
Comparative Analysis of Brain Tumor Classification and Models
Based on VGG16
Yilin Chen
a
Faculty of Data Science, City University of Macao, Macao, China
Keywords: VGG16, Brain Tumor Diagnostics, Convolutional Neural Network.
Abstract: This study delves into the effectiveness of the Visual Geometry Group Network 16 (VGG16) convolutional
neural network (CNN) in the crucial task of classifying brain tumors, a pivotal endeavor aimed at enhancing
diagnostic accuracy and tailoring patient treatment in the field of oncology. Leveraging the renowned VGG16
model, celebrated for its deep architecture and robust feature extraction capabilities, this research seeks to
propel the accuracy of brain tumor diagnostics to new heights. Through a meticulously crafted methodology
encompassing comprehensive image preprocessing, meticulous optimization of the VGG16 model, and
meticulous comparison with other CNN models, the study meticulously evaluates crucial metrics such as
accuracy, sensitivity, and specificity. Drawing upon a rich dataset of brain tumor images for analysis, the
findings underscore VGG16's superior classification performance, highlighting its profound potential to
revolutionize medical imaging practices and elevate the standard of patient care in oncology. These
compelling results not only bolster the utilization of deep learning techniques in medical diagnostics but also
pave the way for future advancements in personalized healthcare methodologies.
1 INTRODUCTION
Brain cancer, a formidable adversary in oncology,
remains a leading cause of cancer-related morbidity
and mortality worldwide (Harachi, 2024). The
heterogeneity of brain tumors, with their complex
biological characteristics, presents a significant
challenge for accurate diagnosis and classification,
crucial for effective treatment planning (Xie, 2024).
The advent of Convolutional Neural Networks
(CNNs) has opened new vistas in medical image
analysis, offering a potential leap forward in the
precision of brain tumor classification (Irgolitsch,
2024). The significance of this research lies in
harnessing the power of CNNs, particularly the
Visual Geometry Group Network 16 (VGG16)
model, to improve the accuracy and efficiency of
brain tumor diagnosis, thereby contributing to
personalized medicine and better patient outcomes.
In the realm of medical image processing, CNNs
have emerged as a transformative force, particularly
in cancer diagnosis, including brain tumors. with
numerous studies demonstrating their efficacy in
classifying various types of cancers, including those
a
https://orcid.org/0009-0000-5833-2153
of the brain. In the domain of medical imaging,
particularly brain tumor classification, significant
strides have been made with the adoption of CNNs.
Research has evolved from traditional image
processing methods to advanced deep learning
models, with VGG16 emerging as a key player due to
its deep architecture and superior feature extraction
capabilitiesVGG16, a deep CNN architecture, has
been particularly noted for its success in image
recognition tasks due to its depth and robust feature
extraction capabilities(Jahannia, 2024). Previous
research has leveraged VGG16 for brain tumor
classification, achieving promising results that
underscore the model's potential in medical
applications(Khaliki, 2024). The VGG16
architecture, known for its depth and robust feature
extraction, has played a crucial role in this progress.
It has been effectively used for brain tumor
classification, showcasing the potential of deep
learning in medical applications. However, the
integration of CNNs in clinical workflows is still in
its infancy, with ongoing debates regarding model
interpretability, data privacy, and the need for large,
annotated datasets(Sachdeva, 2024). This study aims
652
Chen, Y.
Comparative Analysis of Brain Tumor Classification and Models Based on VGG16.
DOI: 10.5220/0012961300004508
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Engineering Management, Information Technology and Intelligence (EMITI 2024), pages 652-658
ISBN: 978-989-758-713-9
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
to build upon the existing body of work, addressing
some of these challenges and pushing the boundaries
of what is currently achievable with CNN-based brain
tumor classification. The exploration of CNNs,
especially VGG16, in brain tumor classification not
only enhances diagnostic precision(Reddya, 2024)
but also paves the way for novel therapeutic
strategies. By accurately categorizing brain tumors,
clinicians can tailor treatments to individual patients,
thereby optimizing outcomes and minimizing adverse
effects(Balajee, 2024). Moreover, the integration of
CNNs into clinical decision-making processes
underscores the convergence of technology and
healthcare, promising a future where medical
interventions are more data-driven and patient-
specific(Yalamanchili, 2024). While the journey
toward fully integrating CNNs into routine clinical
practice is fraught with challenges, the potential
benefits in terms of improved diagnostic accuracy,
patient outcomes, and healthcare efficiency are
immense. The ongoing research and development in
this area are crucial steps toward realizing the full
potential of CNNs in medical imaging and oncology,
signifying a paradigm shift in how brain cancer is
diagnosed and treated(Rahman, 2024).
