
Convolutional Neural Networks (CNNs)-Based for Medical Image
Analysis
Leyan Li
a
Department of Computer Science, University of Liverpool, Liverpool, U.K.
Keywords: CNNs, Medical Image Processing, Residual Network, Transformers.
Abstract: This paper provides an exhaustive examination of convolutional neural networks (CNNs) in medical image
processing, recognizing their pivotal role in healthcare diagnostics. As CNNs continue to evolve, they offer
promising avenues for enhancing accuracy and efficiency in image analysis. The primary objective of this
study is to scrutinize and assess the performance of both classic and contemporary CNN models across a
spectrum of pathological datasets. The methodology entails a comprehensive analysis of various CNN
architectures, ranging from well-established models to more advanced approaches. Emphasis is placed on
their efficacy in disease classification and feature extraction tasks. Experiments conducted on datasets
underscore the models' adeptness in handling intricate medical images. The findings indicate CNNs'
superiority in feature extraction, the proficiency of Residual Network (ResNet) in managing depth and
ensuring robust training, and Transformers' effectiveness in navigating high-dimensional data through their
attention mechanisms. These insights hold profound implications for medical diagnostics, promising
significant advancements in accuracy and timeliness of health interventions.
1 INTRODUCTION
Medical imaging technology is an effective means of
understanding pathological processes that affect
human health (Jannin, 2006). Compared to natural
images, medical images (such as slices or patches
from different modalities) contain richer information
due to their more organized and similar visual
representations of human organs (Dai, 2021). With
the advancement of technology, the main tasks of
medical image processing can be summarized as
generating new images from original ones, computing
features and measurements (known as image
analysis), or extracting high-level descriptions
(referred to as image understanding) (Jannin, 2006).
In the medical field, the quality and accuracy of
image processing have become benchmarks, and
these processing results are crucial for medical
decision-making (Sonka, 2000).
Advances in algorithms in the domain of medical
imaging technology often stem from the need for new
image analysis capabilities (Jannin, 2006). For
instance, since Krizhevsky (Krizhevsky, 2012) and
others introduced the convolutional neural network
a
https://orcid.org/0009-0009-5255-2230
(CNN) with AlexNet winning the ImageNet image
classification championship in 2012, CNNs have
shown tremendous advantages in disease detection
and classification, and local feature extraction from
images. Classic CNN models like AlexNet, Network
in Network (NIN) which reduces the risk of
overfitting through global average pooling (Lin,
2013), and Visual Geometry Group (VGGNet) and
GoogLeNet, which enhanced precision on the
ImageNet dataset (Simonyan, 2014), have all
performed well. In 2015, He and others introduced
Spatial Pyramid Pooling (SPP), which addressed the
strict input size requirements of CNNs (He, 2015),
and in the subsequent year introduced the residual
network (ResNet) to address the issue of model
degradation. Recently, the Transformer has excelled
in tasks needing a deep understanding of visual
contexts and details, thanks to its capability to handle
high-dimensional data and synthesize details from
different image sections.
This paper comprehensively reviews and
summarizes the current research status of utilizing
CNNs for medical image processing. Chapter Two
analyzes and explains classic and current mainstream
546
Li, L.
Convolutional Neural Networks (CNNs)-Based for Medical Image Analysis.
DOI: 10.5220/0012958600004508
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Engineering Management, Information Technology and Intelligence (EMITI 2024), pages 546-552
ISBN: 978-989-758-713-9
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

network models based on CNNs in deep learning. It
delves into the architecture, training strategies, and
applications of these models in medical imaging tasks.
Chapter Three provides a detailed analysis and
comparison of the results obtained by different
models using various pathological datasets. It
evaluates the performance metrics, including
accuracy, sensitivity, and specificity, to assess the
effectiveness of CNN-based approaches in medical
image analysis. In Chapter Four, the paper
summarizes the advantages of CNNs in medical
imaging, highlighting their ability to extract
meaningful features and their reliance on large
datasets for training. Furthermore, it explores
potential future trends, such as the development of
CNN architectures that require less data for training
or the utilization of artificial intelligence techniques
to generate novel models tailored to specific medical
imaging tasks.
