional convo-
lution layers with 128 and 256 filters are used, with
a final FC layer flattening the volume to 7x7x512 and
using 256 and 128 neurons with an activation function
softmax in the final layer.
ResNet50 Model. ResNet50 is a 50-layer CNN
model designed to resolve the vanishing gradient
problem in deep networks through skip connections.
The ResNet50 architecture uses a bottleneck building
block and a stack of three layers, making it compu-
tationally more efficient. The model takes a 224x224
ICSOFT 2024 - 19th International Conference on Software Technologies
272

image as input, and was successfully used on a maize
crop dataset to classify diseased and healthy leaves.
3 RELATED WORK
Classification remains the primary focus of DL-
based plant disease identification in its early stages.
Olawuyi et al. (Olawuyi and Viriri, 2022) used
deep learning and CNNs for detecting and classify-
ing crop (corn and potato) diseases using a pre-trained
resnet50 model. The authors’ model achieved an ac-
curacy of 98.0%. In a similar way, Divyanth et al.
(Divyanth et al., 2023) presented a two-stage deep
learning approach that can precisely identify and es-
timate the severity of three corn diseases using a cus-
tom dataset. The proposed approach uses CNNs for
identification and preprocessing techniques such as
CLAHE and RGB to HSV conversion. The model
achieves an accuracy of 96.76% on the plant village
maize crop dataset.
Chauhan et al. (Chauhan et al., 2022) high-
lighted the challenges of detecting crop diseases in
India, especially for smallholder farmers. They de-
veloped a low-cost solution utilizing feature extrac-
tion using RegNet, dimensionality reduction using
Kernel-PCA, and XGBoost classification. Results in-
dicate an accuracy of 96.74%, demonstrating intelli-
gent systems’ potential to benefit smallholder farm-
ers. Yang et al. (Yang et al., 2023) proposed a solu-
tion, Maize-YOLO, to detect maize pests in real time
with high precision. The solution utilizes YOLOv7
as a backbone network and improves accuracy and
detection speed by integrating CSPResNeXt-50 and
VoVGSCSP modules. When evaluated on a compre-
hensive pest dataset, Maize-YOLO achieved 76.3%
mean average precision (mAP) and 77.3% recall. Us-
ing the same dataset with 15200 images, the authors
of (Kumar et al., 2020) utilized ResNet34 to detect
plant leaf diseases and achieved 99.40% accuracy.
Gayathri et al. (Gayathri et al., 2020) used trans-
fer learning to classify tea leaf diseases using the
pre-trained model LeNet, resulting in an accuracy
of 90.23%. Using ResNet50, the authors suggested
an effective method for identifying and estimating
the degree of biotic agent-induced stress in coffee
leaves from the PlantVillage database. The proposed
method accurately estimated biotic stress at 95.24%
and severity at 86.51% (Esgario et al., 2020).
Huang et al. (Huang et al., 2023) proposed a fully
convolutional switchable normalization dual path net-
works model to identify and detect tomato leaf dis-
eases. The model combines an FCN algorithm based
on the VGG-16 model to segment the target crop im-
ages and an enhanced DPN model to extract the fea-
tures of the crop. Resnet and DesNet layers are com-
bined and adaptive parameters are optimized to op-
timize the network’s versatility for different diseases
and speed of training, achieving thus an accuracy of
97.59%.
The article (Arun and Umamaheswari, 2023)
presents a novel approach to identify and categorize
plant leaf diseases. The proposed method utilizes an
advanced mobile network-based CNN (OMNCNN)
that optimizes detection by incorporating several key
stages, including preprocessing, segmentation, fea-
ture extraction and classification. The experimental
results demonstrate that the OMNCNN model sur-
passes the current state-of-the-art techniques, achiev-
ing a precision rate of 0.985, a recall rate of 0.9892,
an accuracy rate of 0.987.
The authors of (Ramcharan et al., 2019) devel-
oped a transfer-learning solution to identify three dis-
eases and two pests damaging cassava plants, deploy-
able on resource-constrained environments such as
smart phones. Although the solution is computation-
efficient, the achieved accuracy of 80.6% is rather low
compared to the state of the art.
Despite the potential benefits of using machine
learning and deep learning algorithms for maize crop
disease identification, there is still a need for more
efficient and resource-friendly models that can be im-
plemented in low-resource settings. While some ex-
isting models have achieved good accuracy, they often
require large amounts of computational power, mem-
ory, and data storage, making them impractical for use
in resource-constrained environments.
This paper proposes a computation-efficient
model, in alignment with (Arun and Umamaheswari,
2023), to identify crop diseases through features ex-
traction, classification and parameters optimization.
The proposed model achieves a high accuracy and
moderate computation cost compared to the state of
the art models.
4 PROPOSED MULTILAYER CNN
MODEL
Instead of manually extracting features, CNNs can
learn more advanced features from an input im-
age. The performance of traditional feature extrac-
tion methods is inferior to that of automatic feature
extraction (Liu et al., 2020). This section presents an
efficient and effective ML-CNN-based model archi-
tecture, as shown in Figure 1, for features extraction
and classification of maize crop images. The pro-
posed ML-CNN comprises a five-level and 17-layer
A Lightweight, Computation-Efficient CNN Framework for an Optimization-Driven Detection of Maize Crop Disease
273
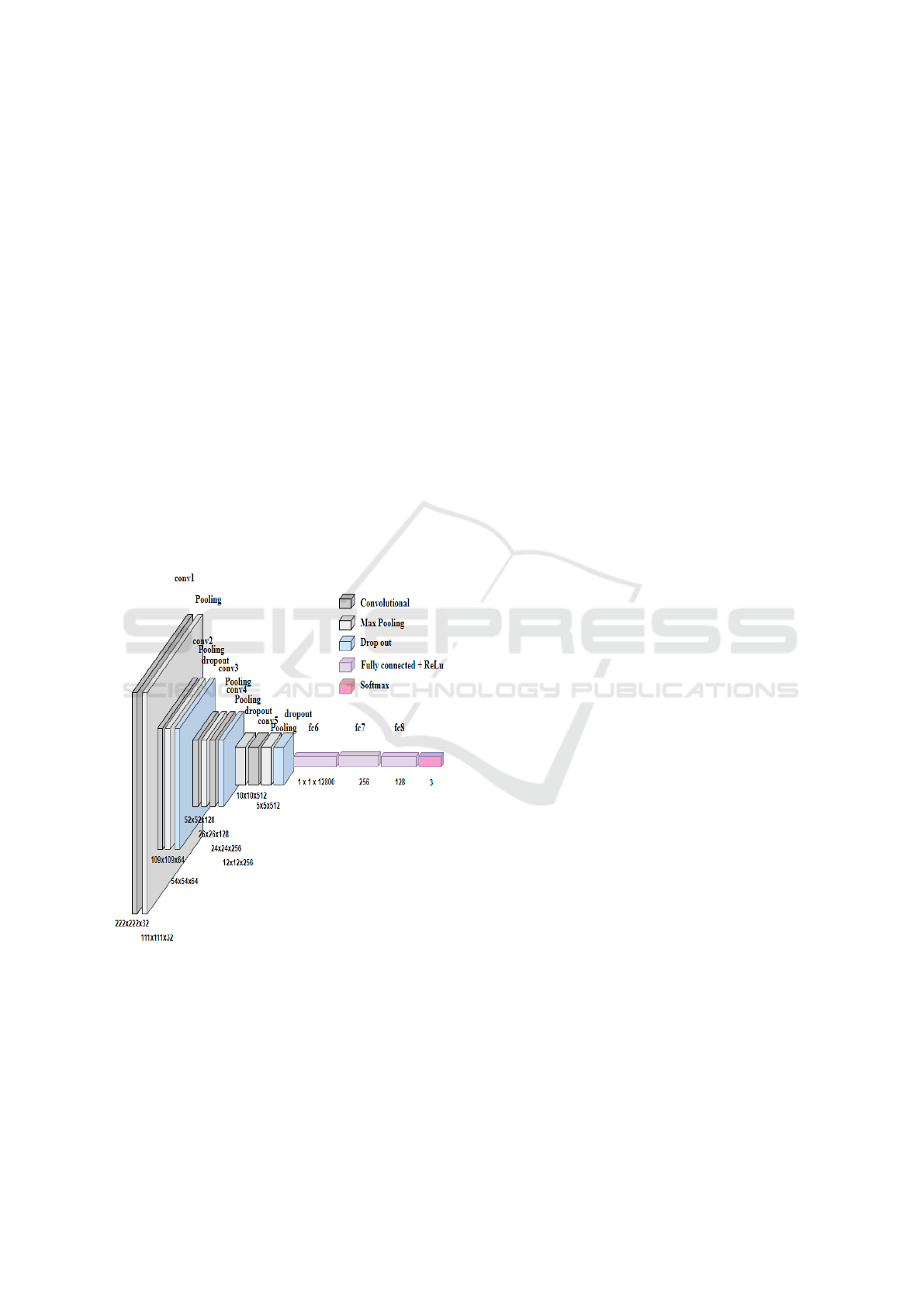
model where convolutional layer is the most impor-
tant layer of the network used for feature extraction.