This study utilizes the VGG16 model to develop
a robust framework for brain cancer detection,
meticulously adjusting its parameters to gauge their
impact on model performance. Renowned for its deep
architecture and remarkable efficacy in feature
extraction, VGG16 plays a pivotal role in accurately
identifying various brain tumor types. To further
enhance the quality of the data, advanced image
preprocessing techniques are incorporated, thereby
augmenting the model's learning capabilities and
predictive accuracy. A comprehensive comparative
analysis with other CNN models is undertaken, with
a keen focus on key metrics such as accuracy,
sensitivity, and specificity in tumor classification.
Furthermore, the research evaluates VGG16's
scalability and consistency across diverse datasets
and operational scenarios, demonstrating its
adaptability in real-world settings. The results
unequivocally demonstrate that VGG16, coupled
with effective preprocessing techniques, surpasses
conventional models, representing a significant leap
forward in medical imaging. This advancement holds
both theoretical and practical implications, enhancing
diagnostic accuracy and potentially improving patient
treatment outcomes. Moreover, these findings
underscore the immense potential of deep learning in
medical diagnostics, paving the way for impactful
future research endeavors and clinical applications in
the realm of healthcare.
2 METHODOLOGIES
2.1 Dataset Description and
Preprocessing
The dataset used in this study is called the brain tumor
dataset and is derived from the Kaggle (Seif, 2024).
The dataset includes brain Magnetic Resonance
Imaging (MRI) scans obtained from patients with and
without brain malignancies. Each image acquire is
labeled with "Yes" or "No" to indicate the presence
or absence of the tumor. The aim here is to determine
the presence of tumors in the patient based on
magnetic resonance imaging. The training data set
contains 253 images, and before model development,
this study applies normalization techniques to
standardize the pixel values in the images.
Furthermore, this study applies the cropping function
to focus on regions of interest in brain MRI images,
which helps to reduce noise and irrelevant
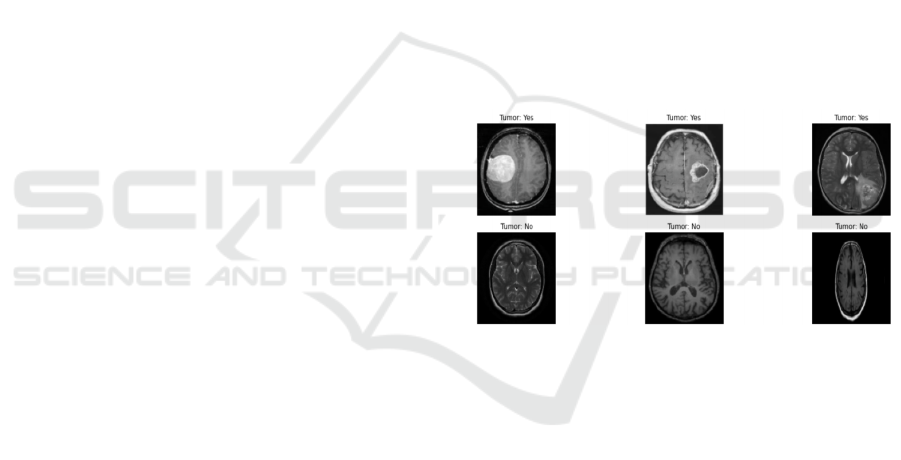
information. Figure 1 shows some instances coming
from this dataset.
Figure 1: Part of the brain_tumor_dataset dataset
(Photo/Picture credit: Original).
2.2 Proposed Approach
Design and train CNN architectures specifically for
brain tumor detection, performing experiments using
different network architectures, regularization
techniques, and hyperparameters to optimize model
performance. This CNN model is a typical deep
learning model used for emotion classification tasks.
It contains multiple convolutional and pooling layers,
as well as fully connected and Dropout layers,
ultimately exporting a sigmoid-activated neuron for
the dichotomy task. The overall flow chart of the
model is shown in Figure 2.
