
Integrated Evaluation of Semantic Representation Learning, BERT, and
Generative AI for Disease Name Estimation Based on Chief Complaints
Ikuo Keshi
1,2
, Ryota Daimon
2
, Yutaka Takaoka
3,5
and Atsushi Hayashi
4,5
1
AI & IoT Center, Fukui University of Technology, 3-6-1, Gakuen, Fukui, Japan
2
Electrical, Electronic and Computer Engineering Course, Department of Applied Science and Engineering,
Fukui University of Technology, 3-6-1, Gakuen, Fukui, Japan
3
Data Science Center for Medicine and Hospital Management, Toyama University Hospital, 2630 Sugitani, Toyama, Japan
4
Department of Ophthalmology, University of Toyama, 2630 Sugitani, Toyama, Japan
5
Center for Data Science and Artificial Intelligence Research Promotion, Toyama University Hospital, 2630 Sugitani,
Toyama, Japan
Keywords:
Generative AI, Electronic Medical Record (EMR), Chief Complaints, Disease Name Estimation, Medical AI,
Medical Diagnostic Support Tool, Semantic Representation Learning, BERT, GPT-4.
Abstract:
This study compared semantic representation learning + machine learning, BERT, and GPT-4 to estimate dis-
ease names from chief complaints and evaluate their accuracy. Semantic representation learning + machine
learning showed high accuracy for chief complaints of at least 10 characters in the International Classifica-
tion of Diseases 10th Revision (ICD-10) codes middle categories, slightly surpassing BERT. For GPT-4, the
Retrieval Augmented Generation (RAG) method achieved the best performance, with a Top-5 accuracy of
84.5% when all chief complaints, including the evaluation data, were used. Additionally, the latest GPT-4o
model further improved the Top-5 accuracy to 90.0%. These results suggest the potential of these methods
as diagnostic support tools. Future work aims to enhance disease name estimation through more extensive
evaluations by experienced physicians.
1 INTRODUCTION
We developed a method for estimating disease names
based on learning semantic representations of medi-
cal terms to improve both accuracy and interpretabil-
ity (Keshi et al., 2022). While semantic representation
learning provides high interpretability for discharge
summaries, it struggles with texts with poor context,
such as a patient’s chief complaint. Therefore, we
aimed to improve the accuracy and interpretability of
disease name estimation by evaluating generative AI
techniques like GPT-4.
This study evaluated semantic representation
learning to determine the conditions of the chief com-
plaint using generative AI. We conducted a reference
evaluation using BERT models (Devlin et al., 2019;
Kawazoe et al., 2021), pretrained on Japanese clinical
texts, and Wikipedia. Finally, we used an integrated
approach to infer disease names from chief com-
plaints, applying zero-shot learning, few-shot learn-
ing, and RAG with GPT-4. We comprehensively eval-
uated these approaches’ accuracy and explored their
potential application for medical diagnosis.
This study highlights the importance of combin-
ing traditional supervised learning and generative AI
techniques to improve the accuracy of disease name
estimation, especially from minimal contextual data
like chief complaints. This combination is crucial to
address the challenges of medical diagnosis and en-
hance accuracy.
2 RELATED RESEARCH
The field of medical AI is rapidly advancing with
the application of large language models. Gen-
erative AI is being widely adopted in the medi-
cal field, and its democratization has the potential
to enhance diagnostic accuracy (Chen et al., 2024).
Google’s Med-PaLM2, fine-tuned with medical texts,
has shown high performance in the US medical li-
censing exam (Singhal et al., 2023). OpenAI’s GPT-
4 can pass the Japanese national medical exam but
still faces challenges in professional medical applica-
294
Keshi, I., Daimon, R., Takaoka, Y. and Hayashi, A.
Integrated Evaluation of Semantic Representation Learning, BERT, and Generative AI for Disease Name Estimation Based on Chief Complaints.
DOI: 10.5220/0012927100003838
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2024) - Volume 1: KDIR, pages 294-301
ISBN: 978-989-758-716-0; ISSN: 2184-3228
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
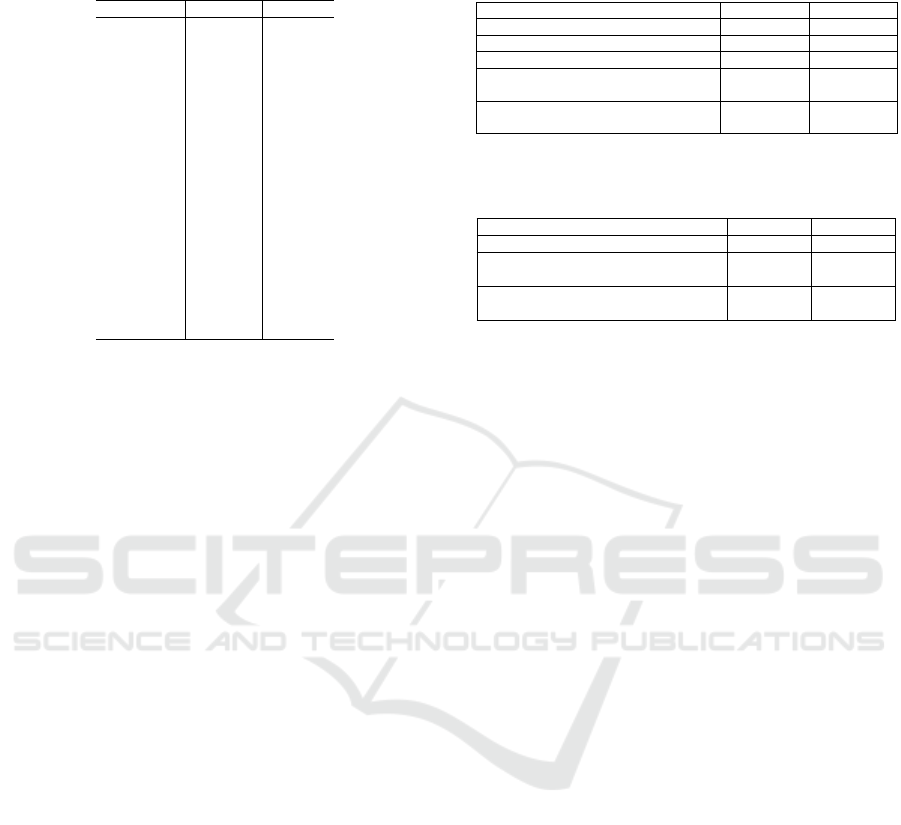
Table 1: Number of cases in the old EMR corresponding to
the top 20 ICD-10 codes in the new EMR.