Our ML-CNN adopts VGGNet with smaller 3x3 con-
volutional filters instead of larger convolutional filters
such as 5x5 and 7x7. This is because a smaller fil-
ter with fewer parameters will have the same effect
as a larger one. As a result, while maintaining high
accuracy, the model enables faster features extraction
because the reduced number of parameters.
Assuming the model’s input layer m−1, input fea-
ture map Y
m−1
, convolutional kernel Ck
m
, the output
of the convolutional layer is Om and bias b
m
. In the
convolutional layer, the output is calculated by con-
volving the input feature map with the convolution
kernel and adding bias, as shown in the equations
(1) and (2). The output is passed into the next layer,
which is the max-pooling layer.
O
m
u,v
=
∞
∑
i=−∞
∞
∑
j=−∞
Y
m−1
i+u, j+v
.Ck
m
.y(i, j) + b
m
(1)
y(i, j) =
(
1 0 ≤ i, j ≤ n
0 otherwise
(2)
Figure 1: Proposed ML-CNN Architecture.
The first convolutional layer has 32 filters of size
3 x 3, and one max-pooling layer with a window of
2x2 size. This represents the first level of the ML-
CNN network. The second convolutional layer with
64 3x3-sized filters, one max-pooling layer, and one
dropout layer with 0.2% forms the second level of the
network. The third level is composed of a convolu-
tional layer, having 128 filters of size 3x3, and one
max-pooling layer having a window of size 2x2. The
fourth convolutional layer with 256 3x3 filters, one
max-pooling layer, and one more layer of dropout
with 0.2% has been added to form the fourth level
of the ML-CNN network. The fifth level of the net-
work consists of one more convolutional layer with
512 filters with 3x3 size, one 2x2 max-pooling layer
and another dropout layer with 0.5% of dropout of
neurons. A flattening layer (fc6) converts the input
data from the convolution layer (conv5) into a vec-
tor. This process is often referred to as ”flattening”
the data. Thereafter, a dense layer (fc7) with 256 neu-
ron, ReLu as an activation function is used for lin-
ear transformation so that when the outcome is below
0, ReLu does not activate the Neurons which reduces
the computations. Finally, another denser layer (fc8)
with 128 neurons and ReLu as an activation function
has been added following the application of a dropout
layer with 0.2%. The last layer applies Softmax with
three neurons, as we have three classes for classifica-
tion. The probabilities of every class and target class
are computed using the softmax activation function,
which ranges from 0 to 1.
The target class for the given input is then deter-
mined using the cascade of the 5 levels and a sparse
categorical cross-entropy as a loss estimation func-
tion. The model has been implemented with a batch
size of 32, an input image size of 224x224, an ac-
tivation function referred to as ”ReLU” at the dense
layer, a function referred to as ”softmax” at the clas-
sification layer, an optimizer referred to as ”Adam”
keeping a learning rate of 0.0001, and filter size of
3x3 at each convolutional layer.
Current crop disease image identification mainly
involves the use of pre-trained highly-parameterized
CNNs. However, these fine-tuned networks tend to
have high complexity due to many parameters, as they
are trained on a large dataset containing a lot of infor-
mation, leading to high bias and low accuracy. This
challenge is tackled in our ML-CNN by having a sim-
pler structure, lower complexity and fewer parameters
as it is trained on a task-specific dataset. Furthermore,
using 3x3 convolutional kernels increases the recep-
tive field of view and decreases the number of param-
eters in the network, which leads to low bias and high
accuracy. This has led to achieve the highest accuracy
level at 31 epochs. Table 1 shows the hyperparameter
of the proposed model.