Comparative Analysis of Brain Tumor Classification and Models Based on VGG16
653
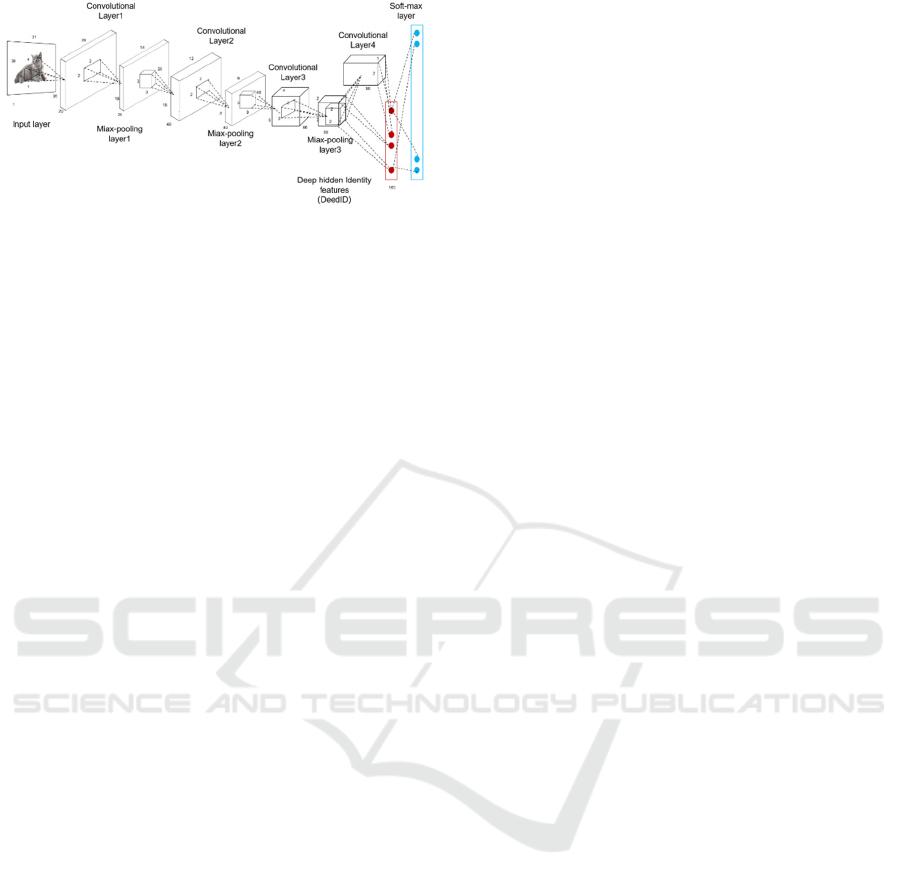
Figure 2: Flow diagram of the CNN network
(Photo/Picture
credit: Original).
The core construction of the model lies in its
clever use of multiple convolution layers and pooling
layers, which are connected and work together to
gradually extract various features from the input
images. These features start with the base edges and
textures and gradually upgrade to more complex and
higher-level feature representations. This hierarchical
feature extraction method enables the model to have
a deeper understanding of the image content, and lays
a solid foundation for the subsequent classification
task. After feature extraction, the model further
integrates and maps these features using the fully
connected layer. The fully connected layer transforms
the extracted features into a form corresponding to the
final output category by learning the weight and bias.
In this way, the model can predict the category of the
input image based on its features.
To prevent overfitting of the model during
training, this paper introduces the dropout layer. The
dropout layer is discarding some connections of
neurons randomly during training, so that the model
does not rely too much on some specific features or
weights to improve its generalization ability. At the
output end of the model, this study employs a neuron
with an s-type activation function. This neuron
transforms the model's predictions into a value
between 0 and 1, representing the probability that the
image belongs to a certain class. This probabilistic
output mode enables the model to show its prediction
results more intuitively and facilitates us to make
subsequent threshold setting and classification
decisions.