2 METHODOLOGIES
2.1 Dataset Description and
Preprocessing
Interstitial Lung Disease (ILD) encompasses various
pulmonary diseases affecting the lung parenchyma,
associated with significant morbidity and mortality
(Stanford, 2024). Recent advances have led to a
substantial collection of genetic and disease data
integrated into the International Lattice Data Grid
(ILDG) database, 2024 version, featuring 20 types of
ILD across four species with over 600 genes and
2,018 entries. This database includes detailed records
of species, disease types, gene symbols, and primary
references. The study utilized image blocks where at
least 75% of pixels are within Regions of Interest
(ROI), involving 16,220 blocks from 92 high-
resolution computed tomography (HRCT) image sets.
The images were divided into ten groups per round,
with one for testing and nine for training, using
random image shifting to enhance diversity and
prevent overfitting. The MRNet dataset includes
1,370 magnetic resonance imaging (MRI) knee
examinations, categorized by conditions like anterior
cruciate ligament (ACL) tears (Touvron, 2021). It
splits into 1,130 training, 120 validation, and 120
testing cases and uses three MRI scan types: T1, T2,
and proton density, with resolutions reformed into 3D
stacks for T1 and T2 scans. The preprocessing
includes the OTSU algorithm for background
separation, image alignment, and formatting into
stacks of 36x448x448. The augmentation techniques
include random flipping and Gaussian noise to
improve dataset robustness.
2.2
Proposed Approach
This study aims to comprehensively review and
analyze the applications of CNN in medical image
processing, with a focus on evaluating the efficacy of
both classic and contemporary mainstream deep
learning models based on CNNs. The performance of
these models is assessed across a range of
pathological datasets, employing detailed
methodologies and comparisons to elucidate each
model's strengths and applicable scopes. Additionally,
the principal flowchart of the main process is outlined
in Figure 1 to provide a visual representation of the
workflow.
Figure 1: The pipeline of the model (Photo/Picture credit:
Original).
2.2.1 Introduction to Basic Techniques
CNNs represent a form of deep learning model,
evolved from the multilayer perceptron, as shown in
the Figure 2. They simplify the learning process by
using smaller kernel filters to incorporate weights,
which speeds up operations and enhances robustness.
Due to their ability to automatically and efficiently
learn intrinsic features from blocks of medical images
and their strong generalization capability, CNNs are
widely used in medical image processing. The main
structures of CNNs include: Data Input Layer: This
layer preprocesses the raw image data, including
mean subtraction (centring the data around zero to
reduce variations between samples), normalization
(aligning the amplitude range across different
dimensions), and Principal Component Analysis
(PCA)/whitening (reducing dimensionality and
normalizing the amplitude of the data feature axes).
Convolutional Layer (CONV layer) and rectified
linear unit (ReLU) Activation Layer: The
convolutional layer applies multiple filters through
local connections and sliding window mechanisms,
with each neuron acting as a filter. Key parameters
include filter size, stride, and padding. The stride
controls the movement distance of the filter over the
input, while zero padding adds zeros at the boundaries
of the input to maintain consistent spatial dimensions
Convolutional Neural Networks (CNNs)-Based for Medical Image Analysis
547
between the input and output. The formula to
calculate the output size is:
Output Size
1(1)
The ReLU activation layer provides a non-linear
mapping, enhancing the model's non-linear
expression capability and convergence speed. The
next is pooling layer, which situated between
successive convolutional layers, its primary function
is to compress data and parameter quantity, thus
reducing overfitting. Typical techniques involve
average pooling and max pooling, with max pooling
choosing the maximum value from each window for
the output. Next is Fully Connected Layer (FC layer).
The last layer of a CNN typically comprises an FC
layer, in which each neuron is connected to every
neuron in the preceding layer, culminating in the
network's ultimate output.
Figure 2: The structure of the FC layer (Photo/Picture
credit: Original).
These components enable CNNs to effectively
process medical images, through this process, CNNs
have successfully achieved efficient processing of
medical images, demonstrating their strong potential
for the medical domain.