Convolutional Layers. The primary method for ex-
tracting features from input images is convolution. A
convolution window can be mapped onto a 2-D image
to calculate 2-dimensional convolution and obtain the
corresponding convolution value by multiplying the
ICSOFT 2024 - 19th International Conference on Software Technologies
274
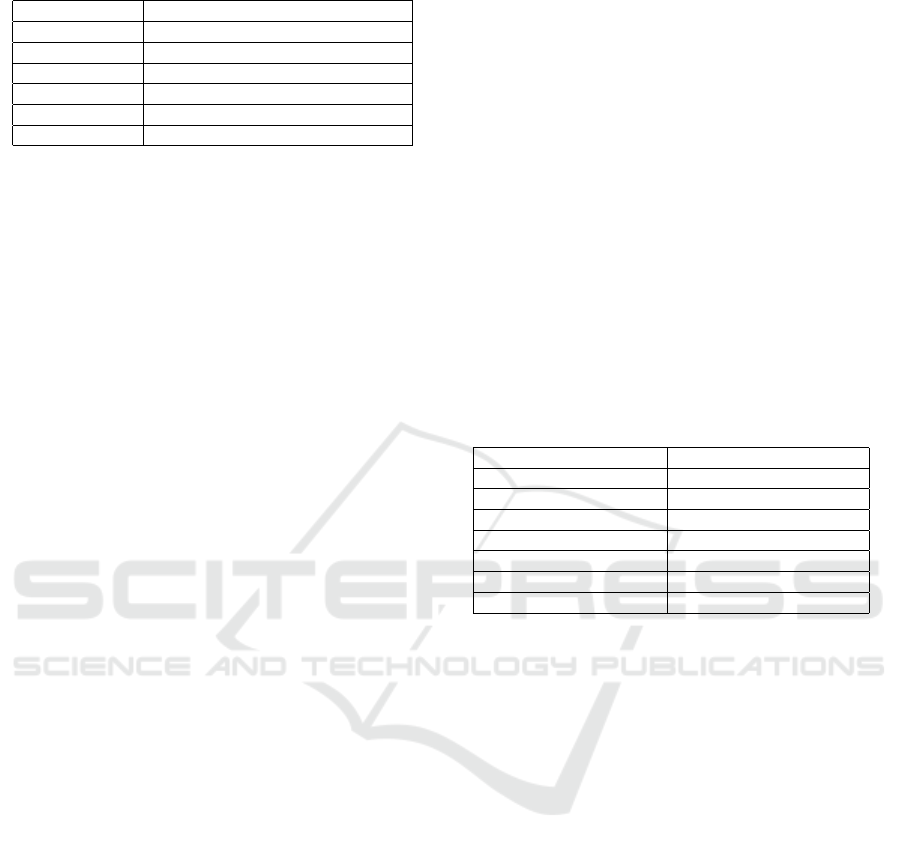
Table 1: Hyper-parameter for Proposed ML-CNN.
Dataset 7:2:1 ratio for train, valid, and test
Pre-processing Resizing at 224x224 pixels
Learning rate 0.0001
Epochs 200
Optimizer ADAM
Batch size 32
Loss Function Sparse categorical cross-entropy
input with a convolution filter (kernel). The convo-
lution outcome is a feature map having a shape com-
puted using Equation 3.
(n + 2p − f + 1)/s ∗ (n + 2p − f + 1)/s ∗ 3 (3)
Where n represents the input image size, p shows
padding, f means the size of the filter, and s is stride.
3 represents the three channels “RGB.”
Max-Pooling Layers. Max pooling is a down-
sampling operation that reduces computational costs
and enhances spatial invariance in an image by select-
ing the maximum value among the elements covered
by the kernel. It summarizes the features produced by
the convolution layer.
Dropout Layer. The primary objective of employ-
ing a dropout layer is to enhance the trained model’s
prediction performance. During the training phase,
ignoring neurons of a randomly selected set is re-
ferred to as a dropout. All neurons are used during
the testing phase but are scaled by factor p.
Fully Connected Layers. A fully connected layer
transforms the outputs of the preceding layer into a
single vector, ”flattening”, that can serve as an in-
put for the subsequent stage in the ML-CNN. The
last layer of our ML-CNN uses the softmax activa-
tion function to calculate the probability of each class
from the hidden layers. Our classification model can
have multiple fully connected layers added depend-
ing on how deep the architecture can be, however this
would require larger and diverse dataset for training
and high computation cost for testing.
ReLU Activation Function. It refers to the Recti-
fied linear unit and primarily implemented within the
neural network’s hidden layers. ReLU function ac-
tivates multiple layers of neurons to back-propagate
the errors. We chose the ReLu function because it
requires fewer mathematical operations compared to
Tanh and Sigmoid, so that to achieve less computa-
tion cost. Furthermore, in our ML-CNN, the network
is sparse with only a few neurons activated at once
depending on the linear transformation, or no neuron
is activated if the linear transformation is below or
equal 0 as shown in Equation (4), making it efficient
and simple to compute.
A(x) = max(0,x) (4)
5 DATASET AND PROCESSING
It is essential to collect a large number of plant images
to achieve a highly accurate classification of maize
crop diseases. For the proposed ML-CNN, the maize
crop dataset is composed of images from the PlantVil-
lage database (GHOSE, 2022) and OSF (Wiesner-
Hanks and Brahimi, 2022). Table 2 shows the hard-
ware configuration and software resources used for
the training.
Table 2: HW and SW resources used for training.
Units Parameters
System NVIDIA-SMI 460.32.03
Graphics processor unit Tesla T4
RAM 32GB
Environment Google Colab pro
Framework Keras with TensorFlow
Operating system Windows 10
Programming language Python
5.1 Dataset
In general, datasets on agricultural concerns are not
widely accessible, and real-time images are a key con-
cern (Redmon and Farhadi, 2018). We acquired the
maize crop dataset from PlantVillage (GHOSE, 2022)
and OSF (Wiesner-Hanks and Brahimi, 2022). Each
image is taken on a solid background, with a single
leaf in a controlled environment. The dataset consists
of two disease classes along with the healthy class.
Two important agricultural diseases that affect the
maize crop, namely Northern leaf blight (NLB) and
Common Rust, were considered. Most commonly,
plants infected with common rust produce brown pus-
tules on the surface of their leaves. The infection also
spreads to the sheaths and other parts of the plant.
Lesions of northern corn leaf blight first appear on
the plant’s lower parts, then spread to the plant’s en-
tire leaves, where they turn a pale gray as they grow.
The dataset description is given in Table 3. We divide
the dataset into three sets of data: train set, validation
set and test set, which represented 70% and 30%, and
10%, respectively of the total images. Some sampled
images from the dataset are shown in Figure 2.
A Lightweight, Computation-Efficient CNN Framework for an Optimization-Driven Detection of Maize Crop Disease
275

Table 3: Maize crop dataset description.
Category No of Samples
Northern leaf blight (NLB) 1146
Common Rust 1306
Healthy 1162
(a) NLB (b) Common Rust (c) Healthy
Figure 2: Sample images from the dataset.
5.2 Data Processing
We performed image processing steps on the
dataset images, including image augmentation, pre-
processing and resizing.
Image Augmentation. Image augmentation refers
to transforming input image samples by minor rota-
tions, reflections, flips zooming, scaling and shifting.
As a result, data augmentation enhances the dataset
by increasing the number of training samples which
can significantly improve deep CNN’s efficiency.
• Rotation: it rotates a training image at random
through different angles.
• Shear: it adjust the shearing range. We adopted a
shearing of 0.2.
• Brightness: it aids the model in adjusting to
changes in lighting by feeding images of varying
brightness during training.
• Flip: an image can be flipped at different posi-
tions.
Image Pre-Processing. In image pre-processing,
digital images are processed before being fed into
computer vision algorithms for further processing.
The pre-processing of images improves the quality of
input data, enhances particular image features, or ex-
tracts meaningful information from images. In our
case, the pre-processing reduces the blurriness and
noise in input images.
As part of the pre-processing, to speed up the con-
volution and classification operations, we first resize
the input samples to 224 x 224 pixels.
6 RESULTS AND DISCUSSION
This section presents the analysis results of the de-
tection of maize crop diseases using our ML-CNN
model. The dataset was divided into three classes:
healthy and two diseases with a total number of 6940
input image samples (after augmentation).