To optimize the training process of the model, this
study chooses the binary cross-entropy as the loss
function, which is well suited for scenarios with
dichotomous tasks. Meanwhile, this study also
adopted the Adam optimizer and set the learning rate
to 1e-4 to ensure that the model can be quickly and
stably converged to the optimal solution. This study
also performs a series of preprocessing steps before
the images enter the network. These steps include
resizing the images, normalizing them, and applying
data augmentation techniques. Through these
preprocessing measures, this study could not only
ensure the consistency and standardization of the
input data, but also improve the robustness of the
model, so that it can better cope with various complex
image changes and challenges.
2.2.1 Training Parameter Setting
During training, 50 training epochs were set and each
batch containing 32 samples to ensure that the model
was adequately learned and adapted to the data.
Meanwhile, to avoid overfitting of the model during
training, an early stopping strategy was used. When
the loss of the validation set does not significantly
improve over the five consecutive epochs, the
training ends early, thus preserving the performance
of the model at the best state. Before the training
started, this study performed a series of preprocessing
operations on the images. These operations include
adjusting the image size to meet the input
requirements of the model, performing normalization
processing to eliminate differences in brightness and
contrast between different images, and applying data
augmentation techniques to increase the diversity of
training samples and improve the robustness of the
model. The core part of the model consists of multiple
convolution layers and pooling layers. These
hierarchies enable automatic learning and extracting
key features in images, ranging from lower-level edge
and texture information to higher-level emotion-
related features. By passing and processing layer by
layer, the model can gradually deepen the
understanding of the image content. After feature
extraction, the model maps these features to the final
output category through a fully connected layer. The
fully connect layer transforms the prediction results
of the model into specific emotion classification
labels by learning and integrating feature information.
To prevent model overfitting, this study introduces
Dropout layers between the fully connected layers.
The Dropout layer randomly discards the connections
of some neurons during training, so that the model
does not rely too much on some specific features or
weights, thus improving its generalization ability.
During training, focus on loss and accuracy changes
in the training and validation sets. By monitoring
these indicators, this study could understand the
training state of the model and find and deal with
possible problems in time. Once the training is
complete, this study could use the model to classify
the new images emotionally and evaluate their
performance. The key to this classification method
lies in the rational design of the model structure and
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
654
the training strategy. By continuously optimizing the
model parameters and the training process, this study
could improve the performance of the model in the
emotion classification task, so as to better understand
and analyze the information about the presence of
tumors in the image.
2.2.2 Loss Function
Loss function plays a crucial role in machine learning
and deep learning, measuring the difference between
model prediction results and actual labels, and is a
key indicator in the model optimization process.
Through the loss function, that can quantify the
performance of the model on the training set, and then
adjust the model parameters through the optimization
algorithm (such as gradient descent), so that the
model can better fit the data. Different tasks and
models may need to use different loss functions, and
common loss functions include mean square error
(MSE), cross-entropy, etc. Choosing the appropriate
loss function can help the model to better learn the
features of the data and improve the generalization
ability and accuracy of the model. Therefore, the loss
function can be regarded as an objective function
guiding the model learning and is an integral part of
the model training process. Here the binary crossover
loss function is used and the formula is as follows:
1
log( ( ))
1
(1 ) log(1 ( ))
N
ii
i
ii
ypy
Loss
ypy
N
=
+
=−
−−
(1)
For the binary label y, the value is not 0 or 1, while
p (y) indicates the probability that the output belongs
to the y label. As a key indicator of the prediction
effect of binary classification models, the binary
cross-entropy loss function plays a crucial role in
evaluating the model performance. In short, when the
label y is 1, if the p (y) value predicted by the model
is close to 1, it means that the prediction of the model
is highly consistent with the true label. At this time,
the value of the loss function should be close to 0,
indicating that the prediction effect of the model is
very good. On the other hand, if the p (y) value tends
to 0, that is, the model mistakenly predicts the sample
with label 1 to 0, the value of the loss function will
become very lase binary cross-entropy loss function
to effectively guide the model optimization, thus
improving the prediction accuracy of the binary
classification task.