ICD-10 code new EMR old EMR
C34.1 1127 210
H25.1 929 123
C61 912 2216
C34.3 893 158
C22.0 864 1501
I20.8 698 75
I35.0 690 70
I50.0 545 166
C16.2 536 231
I67.1 515 387
C25.0 503 111
C15.1 483 253
I48 483 253
C34.9 468 1579
P03.4 432 399
C56 393 1276
M48.06 373 845
H35.3 368 1060
H33.0 361 625
C20 357 343
tions (Kasai et al., 2023). In the 2022 National Med-
ical Examination for Physicians (NMLE) in Japan,
GPT-4 achieved a correct response rate of 81.5%,
significantly higher than GPT-3.5’s 42.8%, and ex-
ceeded the passing standard of 72%, showing its po-
tential to support diagnostic and therapeutic deci-
sions (Yanagita et al., 2023).
Given these advancements, this study focuses on
utilizing these models to establish evaluation criteria
for estimating disease names from chief complaints.
3 DATASET
Developing disease estimation AI models using elec-
tronic medical records faces the challenge of accuracy
drop when applied across different hospitals. This
study aims to create models with high accuracy across
two types of EMRs with different data distributions.
3.1 Progress Summary Dataset
The training data includes discharge summaries from
Toyama University Hospital (2004-2014, 94,083
cases) and the evaluation data from 2015-2019
(61,772 cases). Data cleansing involved excluding
cases with missing values, unused fields, rare disease
names (less than 0.02%), and short progress sum-
maries (less than 50 words).
Table 1 shows the number of cases in both EMRs
for the top 20 disease codes. Despite distribution dif-
ferences, the top 20 disease codes in the new EMR
appear in the old EMR, ensuring sufficient cases for
model training and evaluation.
The records include the ICD-10 code, the first 500
Table 2: The number of cases according to different chief
complaint conditions.
old EMR new EMR
Before data cleansing 94,083 cases 61,772 cases
After data cleansing 73,150 cases 48,911 cases
Subcategories with any chief complaint 35,509 cases 28,787 cases
Subcategories with chief complaints of
more than 10 characters
8,300 cases 5,876 cases
Middle categories with chief complaints
of more than 10 characters
6,766 cases 4,949 cases
Table 3: The number of cases for benchmarks focusing on
the top 20 ICD-10 codes.
old EMR new EMR
Subcategories with any chief complaint 4,205 cases 5,547 cases
Subcategories with chief complaints of
more than 10 characters
1,013 cases 1,054 cases
Middle categories with chief complaints
of more than 10 characters
1,605 cases 1,715 cases
characters of the progress summary, department, gen-
der, and age.
3.2 Chief Complaint Dataset
Chief complaints were extracted from both EMRs.
Table 2 shows the variation in case numbers under
different conditions. Table 3 presents benchmarks for
the top 20 ICD-10 codes in the new EMR.
In the chief complaint dataset, restricting the num-
ber of letters significantly reduces case numbers but
retains sufficient data for machine learning. Records
include the ICD-10 code, chief complaint, depart-
ment, gender, and age.
4 PROPOSED METHOD
We developed a model to estimate disease names from
chief complaints by extending GPT-4 using EMRs.
GPT-4 can pass the Japanese national examination for
physicians, but its performance can be improved us-
ing the chief complaint dataset from Chapter 3. This
study employs supervised learning (semantic repre-
sentation learning + machine learning) and a BERT
model pretrained on medical documents for compar-
ative validation.