6.1 Accuracy
Table 4 depicts a comparison of the train accuracy,
test accuracy, train loss and test loss of state of the
art pre-trained deep learning models such as Incep-
tionV3, VGG16, VGG19, and ResNet50 with our
ML-CNN model. The result demonstrates that our
ML-CNN model outperforms the conventional clas-
sifiers in terms of identification accuracy. Further-
more, the proposed ML-CNN improved identification
accuracy by 16.32%, 1.48%, 1.28%, and 2.26%, re-
spectively. One can see as well that our ML-CNN
achieves the best testing accuracy of 98.16% while
exhibiting lower training, and testing loss of 0.0097%
and 0.0943%.
Table 4: Training accuracy, test accuracy, training loss, and
test loss of InceptionV3, VGG16, VGG19, ResNet50, and
Proposed ML-CNN.
Algorithm Train Acc Train Loss Test Accu Test Loss
InceptionV3 88.75% 18.56 81.84% 24.5
VGG16 98.75% 0.21 96.68% 1.1
VGG19 98.12% 0.47 96.88% 1.6
ResNet50 98.75% 0.20 95.90% 1.7
ML-CNN 99.60% 0.009 98.16% 0.09
Accuracy and loss of both training and test for In-
ceptionV3, VGG16, VGG19, ResNet50 and proposed
ML-CNN are depicted in Figure 3, Figure 4, Figure 5,
Figure 6 and Figure 7 respectively.
The x-axis labeled with the number of epochs can
be seen as the number of times the algorithm will
learn from the complete dataset. The y-axis shows
the models accuracy. Our proposed model achieves
better testing accuracy of 98.16% than other models
such as InceptionV3, VGG16, VGG19 and ResNet50,
gaining 81.84%, 96.68%, 96.88%, 95.90% test accu-
racy respectively. Meanwhile, our ML-CNN achieved
a minimum loss of 0.0943%.
Figure 8 presents a training accuracy compar-
ison between InceptionV3, VGG16, VGG19, and,
ResNet50 with our proposed ML-CNN algorithm.
Similarly, the testing accuracy of InceptionV3,
VGG16, VGG19, and ResNet50 compared to the pro-
posed ML-CNN algorithm is depicted in Figure 9.
One can see from both figures that the proposed ML-
CNN outperforms the considered state of the art alter-
ICSOFT 2024 - 19th International Conference on Software Technologies
276
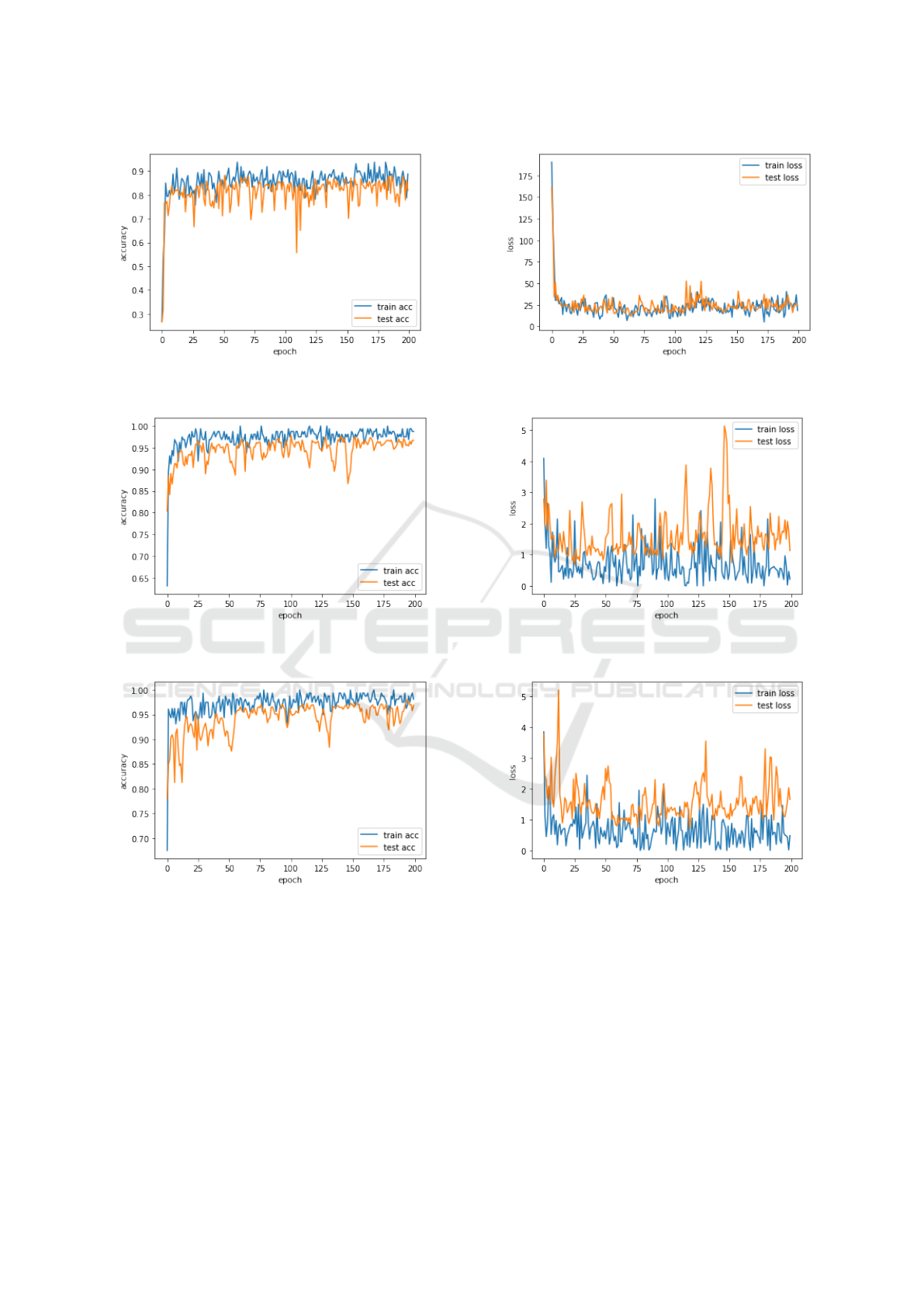
(a) InceptionV3 Accuracy (b) InceptionV3 Loss
Figure 3: Training and Test Accuracy of InceptionV3.
(a) VGG16 Accuracy (b) VGG16 Loss
Figure 4: Training and Test Accuracy of VGG16.
(a) VGG19 Accuracy (b) VGG19 Loss
Figure 5: Training and Test Accuracy of VGG19.
natives for both training and testing accuracy.
6.2 Precision, Recall, and F1-Score
Classification tasks use precision, recall and F1-score
as metrics for evaluation. True positive predictions
are measured by precision among all positive predic-
tions, while the recall measures actual positive pre-
dictions.
F1-score is a balanced method of calculating pre-
cision and recall. Macro averaging involves calculat-
ing each class’s metric separately and unweighting the
mean. The weighted average takes the average of all
classes, where the weight is the number of samples in
each class. It can be useful when the different classes
have different sizes, and one needs to assign much
more weight to larger classes.