3 RESULTS AND DISCUSSION
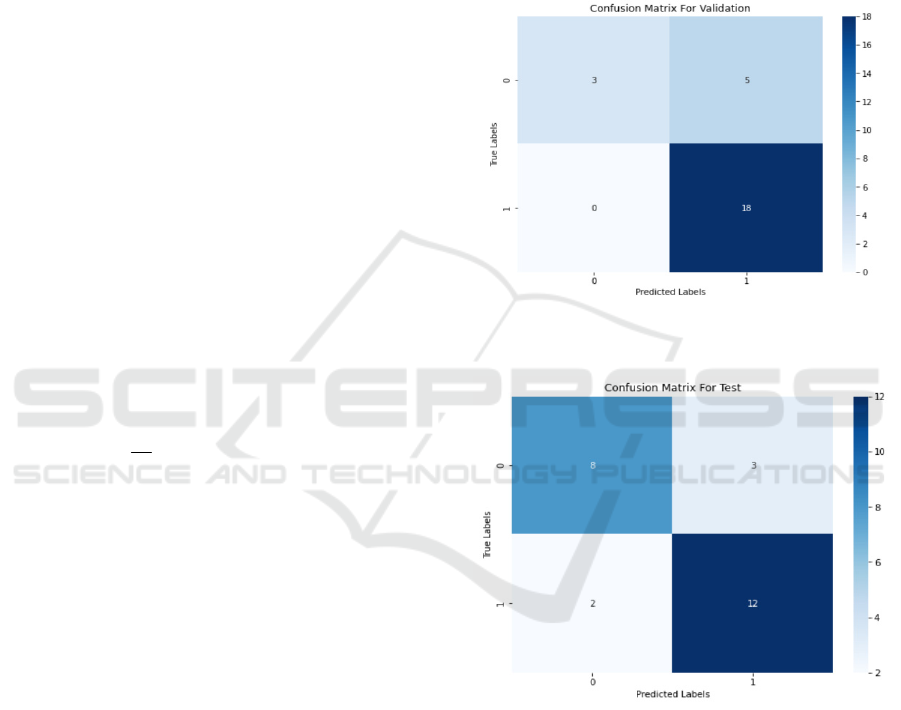
3.1 Confusion Matrix
The confusion matrix represents the correspondence
between the predicted results and the true labels of the
model on the test set. The values of the confusion
matrix are slightly different compared to the
validation set, but the overall trend is similar.
Figure 3: Confusion matrix plot used for the validation
(Photo/Picture credit: Original).
Figure 4: Test the confusion matrix plot (Photo/Picture
credit: Original).
Figure 3 The model performs poorly on the
predicted category 0 (negative class), with a certain
number of false positive classes (misclassifying
negative classes as positive classes). The model
performed well on predictive category 1 (positive),
with most being correctly classified. The model
performs well on the validation set, but there is some
room for improvement in identifying negative
classes. This analysis helps us to understand how the
Comparative Analysis of Brain Tumor Classification and Models Based on VGG16
655
model predicts on different categories, and thus guide
us to further adjustments and improvements.
Figure 4 The overall performance of the model on
the test set is relatively stable, like that on the
validation set. The number of false positive and false
negative classes increased slightly on the test set
compared to the validation set, but the overall
accuracy remained at a high level. Overall, the model
performed well on the test set, effectively
distinguishing between the two classes of samples,
but a small number of samples were still
misclassified. This analysis helps us to evaluate the
practical application of the model and provide a
reference for further improvement.
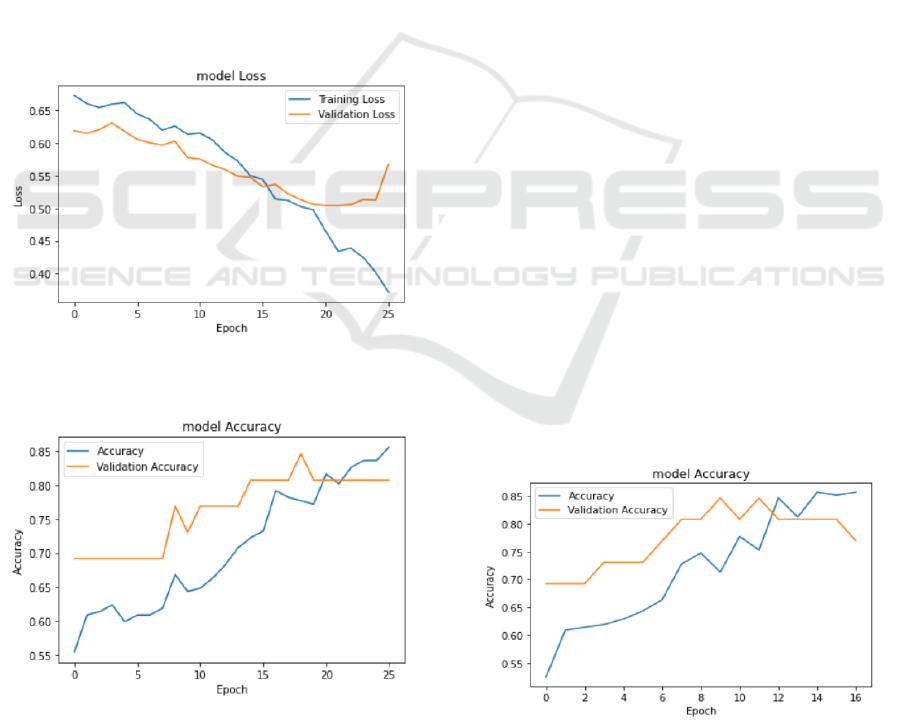
3.2 Plot of the Model Results
During the training process, with the increase of
epoch, the loss on the training set and the validation
set gradually decreases, and the accuracy gradually
improves.