4.1 Semantic Representation Learning
of Medical Terms
The semantic representation learning process (Fig-
ure 1) involves using the first 500 characters of the
progress summary. The step of obtaining a weight
vector of the progress summary includes generating a
paragraph vector (Le and Mikolov, 2014) with initial
Integrated Evaluation of Semantic Representation Learning, BERT, and Generative AI for Disease Name Estimation Based on Chief
Complaints
295
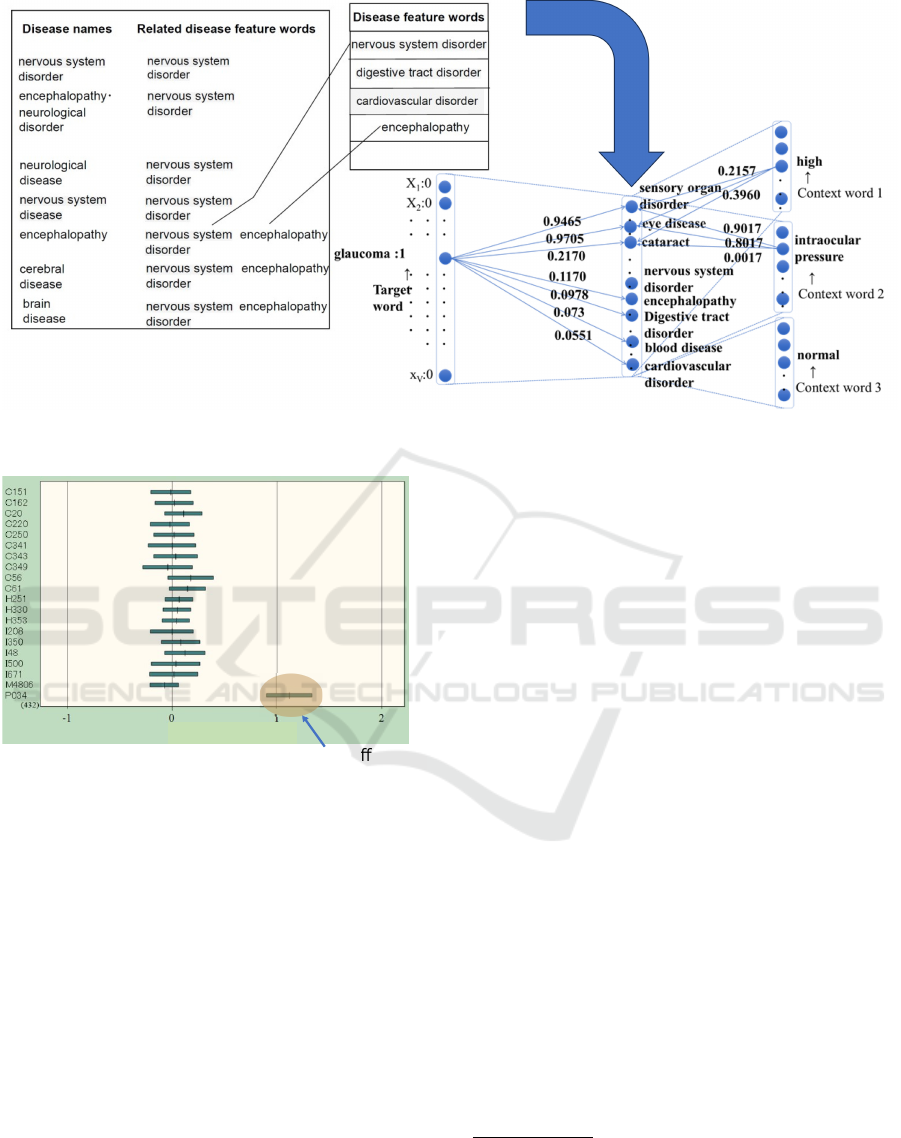
Figure 1: Semantic representation learning process based on the medical-term semantic vector dictionary.
P034… Fetuses and neonates a ected
by cesarean delivery
neonatal disorder
Figure 2: Distribution of weights by ICD-10 code for the
disease feature word ”neonatal disorder”.
weights based on the medical-term semantic vector
dictionary (Keshi et al., 2022). The resulting para-
graph vector, which captures the semantic meaning
of the text, is then combined with other explanatory
variables such as gender, age, and department. The
learning model subsequently uses linear SVM and lo-
gistic regression to classify the ICD-10 codes based
on these features.
4.1.1 Structure of Medical-Term Semantic
Vector Dictionary
The structure of the medical-term semantic vector
dictionary is based on the disease thesaurus named
T-dictionary
*1
. It associates 299 feature words (264
disease feature words + 35 main symptoms) with ba-
sic disease names to provide semantic information for
interpretable disease name estimation (Figure 1).
4.1.2 Classification and Visualization
Figure 2 shows the top 20 ICD-10 codes on the ver-
tical axis and the weight distribution of the disease
feature word ”neonatal disorder” on the horizontal
axis. For ICD-10 code P034, where the mean of the
weight distribution is greater than 1.0, it indicates fea-
tures and neonates affected by cesarean delivery. This
visualization facilitates the interpretation of how the
model arrived at a particular diagnosis by highlight-
ing the significance of specific disease feature words
in the classification process.
4.2 Disease Name Estimation Using
BERT
We evaluated a BERT model pretrained on medical
documents. The BERT model required pre-training
and fine-tuning to achieve accurate disease name esti-
mation.
Table 4 provides information on the BERT models
used in the study.
*1
https://www.tdic.co.jp/products/tdic
*2
https://github.com/cl-tohoku/bert-japanese
*3
https://ai-health.m.u-tokyo.ac.jp/home/research/uth-
bert
*4
https://github.com/ou-medinfo/medbertjp
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
296

Table 4: Information on the BERT Models Used.