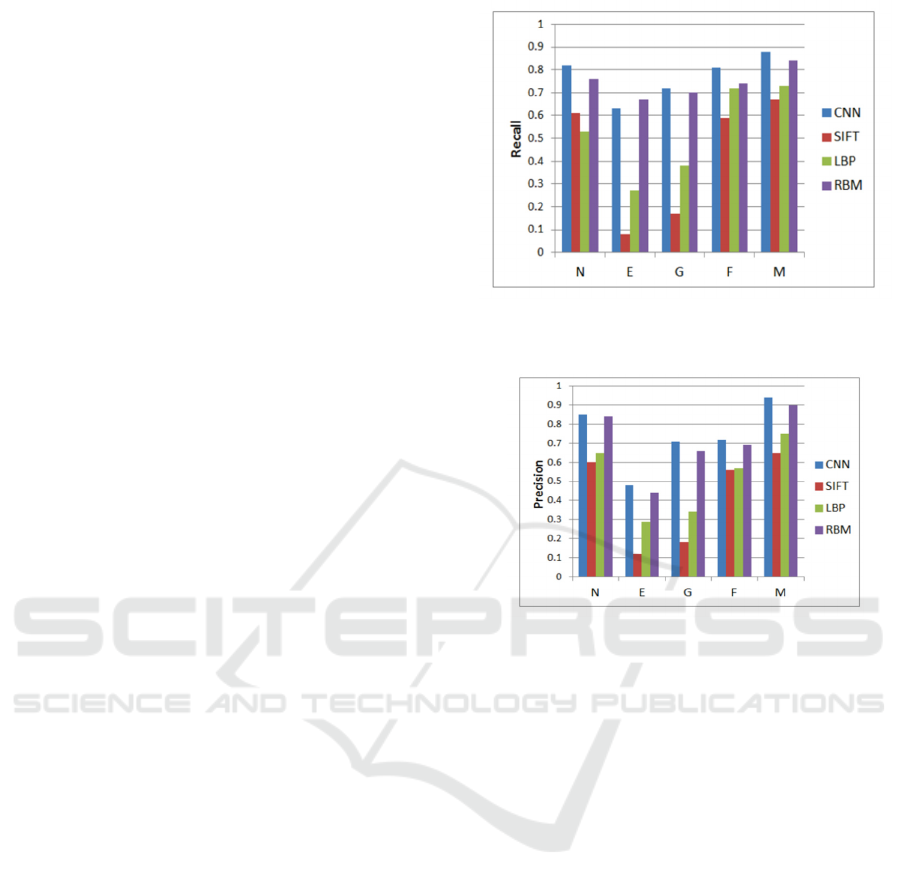
2.2.2 ResNet
CNNs have shown exceptional effectiveness in object
recognition and have gradually become the preferred
method for image analysis, as shown in the Figure 3.
He et al. (ildgdb, 2024) introduced the ResNet, which
effectively addresses challenges related to vanishing
gradients and network degradation stemming from
increased network depth. This network structure
significantly speeds up the training of neural
networks and greatly enhances their generalization
capabilities and robustness.
Comprising multiple residual units, Residual
Neural Networks include a convolutional layer (conv
layer), batch normalization layer (BN), and ReLU in
each unit. At the heart of a residual unit lies the direct
passage of input to output, thereby forming the
foundational result. In the event that the input to the
neural network is denoted as 𝒙 and the anticipated
output as 𝐇𝐱, the residual function is represented
as H(x)-x, the output of the residual unit is 𝐱
𝐇𝐱 𝐱, so the network's learning target becomes
H(x)-x —the residual. By fitting the residual
mapping, ResNet simplifies the learning process,
making the optimization of deep networks easier and
solving the issues of gradient disappearance and
degradation with increased depth. Another key
feature of residual units is the shortcut connection
(identity mapping) that changes the learning target
from the direct mapping 𝐇𝐱 to 𝐇𝐱 𝐱.
When the input and output dimensions are
consistent, the shortcut connections can directly add
the input to the output. If dimensions are inconsistent,
there are two strategies for handling this:
Zero-padding is used to increase dimensions,
usually combined with pooling operations with a
stride of 2 for downsampling. This method does not
add extra parameters. Projection shortcuts are
typically adjusted through 1×1 convolution to change
dimensions, which increases some parameters and
computational load. These shortcut connection
strategies not only maintain the network’s parameters
and computational load but also significantly improve
the model's training speed and efficiency in
Figure 3: The Structure of the ResNet (Photo/Picture credit: Original).