Figure 5: Model loss function (Photo/Picture credit:
Original).
Figure 6: Model accuracy map (Photo/Picture credit:
Original).
Figure 5 With the increase of the epoch, the loss
value of the model gradually decreases in both the
training set and the validation set. This shows that the
model has gradually learned the characteristics of the
data during the training process and has made some
progress. However, it should be noted that the loss
values on the validation set do not always drop and
sometimes fluctuate, which may be caused by some
difficulties in the model during the training process or
noise from the data. Therefore, this study needs to
consider the performance on the training and
validation sets to comprehensively evaluate the
performance of the model.
Figure 6 shows that the accuracy of the model on
the training and validation sets increases with the
epochs. This shows that the model has gradually
learned the characteristics of the data during the
training process and has made some progress.
However, it should be noted that although the
accuracy of the model on the training set is constantly
increasing, the accuracy on the validation set is not
always increased, and sometimes it fluctuates or even
slightly decreases. This may be due to overfitting of
the model during training or noise from the data.
3.3 Adjust the Model Parameters
The original three convolution layers and pooling
layers were increased to three, and the number of
convolution kernels was adjusted to 64,128 and 256,
respectively, keeping the convolution kernel size at
(3,3) and the pooling layer size at (2,2). The number
of neurons in the fully connected layer adjusted the
original 128 neurons to 256.
With the increase of epoch, the accuracy of the
model on the training set was gradually improved,
from about 52.5% to about 85.6%. On the validation
set, the accuracy of the model showed a similar trend,
increasing from the initial ~ 69.2% to the final ~
80.8%, as shown in Figure 7.
Figure 7: Model accuracy chart (Photo/Picture credit:
Original).
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
656

The model has certain generalization ability and can
perform well on unseen data. In later times, although
the model accuracy continued to improve on the
training set, the accuracy on the validation set began
to fluctuate and did not consistently improve. This
may be a sign of model overfitting and can adjust the
model structure or hyperparameters according to its
performance on the validation set to further improve
the model performance and generalization ability.
Possible adjustments include increasing data increase,
adjusting learning rate, adjusting network structure, etc.
3.4 Image Enhancement
Use ImageDataGenerator to perform image data
augmentation and create a data generator for the
training and validation sets. Specific data
enhancement operations include random rotation,
horizontal flip, vertical flip, random width, and height
offset, shear, and random scaling, etc. These
operations can increase the diversity of the data and
help to improve the generalization ability of the
model. The data generator of the training set uses the
data augmentation operation, while the data generator
of the validation set does not use the data
augmentation, maintaining the state of the original
data. In this way, the model can dynamically acquire
the enhanced data during training, further improving
the training effect, showing the partially enhanced
image as shown in Figure 8.
Figure 8: Image enhancement part of the image display
(Photo/Picture credit: Original).
3.5 VGG16 Model
The pre-trained VGG 16 model was used as the base
model, and several layers of global average pooling,
full connectivity and dropout layers were added to the
top, finally exporting a predicted layer of sigmoid
activation.