Model Name Model Size Training Data
TU-BERT
*2
(Tohoku University
BERT)
Base Japanese Wikipedia
(approximately 17 million
sentences)
UTH-BERT
*3
(University of
Tokyo Hospital BERT)
Base Clinical texts (120 million
records)
MedBERTjp
*4
(Osaka
University Graduate School of
Medicine BERT)
Base Japanese Wikipedia + Corpus
scraped from “Today’s Diagnosis
and Treatment: Premium”
4.3 Estimation of Disease Names Using
GPT-4
We used GPT-4 (model version: 1106-Preview)
from Azure OpenAI Service.
*5
, The chief complaint
dataset was selected for training and evaluation pur-
poses to avoid personal information. Additionally,
we conducted an evaluation using the latest GPT-4o
(model version: 2024-05-13) under the same condi-
tions that yielded the best performance in the earlier
evaluation.
4.3.1 Zero-Shot Learning
In zero-shot learning, GPT-4 estimated disease names
based solely on a system prompt, without any specific
training on the target dataset. This approach leverages
the model’s pre-existing knowledge to make predic-
tions, demonstrating its ability to infer disease names
from chief complaints even in the absence of domain-
specific data.
4.3.2 Few-Shot Learning
In few-shot learning, one set of chief complaints and
corresponding ICD-10 codes for each of the top 20
ICD-10 codes in the new EMR was used from the old
EMR, providing 20 sets as example responses to GPT-
4.
4.3.3 RAG
The RAG approach used three databases:
• RAG1: A database of chief complaints and ICD-
10 codes excluding the chief complaints of the top
20 ICD-10 codes in the new EMR.
• RAG2: A database of chief complaints and ICD-
10 codes from the old EMR corresponding to the
top 20 ICD-10 codes from the new EMR.
• RAG3: A database linking all chief complaints
with corresponding ICD-10 codes, including the
evaluation data.
*5
https://portal.azure.com/#view/Microsoft Azure
ProjectOxford/CognitiveServicesHub/
∼
/OpenAI
old EMR
35,509 cases
Seman c
representa on
learning
Machine learning
Training data
Each benchmark
of old EMR
old EMR
learned model
new EMR
28,787cases
Seman c
representa on
learning
Evalua on data
Each benchmark
of new EMR
Disease name
es ma on
Figure 3: Experimental flow of semantic representation
learning.
5 EXPERIMENTAL SETUP
5.1 Semantic Representation Learning
+ Machine Learning
We used vectors of disease feature words from se-
mantic representation learning to create models using
machine learning. Statflex
*6
was employed for inter-
pretability evaluation to graph the variance and mean
of the vectors. Figure 3 shows the experimental flow
of disease name estimation from chief complaints
using semantic representation learning and machine
learning.
The datasets of all chief complaints shown in Ta-
ble 2 (35,509 cases in the old EMR and 28,787 cases
in the new EMR) were used for semantic representa-
tion learning. We evaluated each benchmark shown
in Table 3. Both linear SVM and logistic regression
were evaluated due to the shorter text length of chief
complaints.
We determined the optimal conditions for chief
complaints with the highest accuracy based on over-
all accuracy and macro-average F1 score of the top 20
ICD-10 codes. These conditions were used in subse-
quent BERT and GPT-4 experiments.
5.2 BERT
All training data were taken from the progress sum-
mary dataset in the old EMR for fine-tuning BERT.
The evaluation consisted of two methods:
• Extracting progress summaries related to the top
20 ICD-10 codes from the new EMR and classi-
fying them as evaluation data.
• Extracting chief complaints related to the top 20
ICD-10 codes from the new EMR and classifying
them as evaluation data.
*6
https://www.statflex.net/
Integrated Evaluation of Semantic Representation Learning, BERT, and Generative AI for Disease Name Estimation Based on Chief
Complaints
297

5.3 GPT-4
For GPT-4 experiments, we used the chief complaint
dataset to avoid personal information.
5.3.1 Zero-Shot Learning
GPT-4 estimated disease names based solely on a sys-
tem prompt, without any specific training on the target
dataset.
System Prompt Example
# R ole
You are an e x p eri e n ced d oc tor at a
→ ho s pit a l . You wil l an sw er
→ qu e s ti o n s fro m you ng d oc t or s and
→ me d ic a l staf f in J ap a nes e .
# O b j ec t i ve
Ba se d on the in put of th e pa tient ’ s
→ ch ie f c om pl ai nt , you will
→ pe r fo r m th e f o ll o w in g ta sks :
- E st i mat e t he pati en t ’ s d i sea se and
→ pr o vi d e up to five po s sib l e
→ di a g no s e s a lo ng wi th th eir ICD
→ - 10 c od es of mi d dl e cat e gor i e s .
# D ata S p e cif i c a tio n s
For each c hie f com pl ai nt , d i spl ay the
→ ICD -10 c od e of the mi d dl e
→ cat e gor i es an d the top f iv e
→ ca n d id a t e d i a gn o s es .
# O utp ut F or mat
The ou tpu t s h ou ld be in t he fo l low i ng
→ J SO N f orm at :
( for mat det ail s om i tt e d )
5.3.2 Few-Shot Learning
Few-shot learning involved providing example sen-
tences to GPT-4 to enable in-context learning.