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
548

deeper network structures, effectively preventing
performance degradation. With these innovative
designs, ResNet has shown immense potential and
practical utility, particularly in deep medical image
processing in the field of deep learning.
2.2.3 Application of Transformers in
Medical Imaging
Transformers, initially designed for Natural
Language Processing, effectively capture long-range
dependencies using self-attention mechanisms. This
technology has been adapted for visual tasks such as
object detection with Detection transformer (DETR),
semantic segmentation, and image classification with
vision in transformer (ViT) (Touvron, 2021). In
multimodal medical imaging, where capturing long-
range interactions is essential, Transformers enhance
deep learning models by effectively integrating
multimodal data, outperforming traditional CNNs
that excel in local feature extraction but struggle with
distant relationships (Touvron, 2021). The
fundamental element of the Transformer are Self-
Attention Mechanism (SA): SA is the foundation of
the Transformer, allowing the model to enhance its
predictions by using other parts of a data sample
during processing. In the self-attention layer, the
input vector X undergoes the transformation into
three distinct vectors: Query matrix Q, Key matrix K,
and Value matrix V, as shown in the Figure 4.
Weights are assigned based on the dot product of
queries and their respective keys. The attention
function is calculated as follows:
AttentionQ, K, V Softmax
V (2)
where 𝐝
𝐤
is the dimension of the key vectors, and this
normalization helps stabilize the gradients. Multi-
Head Self-Attention (MSA) (Figure 5): MSA is
central to the Transformer architecture, enhancing the
model's ability to learn information from different
representational subspaces by splitting the input into
multiple parts and processing them in parallel. The
computation of MSA is expressed as:
head
AttentionQW
,KW
,VW
MSAQ, K, VConcathead
,head
,...,
head
W
(3)
where projection matrices 𝐖
𝐢
𝐐
, 𝐖
𝐢
𝐊
, 𝐖
𝐢
𝐕
, 𝐖
𝐎
are
trainable parameters.
Multi-Layer Perceptron (MLP): Located above
the MSA layer, composed of linear layers and
activation functions (like GeLU), providing the
model with non-linear processing capabilities.
Similar to ResNet, MLP and MSA, integrate layer
normalization and skip connections techniques to aid
in training deep networks.
Figure 4: The structure of the SA (Photo/Picture credit: Original).
Figure 5: The structure of the MSA (Photo/Picture credit: Original).
Convolutional Neural Networks (CNNs)-Based for Medical Image Analysis
549
The input layer includes several embeddings and
tokens: patch embeddings (from CNNs), positional
embeddings (encoding spatial information), class
embeddings (training vectors), and patch and class
tokens. The class token, atop the patch tokens, moves
through Transformer layers and is outputted by a fully
connected layer for classification. This design allows
the Transformer to excel in handling multimodal
medical images with complex, long-range
dependencies.
3 RESULTS AND DISCUSSION
This section analyzes and compares the performance
of different deep learning models such as CNN,
ResNet, and Transformer on various pathological
datasets, revealing the characteristics and advantages
of each model.
3.1 Performance of CNN Compared
with Different Models
In this study, the ILD database (Stanford, 2024) was
used, consisting of 113 HRCT lung image sets with
2062 2D regions of interest (ROI) classified into five
ILD types: Normal (N), Emphysema (E), Ground
Glass (G), Fibrosis (F), and Micronodules (M). CT
slices were segmented into 32×32-pixel semi-
overlapping blocks, using only those where at least
75% of pixels were within ROIs, totaling 16220
blocks. Three feature extraction methods—Scale
Invariant Feature Transform (SIFT), which identifies
central key points; rotation-resistant Local Binary
Patterns (LBP) at varying resolutions; and Restricted
Boltzmann Machine (RBM) for unsupervised
learning—were compared using an supported vector
machine (SVM) classifier. In contrast, CNN directly
classifies through three neural network layers,
optimizing performance via parameter fine-tuning
and backpropagation, without needing a separate
classifier.