The pre-trained VGG 16 model was used, where
include_top=False represents the full connected layer
without the top, and input_shape= (128,128,3)
specifies the size of the input image as 128x128 pixels,
3 channels. All layers of VGG 16 were set to be
untrainable, that is, base_model.layers[0]. trainable =
False, so that the weights of these layers are not
updated during the training process, but only the
weights of the new layers. A global average pooling
layer GlobalAveragePooling2D () was added to the
output of the underlying model to transform the
feature map into a fixed length vector. Then comes a
fully connected layer, Dense (128, activation=
'relu',kernel_regularizer=regularizers.l2 (0.001)),
using the ReLU activation function and L2
regularization. A dropout layer Dropout (0.5) was
added to reduce overfitting. Finally, a Dense layer
was added as the output layer, using the sigmoid
activation function, for the deodorization task. The
model was compiled using the model. Compile
method, assigning the loss function as dichotomy
cross-entropy binary crossentropy, the optimizer as
Adam, with a learning rate of 1e-5, and the evaluation
index as accuracy.
EarlyStopping Used to stop training when the
validation set loss is no longer reduced to avoid
overfitting. The parameter monitor= 'val_loss'
represents the loss value of the monitored validation
set, patience=5 means that it stops training if the loss
is not reduced for five consecutive epoch validation
sets and restore_best_weights=True indicates the
weight restored to the best model when training is
stopped. ReduceLROnPlateau Is used to lower the
learning rate when the validation set loss is no longer
reduced to help the model converge better. The
parameter monitor= 'val_loss' represents the loss
value of the monitored validation set, factor=0.1
represents the factor by which the learning rate will
be reduced, patience=5 reduces the learning rate if the
loss of five consecutive epoch validation sets is not
reduced, and min _ lr = 1 e-7 represents the lower
limit of the learning rate. The model training was
performed using the model.fit method, assigning the
training set data generator train_generator and the
validation set data generator val_generator, training
20 epochs, and assigning the callback function as
[early_stopping, reduce _ lr]. The callback function
set like this can effectively control the training
process of the model, avoid overfitting, and achieve
better performance on the validation set.
Comparative Analysis of Brain Tumor Classification and Models Based on VGG16
657
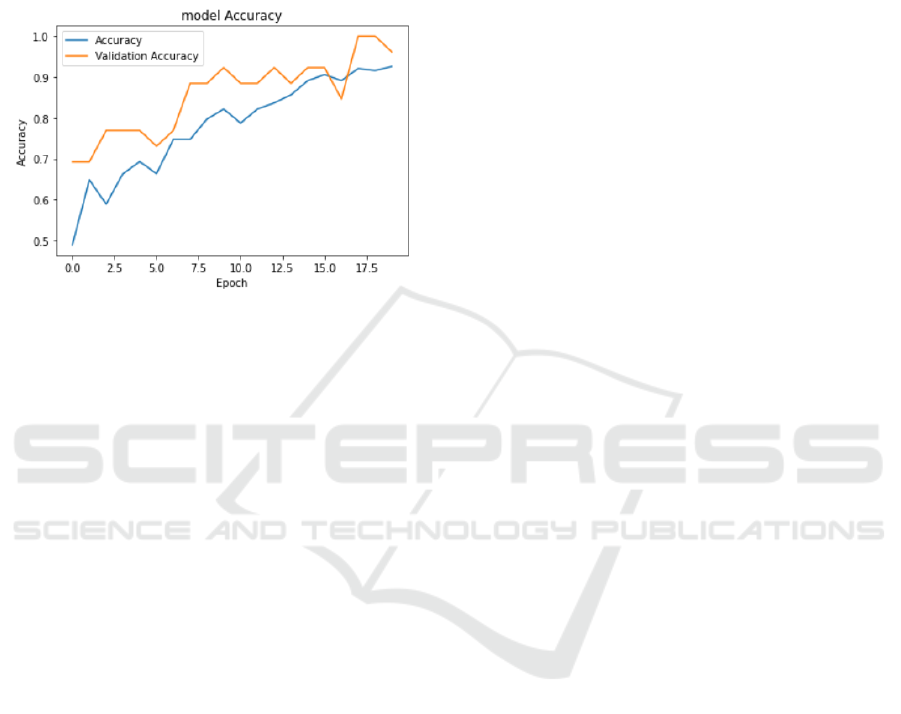
The accuracy of the model gradually improved during
the training process, finally achieving an accuracy of
about 92.6% on the training set and the highest
accuracy of about 96.2% on the validation set. This
shows that the model effectively learns the features of
the data during training and achieves good
performance on the validation set. As shown in Fig. 9.