Figure 10 shows precision, recall, F1-score, macro
A Lightweight, Computation-Efficient CNN Framework for an Optimization-Driven Detection of Maize Crop Disease
277
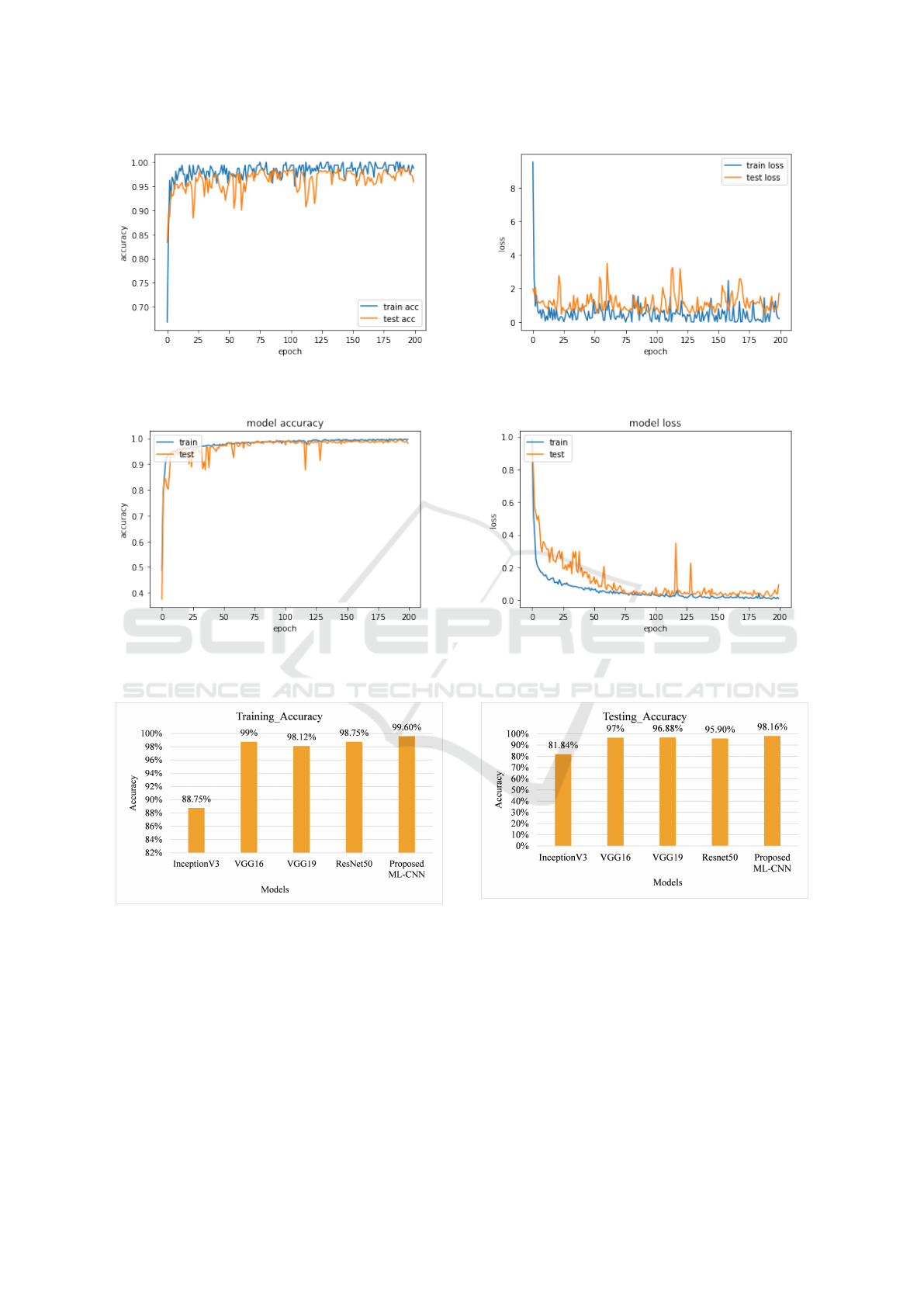
(a) ResNet50 Accuracy (b) ResNet50 Loss
Figure 6: Training and Test Accuracy of ResNet50.
(a) ML-CNN Accuracy (b) ML-CNN Loss
Figure 7: Training and Test Accuracy of the Proposed ML-CNN.
Figure 8: Training Accuracy Comparison of InceptionV3,
VGG16, VGG19, and ResNet50 with our proposed ML-
CNN.
average value, and weighted average of each class for
InceptionV3, VGG16, VGG19, ResNet50, and pro-
posed ML-CNN.
Clearly, the proposed ML-CNN model outper-
forms the state of the art models evaluated in this
study with exceptional scores in precision, recall, F1-
score, macro average, and weighted average.
Figure 9: Testing Accuracy Comparison of InceptionV3,
VGG16, VGG19, and ResNet50 with our proposed ML-
CNN.
6.3 Confusion Matrix
The confusion matrix is widely used for analyzing
classification models performance. The model’s pre-
dictions are presented in a tabular format, displaying
the number of true positive predictions, true nega-
tive predictions, false positive predictions, and false
negative predictions. Figure 11 depicts the confusion