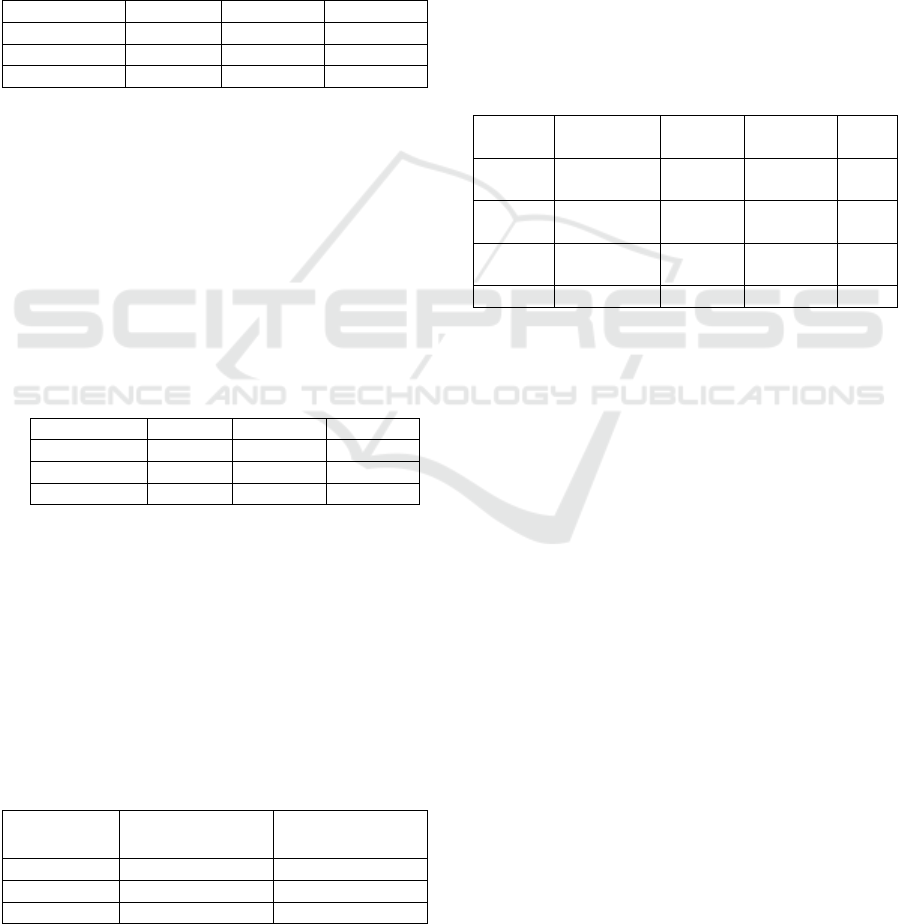
The classification outcomes were assessed
through precision and recall metrics. Figures 6 and 7
illustrate that the CNN method delivered superior
classification performance, surpassing both SIFT and
LBP, demonstrating CNN's clear advantage in
automatic feature learning in medical imaging.
Despite challenges such as ambiguous visual
structures and limited training data, overfitting issues
can be effectively mitigated by designing appropriate
network architectures and applying techniques like
dense dropout and input distortion.
Figure 6: Classification results focused on recall metrics
(Li, 2014).
Figure 7: Classification results focused on Precision metrics
(Li, 2014).
3.2 Different Configurations of ResNet
Models in Medical Image
Processing
This section aims to evaluate the performance of
different configurations of ResNet models in medical
image processing. This paper used three different data
allocation strategies to train and test the ResNet
approaches: Approach 1 employs a data split of 60%
for training and 40% for testing; Approach 2 allocates
75% for training and 25% for testing; Approach 3
utilizes an 80% training and 20% testing data split. At
the start of the experiments, all images were
converted to grayscale and enhanced for contrast
using the Contrast Limited Adaptive Histogram
Equalization (CLAHE) algorithm to standardize
initial inputs. During the learning process of the
ResNet model, in addition to the basic convolutional
layers, batch normalization layers, ReLU activation
layers, and pooling layers were included. Each
architecture of ResNet contains multiple residual
blocks, each divided into 5 layers, with pooling layers
primarily used at the feature extraction and before the
classification layers.