Few-shot Learning Example
{" rol e ": " user " , " co nte n t ": " Loss of
→ a pp et it e , gen e r ali z e d fati gu e ,
→ p ai n in da rk s urr o u n di n g s "} ,
{" rol e ": " as s is t a nt " , " con t ent ":"[ {"
→ Es t i ma t e d D i se a se ": " C 25 ", "
→ Di a g no s i s ": " Canc er of the
→ pa n cre a s "] "}
5.3.3 RAG
In the experiment, the three configurations RAG1,
RAG2, and RAG3 described in the proposed method
were used to evaluate the performance of the model.
Each configuration was designed to test the model un-
der different conditions, focusing on the availability
and relevance of reference data.
RAG External Data Example
Di a g no s is C ode : C34
C34 , B ac k pain , a b dom i nal pain , liv er
→ dys f u nc t i on
C34 , Ab n or m al s e n sa t ion in the ri gh t
→ up pe r arm , sw e ll i ng in the ri gh t
→ s upr a c l avi c u l a r f oss a
In the RAG, new and old EMR chief complaints
were entered into text files for each ICD-10 code of
the middle categories and managed in an Azure stor-
age Blob container. Data was chunked into 512-token
segments with 128-token overlap. The search used
Azure AI Search’s hybrid (keyword + vector) search
and semantic ranking features (Berntson et al., 2023).
For evaluation, the Zero-shot learning, Few-shot
learning, and RAG methods used the same 200 sets
of evaluation data, which consisted of 200 chief com-
plaints randomly selected from the top 20 ICD-10
codes in the new EMR. The results of these evalua-
tions are presented in the following sections. Based
on the results of the semantic representation learn-
ing experiments, RAG was constructed targeting chief
complaints of more than 10 characters in the ICD-10
middle categories. RAG1 and RAG3 included 872
types of ICD-10 codes, while RAG2 focused on the
top 20 ICD-10 codes from the new EMR. To align the
evaluation with the other two methods, 200 evalua-
tion data sets were constructed by randomly selecting
10 chief complaints from each of the top 20 ICD-10
codes. Each evaluation data set had only one correct
ICD-10 code.
6 EVALUATION RESULTS
6.1 Semantic Representation Learning
+ Machine Learning
The evaluation results of disease name estimation
using semantic representation learning and machine
learning (logistic regression and linear SVM) based
on the chief complaint benchmarks are shown in the
first six rows of Table 5. The regularization parame-
ter C was determined using a grid search. The high-
est overall accuracy was 62.0% when the chief com-
plaint had more than 10 characters and the ICD-10
codes were categorized at the middle level. The high-
est macro-average F1 score was 51.7 points when the
chief complaints had more than 10 characters and the
ICD-10 codes were categorized at the subcategory
level, using logistic regression. Linear SVM showed
the best results (the accuracy: 56.1 %, the F1-score:
49.1) with chief complaints of more than 10 charac-
ters and ICD-10 codes categorized at the middle level.
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
298
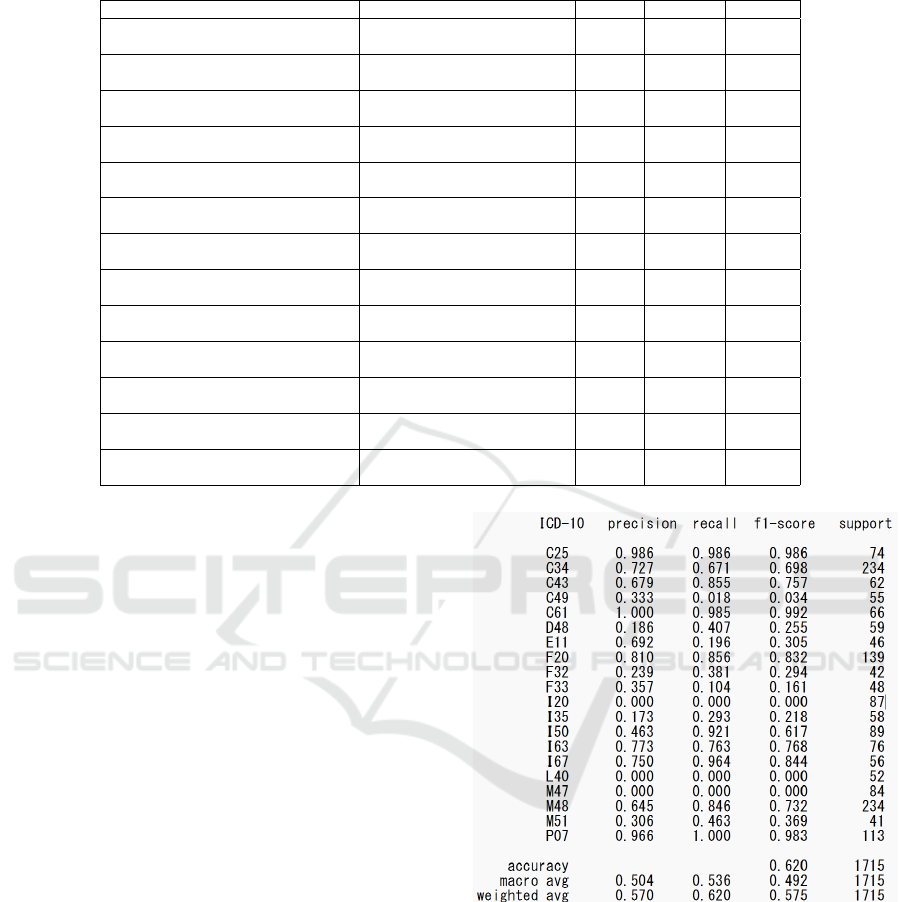
Table 5: Evaluation results of disease name estimation from chief complaints and progress summaries.