matrix for InceptionV3, VGG16, VGG19, ResNet50,
ICSOFT 2024 - 19th International Conference on Software Technologies
278
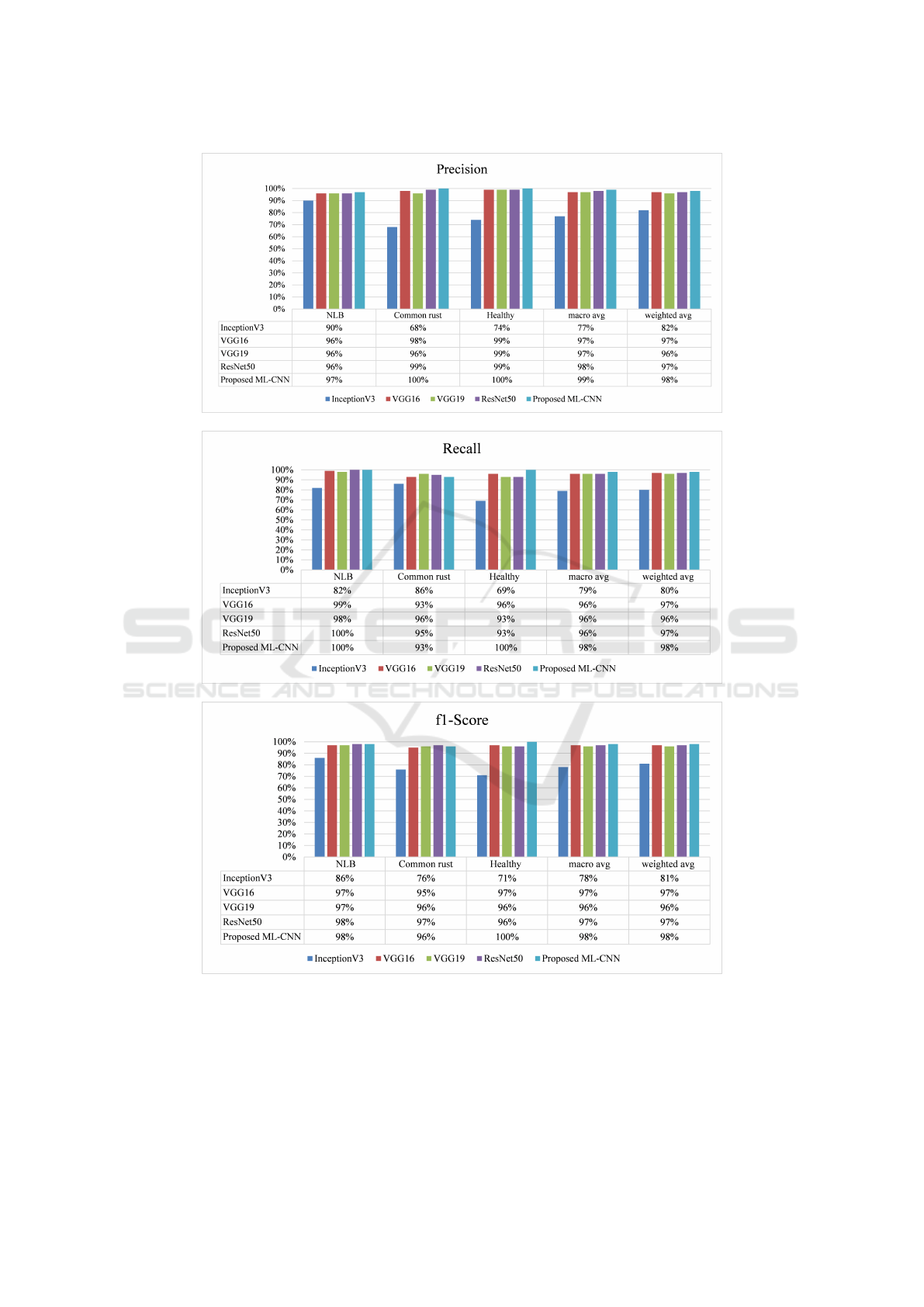
Figure 10: Precision, recall F1-score for InceptionV3, VGG16, VGG16, ResNet50 and Proposed ML-CNN.
and proposed ML-CNN.
The proposed ML-CNN shows a high level of ac-
curacy. This is evident by the concentration of accu-
racy on the diagonal- and the high prediction accuracy
for the three classes. This performance is superior to
that of the other pre-trained models, indicating that
ML-CNN offers excellent identification results.
6.4 ROC-AUC
The proposed ML-CNN was evaluated using the Re-
ceiver Operating Characteristic (ROC) function. ROC
A Lightweight, Computation-Efficient CNN Framework for an Optimization-Driven Detection of Maize Crop Disease
279
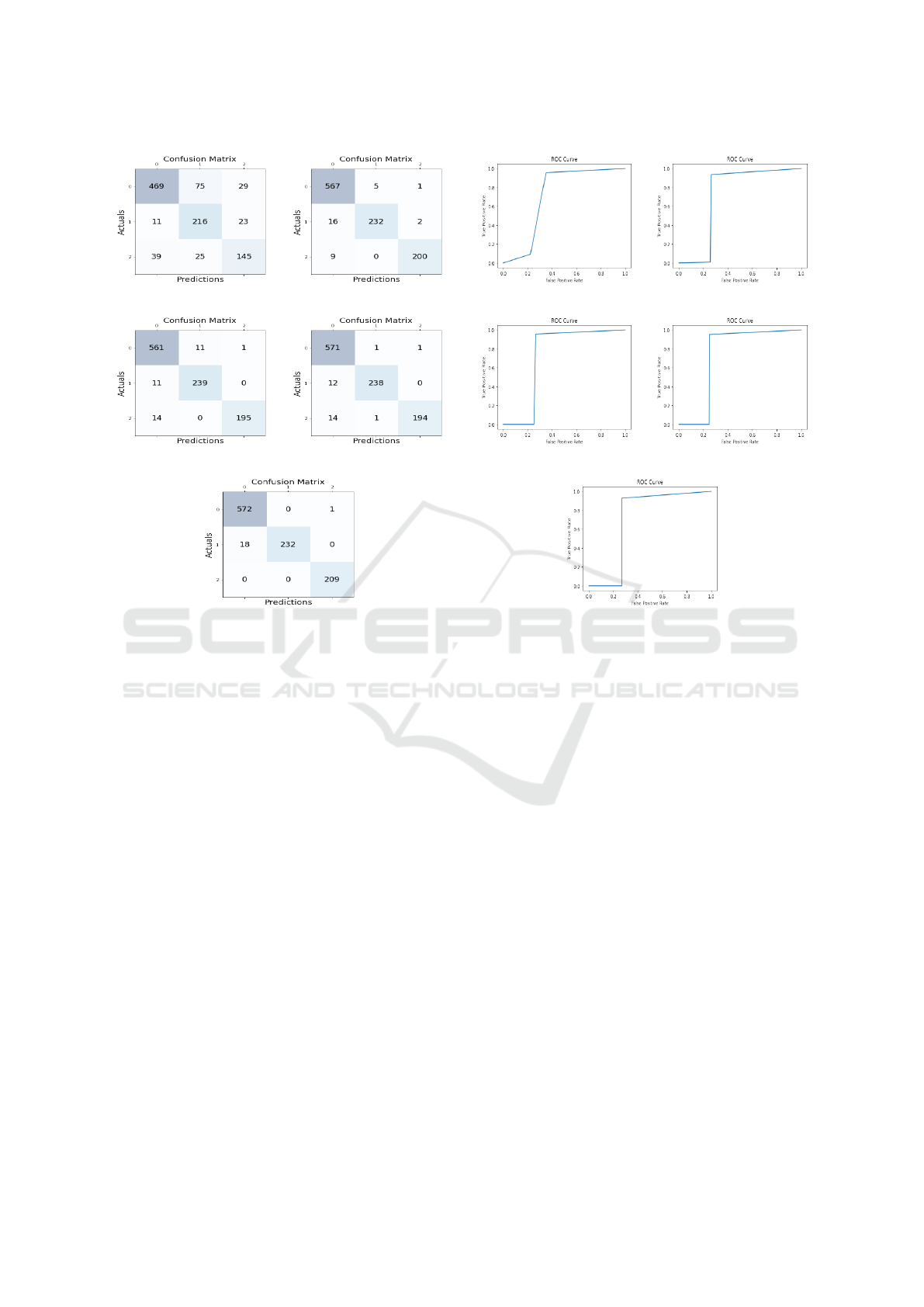
(a) InceptionV3 (b) VGG16
(c) VGG19 (d) ResNet50
(e) Proposed ML-CNN
Figure 11: Confusion Matrix of InceptionV3, VGG16,
VGG16, ResNet50 and Proposed ML-CNN.
for InceptionV3, VGG16, VGG16, ResNet50, and
Proposed ML-CNN are illustrated in Figure 12.
Figure 13 shows the area under the curve for In-
ceptionV3, VGG16, VGG19, ResNet50 and our ML-
CNN of the NLB, common rust and, healthy leaves.