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
550
All models were optimized using the SGD
optimizer with momentum as the optimization
function, accompanied by adjustments to the learning
rate and utilizing binary cross-entropy as the
objective function. Table 1 shows the performance
using the ResNet-18 architecture under different
training-testing ratios. The configuration that
Approach 3 achieved the highest accuracy of 85%.
And the configuration using Approach 2 achieved the
highest sensitivity of 96%.
Table 1: ResNet-18 Evaluation Measures Across Various
Test Datasets (Sarwinda, 2021).
Confi
g
uration Accurac
y
S
p
ecificit
y
Sensitivit
y
A
pp
roach 1 73% 83% 64%
A
pp
roach 2 81% 63% 96%
Approach 3 85% 87% 83%
Table 2 illustrates the efficacy of the approach of
ResNet-50, where the peak accuracy achieved is 88%,
stemming from the training data configurations of
75% and 80%. In the Approach 1 configuration, the
highest sensitivity reached 92%. Comparing the
results of ResNet-18 and ResNet-50 shows that
ResNet-50 has better accuracy and sensitivity on the
same dataset, indicating that stacking more
convolutional layers can enhance the ability to learn
features.
Table 2: Evaluation Measures Across Various Test Datasets
for ResNet-50 (Sarwinda, 2021).
Configuration Accurac
y
Specificit
y
Sensitivit
y
Approach 1 77% 92% 60%
Approach 2 88% 87% 89%
A
pp
roach 3 88% 83% 93%
Data from Table 3 indicate that the training and
testing times for ResNet-18 are generally lower than
for ResNet-50, primarily due to differences in the
number of layers in the architecture. Additionally,
performance analysis of both models shows that the
ResNet variants achieve an accuracy range from 73%
to 88% and a sensitivity range from 64% to 96% in
colorectal cancer detection, proving the effectiveness
of the ResNet architecture in such applications.
Table 3: Evaluation of Execution Time for Each Epoch
Between ResNet-18 and ResNet-50 (Sarwinda, 2021).
Configuration
𝑇
(seconds)
𝑇
(seconds)
Approach 1 77% 60%
A
pp
roach 2 88% 89%
A
pp
roach 3 88% 93%
This detailed performance assessment of ResNet
models underscores their adaptability and efficiency
in handling complex medical imaging tasks across
different configurations and datasets.
3.3 Study on the Use of Transformers
in CT Medical Imaging
This section explores the application of Transformer
models in medical imaging processing, particularly
focusing on their performance in Computer
Tomography (CT) image analysis. CT, especially for
diagnosing chest diseases, provides an ideal scenario
for Transformers due to the high contrast between
gases and tissues.
Table 4: Evaluation of Transformer for Computer
Tomography (He, 2023).
Citations Datasets Accuracy
(
%
)
illness Body
p
art
COVID-
VIT
COV19.CT-
DB
96.0 COVID-19 Lung
Zhang et
al.
COV19.CT-
DB
76.6 COVID-19 Lung
Than et
al.
COVID-
CTset
- COVID-19 Lung
Li et al. - 98.0 COVID-19 Lun
g
As shown in the Table 4, Than et al. studied the
impact of patch size on ViT's performance in
classifying COVID-19 and other lung pathologies.
They found that a patch size of 32x32 achieved the
best accuracy, revealing a trade-off between patch
size and model performance. Li et al. developed a
ViT-based COVID-19 diagnostic platform that
converts CT images into streamlined patches suitable
for ViT input requirements. Using a teacher-student
model strategy, they enhanced the model's diagnostic
capabilities by distilling knowledge from CNNs pre-
trained on natural images. Zhang et al. first
segmented the lung areas in CT images using Unet,
then inputted the segmented lung regions into Swin-
Transformer for feature extraction. This strategy
significantly reduced the computational load of the
Transformer model. The above studies highlight the
role of pretraining in CT image classification
processing. Using attention mechanisms to reduce
computational complexity is particularly crucial for
processing large-volume images.