Model Name Type of Evaluation Data C value Accuracy F1-score
Semantic Representation Learning +
Logistic Regression
Chief Complaints (Any chars,
Subcategories)
60.0 36.0% 29.5
Semantic Representation Learning +
Logistic Regression
Chief Complaints (10+ chars,
Subcategories)
49.0 49.4% 51.7
Semantic Representation Learning +
Logistic Regression
Chief Complaints (10+ chars,
Middle Categories)
34.0 62.0% 49.2
Semantic Representation Learning +
Linear SVM
Chief Complaints (Any chars,
Subcategories)
250 26.2% 22.7
Semantic Representation Learning +
Linear SVM
Chief Complaints (10+ chars,
Subcategories)
130 44.5% 48.6
Semantic Representation Learning +
Linear SVM
Chief Complaints (10+ chars,
Middle Categories)
41.0 56.1% 49.1
Semantic Representation Learning +
Linear SVM
Progress Summaries (500 chars,
Subcategories)
N/A 69.5% 72.1
TU-BERT Progress Summaries (500 chars,
Subcategories)
N/A 77.5% 80.0
UTH-BERT Progress Summaries (500 chars,
Subcategories)
N/A 83.8% 85.3
MedBERTjp Progress Summaries (500 chars,
Subcategories)
N/A 77.1% 80.4
TU-BERT Chief Complaints (10+ chars,
Middle Categories)
N/A 52.2% 44.1
UTH-BERT Chief Complaints (10+ chars,
Middle Categories)
N/A 61.1% 53.7
MedBERTjp Chief Complaints (10+ chars,
Middle Categories)
N/A 53.4% 45.7
Figures 4 and 5 show the evaluation results of
ICD-10 codes categorized at the middle and subcat-
egory levels for chief complaints with more than 10
characters when using logistic regression. For the
middle categories, three ICD-10 codes (I20, L40,
M47) had an F1 score of 0, while no subcategory dis-
ease names had an F1 score of 0. This suggests a
higher overfitting risk for subcategories. Therefore,
the condition of chief complaints with more than 10
characters at the middle category level will be used
for BERT and GPT-4 evaluations.
6.2 BERT
The four rows starting from the middle of Table 5
shows the evaluation results of classifying progress
summaries (up to 500 characters) extracted from the
top 20 ICD-10 codes (subcategories) in the new EMR
as evaluation data. The macro-average F1-score for
semantic representation learning was 72.1, while the
fine-tuned large language model using UTH-BERT
achieved a macro-average F1-score of 85.3, sur-
passing semantic representation learning by over 10
points.
For the evaluation based on chief complaints, as
shown in the last three rows of Table 5, UTH-BERT
had the highest accuracy and macro-average F1 score
among the BERT models. However, the accuracy of
semantic representation learning combined with lo-
gistic regression slightly exceeded that of the BERT
Figure 4: Disease name estimation using semantic represen-
tation learning and logistic regression for ICD-10 codes cat-
egorized at the middle level with chief complaints of more
than 10 characters.
models.
6.3 GPT-4
Table 6 shows the evaluation results of GPT-4 in es-
timating disease names from chief complaints (200
sets of evaluation data). The Top-5 accuracy was
measured, considering a result correct if the cor-
Integrated Evaluation of Semantic Representation Learning, BERT, and Generative AI for Disease Name Estimation Based on Chief