According to the findings, the proposed ML-CNN
achieved an AUC of 98% for NLB, 96% for common
rust, and 100% for healthy leaves, and a strong abil-
ity to distinguish between positive and negative cases.
In general, this demonstrates the efficacy of the pro-
posed ML-CNN model in identifying and classifying
the target variable with high precision.
6.5 Feature Visualization
CNNs use raw image pixels to learn abstract concepts
and features. Activation maximization is used to show
the learned features in feature visualization. Figure
14 depicts the feature visualization when an input im-
age of Northern leaf blight is fed to the trained net-
work. The first ML-CNN layer extracts an image’s
low-level features like edges, blobs and orientation.
Features like more intricate patterns and textures are
learned in 2nd convolutional layers. In the 3rd layer,
(a) InceptionV3 (b) VGG16
(c) VGG19 (d) ResNet50
(e) Proposed ML-CNN
Figure 12: ROC curves for InceptionV3, VGG16, VGG19,
ResNet50, and Proposed ML-CNN.
the filters learn to detect more complex combinations
of edges and textures that are specific to certain ob-
jects or parts of an object. In the 4th layer, the filters
learn abstract patterns such as the parts of an object
or the presence of specific objects in the image. The
final convolutional layer learns features like entire ob-
jects. The fully connected layers learn to link activa-
tion from high-level features to predicted classes.
7 CONCLUSIONS
In this paper, an efficient model based on a deep
convolutional neural network was proposed to clas-
sify healthy and diseased maize crop leaves. A to-
tal of 5,487 training, 965 validation, and 488 test
images (after augmentation) were collected from the
PlantVillage dataset. Northern leaf blight (NLB),
Common rust, and healthy images are included in this
dataset.
Compared to the state of the art pre-trained CNN
models such as InceptionV3, VGG-16, VGG19 and
ResNet50, our ML-CNN model improved identifica-
tion accuracy by 16.32%, 1.48%, 1.28%, and 2.26%
ICSOFT 2024 - 19th International Conference on Software Technologies
280
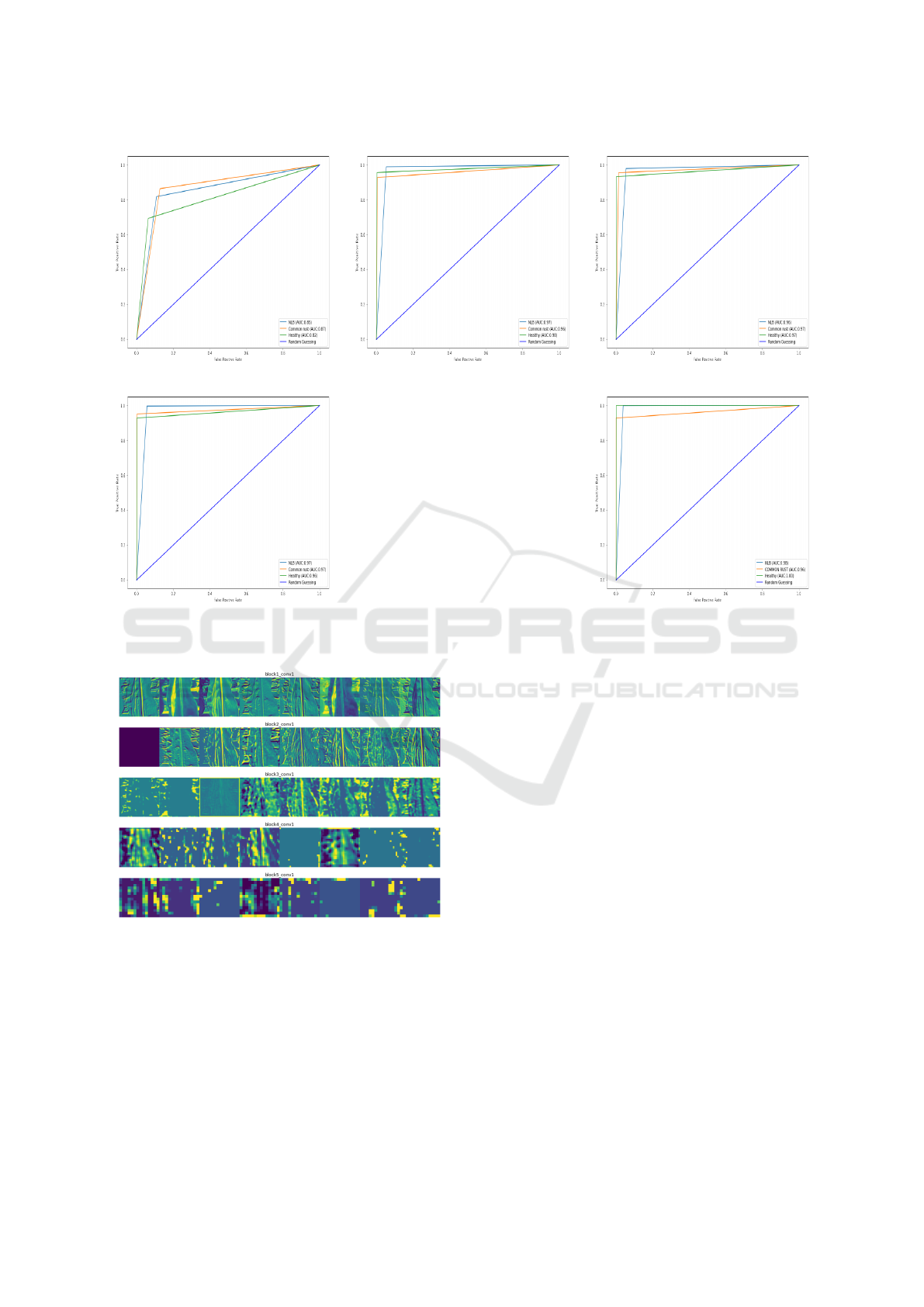
(a) InceptionV3 (b) VGG16 (c) VGG19
(d) ResNet50 (e) Proposed ML-CNN
Figure 13: AUC of InceptionV3, VGG16, VGG16, ResNet50 and Proposed ML-CNN.
Figure 14: Visual representation of each Convolution Layer.
respectively, and achieved a much better test and train
accuracy of 99.60%, and 98.16%, respectively. Glob-
ally, the proposed ML-CNN was proven to be efficient
by a large number of our experiments including pre-
cision, f1-score, recall, and AUC-ROC.
The proposed ML-CNN not only achieves high
accuracy but also significantly reduces the computa-
tional cost and memory footprint, making it a promis-
ing solution for embedded systems. This feature en-
ables the model to be utilized in devices with limited
resources, such as drones or smartphones, allowing
farmers to identify crop diseases in real time.
As a future work, we plan to extend the set of fea-
tures to be identified to detect multiple diseases on
a single maize leaf and estimate their severity. Ad-
ditionally, a user-friendly mobile application will be
developed to aid farmers in identifying crop diseases
as early as possible.
REFERENCES
Al Bashish, D., Braik, M., and Bani-Ahmad, S. (2010).