This chapter reviews three significant deep
learning models—CNN, ResNet, and Transformer—
in medical imaging. CNNs are superior in automatic
feature extraction, outperforming traditional
techniques in classifying complex lung images, with
network enhancements like dense dropout improving
Convolutional Neural Networks (CNNs)-Based for Medical Image Analysis
551

accuracy and recall. ResNet, using deep architectures
and residual learning, excels in tasks requiring the
detection of subtle differences, benefiting from its
efficient data use. Transformers handle complex CT
scans effectively, including those for COVID-19, by
managing long-range dependencies with attention
mechanisms and adaptability to various patch sizes
and pretraining approaches. This analysis highlights
the distinct advantages and contributions of each
model to medical imaging technology.
4 CONCLUSIONS
This research focuses on evaluating deep learning
models such as CNN, ResNet, and Transformer in
medical image processing, with the objective of
enhancing diagnostic accuracy across various
imaging modalities. The study involves
methodological applications and analyses of each
model on different pathological datasets, including
interstitial lung diseases and knee joint injuries,
through ILD and MRI scans.
Extensive experiments were conducted to
evaluate the proposed methods. The experimental
results revealed that CNN excels in automatic feature
extraction, particularly in environments with limited
data and ambiguous visual structures. ResNet
demonstrated superior performance in managing
depth and complexity, significantly enhancing the
model's training and generalization capabilities in
deeper network architectures. Meanwhile,
Transformers displayed their advantage in handling
complex, high-dimensional image data, utilizing their
attention mechanisms to enhance model predictive
capabilities on large and diverse datasets.
Future research will explore integrating
multimodal imaging data to analyze the combined
effects of various imaging modalities using advanced
machine learning frameworks. This aims to enhance
diagnostic precision and robustness, addressing the
limits of single-modality analysis and advancing AI-
driven diagnostic tools in clinical settings, potentially
improving patient outcomes and healthcare efficiency.
REFERENCES
Dai, Y., Gao, Y., & Liu, F. 2021. Transmed: Transformers
advance multi-modal medical image classification.
Diagnostics, 11(8), 1384.
He, K., Gan, C., Li, Z., Rekik, I., Yin, Z., Ji, W., ... & Shen,
D. 2023. Transformers in medical image analysis.
Intelligent Medicine, 3(1), 59-78.
He, K., Zhang, X., Ren, S., & Sun, J. 2015. Spatial pyramid
pooling in deep convolutional networks for visual
recognition. IEEE transactions on pattern analysis and
machine intelligence, 37(9), 1904-1916.
Ildgdb. 2024. Dataset. http://ildgdb.org/
Jannin, P., Krupinski, E., & Warfield, S. K. 2006.
Validation in medical image processing. IEEE
Transactions on Medical Imaging, 25(11), 1405-9.
Krizhevsky, A., Sutskever, I., & Hinton, G. E. 2012.
Imagenet classification with deep convolutional neural
networks. Advances in neural information processing
systems, 25.
Lin, M., Chen, Q., & Yan, S. 2013. Network in network.
arXiv preprint arXiv:1312.4400.
Li, Q., Cai, W., Wang, X., Zhou, Y., Feng, D. D., & Chen,
M. 2014. Medical image classification with
convolutional neural network. In 2014 13th
international conference on control automation robotics
& vision (ICARCV) (pp. 844-848). IEEE.
Sarwinda, D., Paradisa, R. H., Bustamam, A., & Anggia, P.
2021. Deep learning in image classification using
residual network (ResNet) variants for detection of
colorectal cancer. Procedia Computer Science, 179,
423-431.
Simonyan, K., & Zisserman, A. 2014. Very deep
convolutional networks for large-scale image
recognition. arXiv preprint arXiv:1409.1556.
Sonka, M., & Fitzpatrick, J. M. 2000. Handbook of medical
imaging: Volume 2, Medical image processing and
analysis. SPIE.
Stanford. 2024. mrnet. https://stanfordmlgroup.github.io/
competitions/mrnet/
Touvron, H., Cord, M., Douze, M., Massa, F., Sablayrolles,
A., & Jégou, H. 2021. Training data-efficient image
transformers & distillation through attention. In
International conference on machine learning (pp.
10347-10357). PMLR.
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
552