Complaints
299
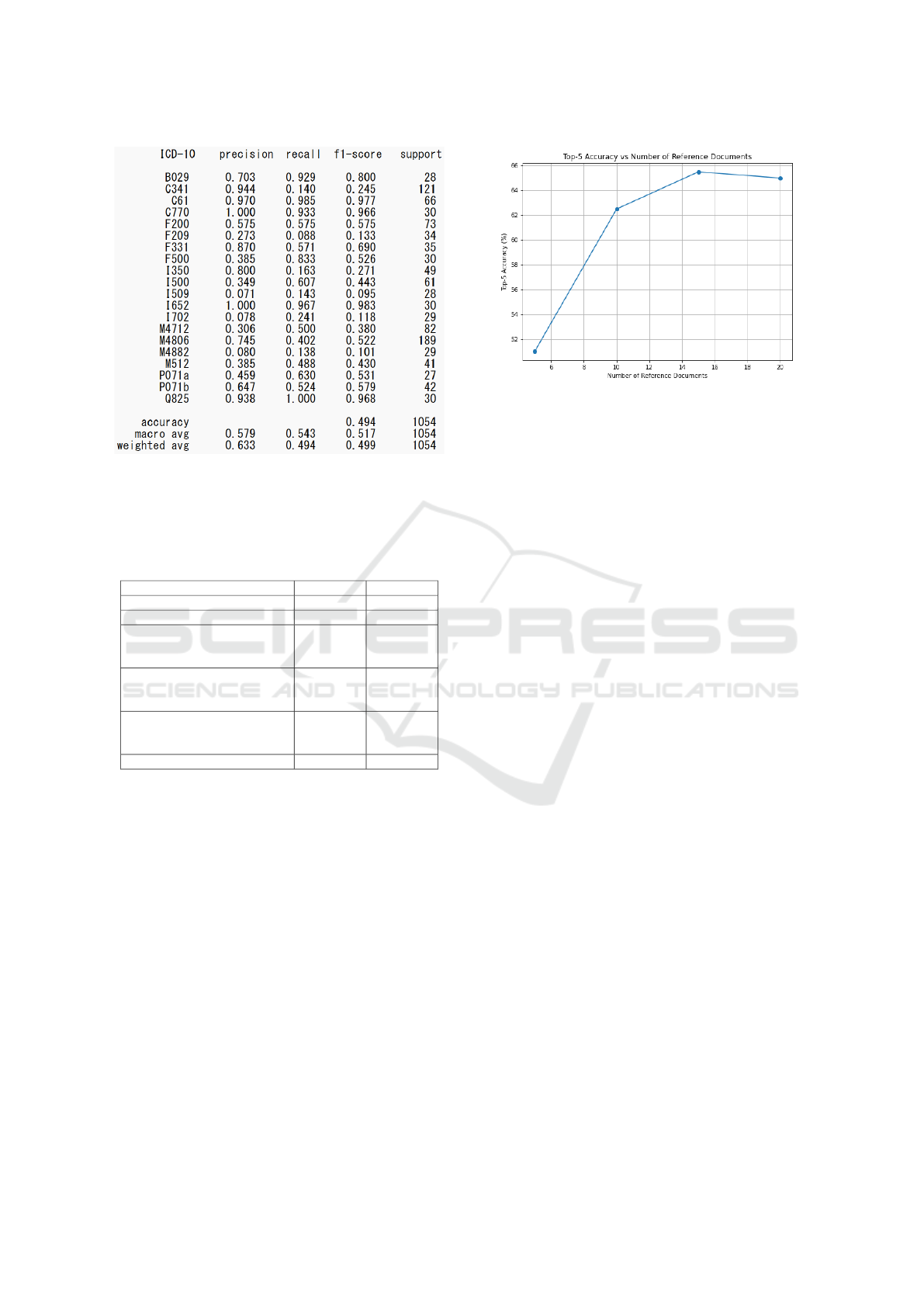
Figure 5: Disease name estimation using semantic repre-
sentation learning and logistic regression for ICD-10 codes
categorized at the subcategory level with chief complaints
of more than 10 characters.
Table 6: Evaluation results of disease name estimation from
chief complaints (200 sets of evaluation data).
Top-5 Acc. Top-1 Acc.
Zero-shot Learning 52.5% 22.0%
Few-shot Learning 61.0% 20.0%
RAG1: All cases except the
benchmark cases in the new
EMR (15 reference documents)
65.5% 19.5%
RAG2: Only the benchmark
cases in the old EMR (5
reference documents)
82.5% 24.0%
RAG3: All cases, including the
benchmark cases in the new
EMR (15 reference documents)
84.5% 25.0%
RAG3: GPT-4o 90.0% 26.5%
rect ICD-10 code was among the top five candi-
dates. Zero-shot learning achieved a Top-5 accu-
racy of 52.5%, while few-shot learning improved it
to 61.0%. RAG1 achieved 65.5% with 15 reference
documents, RAG2 reached 82.5% with 5 reference
documents, and RAG3 achieved the highest Top-5 ac-
curacy of 84.5% with 15 reference documents.
Additionally, the latest GPT-4o was evaluated un-
der the same conditions as RAG3, achieving the high-
est Top-5 accuracy of 90.0%. Excluding one chief
complaint where a response was not generated due
to content filtering, GPT-4o’s Top-5 accuracy reached
90.5%.
Figure 6 illustrates the relationship between the
number of reference documents and the Top-5 accu-
racy for RAG1. The accuracy improves as the num-
ber of reference documents increases, with the best
performance achieved at 15 reference documents.
Figure 6: Top-5 Accuracy vs Number of Reference Docu-
ments.
7 DISCUSSION
This study confirmed that the accuracy of disease
name estimation significantly decreases when chang-
ing the target from progress summaries to chief
complaints. However, using semantic representa-
tion learning, logistic regression achieved an accu-
racy of 62.0% for chief complaints of more than
10 characters classified at the middle category level.
This slightly exceeded the accuracy of UTH-BERT,
which was fine-tuned with over 10,000 progress sum-
maries, while semantic representation learning used
only 1,605 chief complaints. However, for 3 out of the
20 ICD-10 codes, the estimation accuracy was 0%.
This is because chief complaints often consist of gen-
eral symptoms like “fever” or “dizziness,” which do
not include disease names registered in the medical-
term semantic vector dictionary. If the chief com-
plaint does not include a disease name, the feature
vector does not change, leading to estimation failure.
In cases where the data is rich in context, such
as progress summaries of up to 500 characters, SVM
tends to perform better due to its ability to capture
complex relationships within the data. However, for
datasets like chief complaints, which are often lack-
ing in context, logistic regression may be more suit-
able. This is because logistic regression is a simpler
model that is less prone to overfitting, making it bet-
ter suited to handle sparse and less informative data.