A framework for detection and classification of plant
leaf and stem diseases. In 2010 international confer-
ence on signal and image processing, pages 113–118.
IEEE.
Arun, R. A. and Umamaheswari, S. (2023). Effective multi-
crop disease detection using pruned complete concate-
nated deep learning model. Expert Systems with Ap-
plications, 213:118905.
Barbedo, J. G. A. (2016). A review on the main challenges
in automatic plant disease identification based on vis-
ible range images. Biosystems Engineering, 144:52–
60.
A Lightweight, Computation-Efficient CNN Framework for an Optimization-Driven Detection of Maize Crop Disease
281

Chauhan, T., Katkar, V., and Vaghela, K. (2022). Corn leaf
disease detection using regnet, kernelpca and xgboost
classifier. In International Conference on Advance-
ments in Smart Computing and Information Security,
pages 346–361. Springer.
Demilie, W. B. (2024). Plant disease detection and classifi-
cation techniques: a comparative study of the perfor-
mances. Journal of Big Data, 11 (5).
Divyanth, L., Ahmad, A., and Saraswat, D. (2023). A
two-stage deep-learning based segmentation model
for crop disease quantification based on corn field im-
agery. Smart Agricultural Technology, 3:100108.
Esgario, J. G., Krohling, R. A., and Ventura, J. A. (2020).
Deep learning for classification and severity estima-
tion of coffee leaf biotic stress. Computers and Elec-
tronics in Agriculture, 169:105162.
FAO (2020(accessed April 11, 2020)). Fao. https://www.
fao.org/india/fao-in-india/india-at-a-glance/en/.
Gayathri, S., Wise, D. J. W., Shamini, P. B., and Muthuku-
maran, N. (2020). Image analysis and detection of tea
leaf disease using deep learning. In 2020 International
Conference on Electronics and Sustainable Communi-
cation Systems (ICESC), pages 398–403. IEEE.
GHOSE, S. (2022). Corn or Maize Leaf Disease Dataset
— kaggle.com. https://www.kaggle.com/datasets/
smaranjitghose/corn-or-maize-leaf-disease-dataset.
[Accessed 19-04-2024].
Haque, M. A., Marwaha, S., Deb, C. K., Nigam, S., and
Arora, A. (2023). Recognition of diseases of maize
crop using deep learning models. Neural Computing
and Applications, 35(10):7407–7421.
Huang, X., Chen, A., Zhou, G., Zhang, X., Wang, J., Peng,
N., Yan, N., and Jiang, C. (2023). Tomato leaf disease
detection system based on fc-sndpn. Multimedia tools
and applications, 82(2):2121–2144.
Jasrotia, S., Yadav, J., Rajpal, N., Arora, M., and Chaud-
hary, J. (2023). Convolutional neural network based
maize plant disease identification. Procedia Computer
Science, 218:1712–1721.
Jensen, M., Jakobsen, J. T., Sharifirad, I., and Boudjadar,
J. (2023). Advanced acceleration and implementation
of convolutional neural networks on fpgas. In 2023
IEEE International Conference on High Performance
Computing and Communications HPCC.
Ji, M., Zhang, K., Wu, Q., and Deng, Z. (2020). Multi-label
learning for crop leaf diseases recognition and sever-
ity estimation based on convolutional neural networks.
Soft Computing, 24:15327–15340.
Karlekar, A. and Seal, A. (2020). Soynet: Soybean leaf
diseases classification. Computers and Electronics in
Agriculture, 172:105342.
Kaur, H., Kumar, S., Hooda, K., Gogoi, R., Bagaria, P.,
Singh, R., Mehra, R., and Kumar, A. (2020). Leaf
stripping: an alternative strategy to manage banded
leaf and sheath blight of maize. Indian Phytopathol-
ogy, 73(2):203–211.
Kumar, V., Arora, H., Sisodia, J., et al. (2020). Resnet-
based approach for detection and classification of
plant leaf diseases. In 2020 international conference
on electronics and sustainable communication sys-
tems (ICESC), pages 495–502. IEEE.
Liu, B., Ding, Z., Tian, L., He, D., Li, S., and Wang, H.
(2020). Grape leaf disease identification using im-
proved deep convolutional neural networks. Frontiers
in Plant Science, 11:1082.
Manzoor, S., Manzoor, S. H., Islam, S. u., and Boudjadar, J.
(2023). Agriscannet-18: A robust multilayer cnn for
identification of potato plant diseases. In Intelligent
Systems and Applications.
Mohanty, S. P., Hughes, D. P., and Salath
´
e, M. (2016). Us-
ing deep learning for image-based plant disease detec-
tion. Frontiers in plant science, 7:215232.
Olawuyi, O. and Viriri, S. (2022). Plant diseases detec-
tion and classification using deep transfer learning. In
Pan-African Artificial Intelligence and Smart Systems
Conference, pages 270–288. Springer.
Ramcharan, A., McCloskey, P., Baranowski, K., Mbilinyi,
N., Mrisho, L., Ndalahwa, M., Legg, J., and Hughes,
D. P. (2019). A mobile-based deep learning model for
cassava disease diagnosis. Frontiers in plant science,
10:272.
Redmon, J. and Farhadi, A. (2018). Yolov3: An incremental
improvement. arXiv preprint arXiv:1804.02767.
Saleem, R., Yuan, B., Kurugollu, F., Anjum, A., and Liu, L.
(2022). Explaining deep neural networks: A survey on
the global interpretation methods. Neurocomputing,
513:165–180.
Tirkey, D., Singh, K. K., and Tripathi, S. (2023). Per-
formance analysis of ai-based solutions for crop dis-
ease identification, detection, and classification. Smart
Agricultural Technology, 5.
Uchida, S., Ide, S., Iwana, B. K., and Zhu, A. (2016). A
further step to perfect accuracy by training cnn with
larger data. In 2016 15th International Conference on
Frontiers in Handwriting Recognition (ICFHR).
Vallabhajosyula, S., Sistla, V., and Kolli, V. K. K. (2022).
Transfer learning-based deep ensemble neural net-
work for plant leaf disease detection. Journal of Plant
Diseases and Protection, 129(3):545–558.
Waheed, H., Akram, W., Islam, S. u., Hadi, A., Boudjadar,
J., and Zafar, N. (2023). A mobile-based system for
detecting ginger leaf disorders using deep learning.
Future Internet, 15(3):86.
Wiesner-Hanks, T. and Brahimi, M. (2022). OSF — osf.io.
https://osf.io/arwmy/. [Accessed 19-04-2024].
Yang, S., Xing, Z., Wang, H., Dong, X., Gao, X., Liu, Z.,
Zhang, X., Li, S., and Zhao, Y. (2023). Maize-yolo:
a new high-precision and real-time method for maize
pest detection. Insects, 14(3):278.
Zimmermann, A., Webber, H., Zhao, G., Ewert, F., Kros, J.,
Wolf, J., Britz, W., and de Vries, W. (2017). Climate
change impacts on crop yields, land use and environ-
ment in response to crop sowing dates and thermal
time requirements. Agricultural Systems, 157:81–92.
ICSOFT 2024 - 19th International Conference on Software Technologies
282