Figure 9: Training accuracy of the VGG 16 model
(Photo/Picture credit: Original).
4 CONCLUSIONS
Firstly, this study embarked on a sentiment
classification project employing a CNN, commencing
with the utilization of a simple CNN model and
evaluating its performance on both training and
validation datasets. Subsequently, the focus shifted
towards leveraging the VGG16 model and fine-
tuning it, while integrating data augmentation
techniques to enhance the model's generalization
capabilities. For the simple CNN model, the observed
accuracy on the training and validation sets attained
approximately 85.6% and 80.8%, respectively.
Following the adoption of the VGG16 model and
fine-tuning approach, the accuracy on the training and
validation sets surged to around 92.6% and 96.2%,
respectively. Through the adjustment of the CNN
model's structure and parameters, alongside the fine-
tuning of the VGG16 model, endeavors were made to
bolster the model's performance. Noteworthy
callback functions such as Early Stopping and Reduce
learning rate On Plateau are deployed to monitor
model performance and dynamically adjust during
training iterations. While commendable results were
achieved with both the simple CNN and VGG16
models, superior performance was evident with the
VGG16 model, particularly in terms of accuracy on
the validation set.
REFERENCES
Balajee, A., Bharat, B., Mounica, N., & Mandava, S. (2024,
March). Brain tumour detection using deep learning. In
AIP Conference Proceedings, vol. 2966(1).
Harachi, M., Masui, K., Shimizu, E., Murakami, K.,
Onizuka, H., Muragaki, Y., ... & Shibata, N. (2024).
DNA hypomethylator phenotype reprograms
glutamatergic network in receptor tyrosine kinase gene-
mutated glioblastoma. Acta Neuropathologica
Communications, vol. 12(1), p: 40.
Irgolitsch, F., Huppé-Marcoux, F., Lesage, F., & Lefebvre,
J. (2024, March). Slice to volume registration using
neural networks for serial optical coherence
tomography of whole mouse brains. In Computational
Optical Imaging and Artificial Intelligence in
Biomedical Sciences, vol. 12857, pp: 107-115.
Jahannia, B., Ye, J., Altaleb, S., Peserico, N., Asadizanjani,
N., Heidari, E., ... & Dalir, H. (2024, March). Low-
latency full precision optical convolutional neural
network accelerator. In AI and Optical Data Sciences V,
vol. 12903, pp: 26-39.
Khaliki, M. Z., & Başarslan, M. S. (2024). Brain tumor
detection from images and comparison with transfer
learning methods and 3-layer CNN. Scientific Reports,
vol. 14(1), p: 2664.
Rahman, A. (2024). Brain tumour detection and
classification by using CNN (Master's thesis, Itä-
Suomen yliopisto).
Reddy, L. C. S., Elangovan, M., Vamsikrishna, M., &
Ravindra, C. (2024). Brain Tumor Detection and
Classification Using Deep Learning Models on MRI
Scans. EAI Endorsed Transactions on Pervasive Health
and Technology, vol. 10.
Sachdeva, J., Sharma, D., & Ahuja, C. K. (2024).
Comparative Analysis of Different Deep Convolutional
Neural Network Architectures for Classification of
Brain Tumor on Magnetic Resonance Images. Archives
of Computational Methods in Engineering, pp: 1-20.
Seif W., (2024). Brain Tumor Detection. https://www.
kaggle.com/code/seifwael123/brain-tumor-
classification-cnn-vgg16
Xie, H., Zhang, B., Xia, T., Cui, J., Pan, F., Li, Y., ... & Liu,
Y. (2024). Ezrin Thr567 phosphorylation participates in
mouse oocyte maturation, fertilization, and early
embryonic development. Fertilization, and Early
Embryonic Development.
Yalamanchili, S., Yenuga, P., Burla, N., Jonnadula, H., &
Bolem, S. C. (2024). MRI Brain Tumor Analysis on
Improved VGG-16 and Efficient NetB7 Models.
Journal of Image and Graphics, vol. 12(1).
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
658