The results suggest that logistic regression was bet-
ter suited for the chief complaint dataset due to its
simplicity and robustness. Similarly, this may also
explain why semantic representation learning slightly
outperformed BERT, as the former was better able to
handle the limited context and information present in
the chief complaints.
GPT-4 showed significant improvement in Top-5
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
300

accuracy with few-shot learning, providing 20 sets of
example sentences, and RAG, using only the chief
complaints and ICD-10 codes from the old EMR as
external data. The contextual limitation likely con-
tributed to this improvement. For RAG without cor-
rect cases, fewer reference documents resulted in
lower accuracy than few-shot learning, highlighting
the importance of data quality over quantity.
The evaluation set was limited to the top 20 dis-
ease names, and GPT-4 generated 5 candidate disease
names. Expanding the evaluation set to a wider range
of disease names and conducting evaluations using
external data is necessary. Additionally, subjective
evaluation of the validity and diagnostic reasons by
veteran physicians is important.
8 CONCLUSIONS
This study compared disease name estimation meth-
ods using semantic representation learning + machine
learning, BERT, and GPT-4, and evaluated their ac-
curacy. Despite being trained on only 1,605 chief
complaints, semantic representation learning + ma-
chine learning showed slightly higher accuracy than
BERT, which was fine-tuned on over 10,000 progress
summaries, under certain conditions. However, it was
found to have limitations in disease name estimation
based on chief complaints.
For GPT-4, evaluation data were created based on
the top 20 disease names with the highest occurrence
frequency in the new EMR, targeting cases with chief
complaints of more than 10 characters. Evaluations
using zero-shot learning, few-shot learning, and RAG
demonstrated that RAG achieved the highest perfor-
mance. When all chief complaints, including the eval-
uation data, were used, the highest Top-5 accuracy
of 84.5% was achieved, while excluding the evalua-
tion data decreased the accuracy to 65.5%. The op-
timal number of reference chunks for RAG was 15.
Even when excluding the evaluation data, limiting the
database to the 20 diagnostic disease names improved
the Top-5 accuracy to 82.5%. Furthermore, the latest
GPT-4o model was evaluated under the same condi-
tions as RAG, and it further improved the Top-5 ac-
curacy to 90.0%.
In the future, we aim to expand the benchmark to
cover additional middle categories of ICD-10, con-
duct more extensive evaluations, and perform subjec-
tive evaluations by experienced physicians. This aims
to implement disease name estimation from chief
complaints as a practical diagnostic support tool in
medical settings.
ACKNOWLEDGMENTS
Part of this study was conducted by Shuta Asai, Tat-
suki Sakata as their graduation research in 2023, and
is currently being conducted by Mikio Osaki as part
of his ongoing graduation research in 2024, all from
Fukui University of Technology. We thank them for
their contributions. This work was supported by JSPS
KAKENHI Grant Number 24K14964 and 20K11833.
This study was approved by the Ethical Review Com-
mittee of the Fukui University of Technology and the
Toyama University Hospital.
REFERENCES
Berntson, A. et al. (2023). Azure ai search: Outperforming
vector search with hybrid retrieval and ranking ca-
pabilities. https://techcommunity.microsoft.com/
t5/ai-azure-ai-services-blog/azure-ai-search-
outperforming-vector-search-with-hybrid/ba-p/
3929167. Accessed: 2024-05-18.
Chen, A., Liu, L., and Zhu, T. (2024). Advancing the
democratization of generative artificial intelligence in
healthcare: a narrative review. Journal of Hospital
Management and Health Policy, 8(0).
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2019). Bert: Pre-training of deep bidirectional trans-
formers for language understanding.
Kasai, J., Kasai, Y., Sakaguchi, K., Yamada, Y., and Radev,
D. (2023). Evaluating gpt-4 and chatgpt on japanese
medical licensing examinations.
Kawazoe, Y., Shibata, D., Shinohara, E., Aramaki, E., and
Ohe, K. (2021). A clinical specific bert developed us-
ing a huge japanese clinical text corpus. PLoS One,
16(11)(9).
Keshi, I., Daimon, R., and Hayashi, A. (2022). Interpretable
disease name estimation based on learned models us-
ing semantic representation learning of medical terms.
In Coenen, F., Fred, A. L. N., and Filipe, J., editors,
Proceedings of the 14th International Joint Confer-
ence on Knowledge Discovery, Knowledge Engineer-
ing and Knowledge Management, IC3K 2022, Volume
1: KDIR, Valletta, Malta, October 24-26, 2022, pages
265–272. SCITEPRESS.
Le, Q. V. and Mikolov, T. (2014). Distributed representa-
tions of sentences and documents. In Proc. of ICML,
pages 1188–1196.
Singhal, K. et al. (2023). Towards expert-level medical
question answering with large language models.
Yanagita, Y., Yokokawa, D., Uchida, S., Tawara, J., and
Ikusaka, M. (2023). Accuracy of chatgpt on medi-
cal questions in the national medical licensing exam-
ination in japan: Evaluation study. JMIR Form Res,
7:e48023.
Integrated Evaluation of Semantic Representation Learning, BERT, and Generative AI for Disease Name Estimation Based on Chief
Complaints
301
