
The Comparison of Diabetes Risk Prediction Accuracy Across
Different Models
Jing Yang
1a
and Ruolan Zheng
2b
1
School of Software Engineering, Shan Dong University, Jinan, 250101, China
2
School of Mathematics and Statistics, Nanjing University of Science and Technology, Nanjing, 210094, China
Keywords: Disease Prediction, Support Vector Machine, Random Forest, Multilayer Perceptron.
Abstract: The integration of machine learning technologies, particularly deep learning models, has significantly
advanced disease prediction within the healthcare industry. People have begun leveraging large-scale medical
data for research, combining various models aimed at enhancing the accuracy of medical disease prediction.
This study focuses on diabetes, a severe disease, and employs four different machine learning algorithms:
logistic regression, multilayer perceptron, support vector machine, and random forest. The author utilizes a
dataset obtained through direct questionnaire surveys of patients at the Sirhaj Diabetic Hospital in Bangladesh,
and conducts systematic data processing and visualization using the Python language to compare the strengths,
weaknesses, and effectiveness of these four models in disease prediction. This research aims to provide more
accurate and reliable tools for predicting the risk of diabetes in the healthcare field. Not only can this help
doctors better understand the health status of patients, but it can also provide crucial reference for personalized
treatment plans and preventive measures, thereby improving the cure rate of various major diseases.
1 INTRODUCTION
The healthcare sector has consistently been a frontier
for the remarkable success of deep learning
technology. The potent capabilities of deep learning
in pattern recognition and data analysis have
positioned it as a robust tool for addressing medical
challenges and enhancing patient care. In the realm of
healthcare, early prediction of severe diseases holds
paramount significance. Firstly, forecasting the future
progression of diseases aids in the timely
identification and preemptive intervention for
patients, thereby augmenting the chances of recovery.
Secondly, predictive models assist healthcare teams
in tailoring more precise and personalized medical
interventions, maximizing the efficacy of treatments.
Over the past few years, the area of disease
prediction has seen notable developments, with
numerous researchers achieving noteworthy
breakthroughs. In their investigation, Hosseininian
and colleagues (2024) employed a feature selection
approach grounded in ANOVA, or analysis of
variance to pinpoint the most pertinent features
a
https://orcid.org/0009-0004-6618-3257
b
https://orcid.org/0009-0008-4454-0371
concerning the prediction of Parkinson's illness by
means of using voice-based characteristics.
Subsequently, the Echo State Network (ESN) is
evaluated as a predictive model for this purpose.
Multiple models, encompassing the Echo State
Network, Decision trees, Support Vector Classifiers,
Random Forests, K-Nearest Neighbors, and Extreme
Gradient Boosting, were meticulously evaluated and
compared within the framework of the study. The
analysis of results highlights the exceptional
performance of the Echo State Network (ESN) among
the employed models (Hosseininian et al, 2024). Niu
et al. introduced an adaptive algorithm that improved
upon the conventional Grey Wolf Optimization
(GWO) with a sigmoid function, resulting in an
enhanced version of the Grey Wolf Optimization
algorithm for heart disease prediction (Niu et al,
2024). Wang et al. (2024) introduced an innovative
approach termed Hybrid Ordinal Prototype
Embedding (HOPE) in their research, aiming to
delineate the sequential advancement of Alzheimer's
disease for the prediction of mild cognitive
impairment progression. Mary studied supervised
Yang, J. and Zheng, R.
The Comparison of Diabetes Risk Prediction Accuracy Across Different Models.
DOI: 10.5220/0012968900004508
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Engineering Management, Information Technology and Intelligence (EMITI 2024), pages 697-703
ISBN: 978-989-758-713-9
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
697

machine learning algorithms for predicting heart
disease (Kanchan 2016). Frasca and colleagues
(2023) conducted an investigation utilizing the
dataset from the "Parkinson's Progression Markers
Initiative." Their study involved the development of a
deep learning model dedicated to discerning the
various stages of progression in Parkinson's disease.
The conclusive 3DCNN + LSTM model, trained and
assessed on MRI images, demonstrated exceptional
performance in accurately classifying the four distinct
levels of Parkinson's disease progression according to
the Hoehn and Yahr scale. This state-of-the-art model
achieved a macro-averaged One-Versus-Rest Area
Under the Curve (OVR AUC) of 91.90%.
However, the domain of disease prediction
confronts several difficulties and challenges. Issues
such as limited sample sizes for rare diseases leading
to reduced prediction accuracy, the complexity and
uncertainty inherent in disease prediction data, and
the inability of existing models to provide robust
predictions for unknown diseases pose significant
hurdles. Scholars have undertaken targeted research
efforts to address these challenges. The use of bio-
inspired optimization methods, such as the Genetic
Algorithm, Particle Swarm Optimization, and Whale
Optimization Algorithm, was examined by Dyoub et
al, for the purpose of selecting relevant features in
predicting chronic diseases. The primary aim of their
study was to improve the models' ability to anticipate
outcomes, simplify dimension of the data, and make
the resulting predictions more comprehensible and
applicable in a practical context (Dyoub and Letteri,
2023). Saba et al. (2024) introduced a novel
Continual Learning (CEL) model based on domain
adaptation and Elastic Weight Consolidation (EWC),
aiming to alleviate errors caused by catastrophic
forgetting in deep neural networks, such as LSTM, in
the context of incremental domain settings. In their
2019 study, Uddin and colleagues delved into the
predominant trends characterizing diverse categories
of supervised machine learning algorithms,
elucidating both their performance metrics and
practical applications in the realm of disease risk
prediction. Conclusions were drawn through a
comparative analysis. Dahiwade et al. (2019)
discovered that, in this general disease prediction,
considering individuals' lifestyle habits and
examination information enables accurate
forecasting. Patro et al. (2021) introduced an
innovative framework for predicting heart disease,
focusing on key risk factors and employing a variety
of classifiers, including Naïve Bayes (NB), Bayesian
Optimized Support Vector Machine (BO-SVM), K-
Nearest Neighbors (KNN), and Salp Swarm
Optimized Neural Network (SSA-NN). In their
investigation, the Convolutional Neural Network
(CNN) demonstrated a superior accuracy of 84.5% in
general disease prediction compared to the
performance of the KNN algorithm, or K-Nearest
Neighbors. The proposed optimization algorithm in
this framework aims to establish an effective
healthcare monitoring system for the early detection
of heart disease. This paper employs four algorithms
– Random Forest, Support Vector Machine (SVM),
Multilayer Perceptron (MLP), and logistic
regression-to foresee the likelihood of an individual
developing a specific illness. The accuracy of these
four algorithms will be compared to explore ways to
enhance the prediction capabilities for severe
diseases, consequently elevating the potential for
early detection and intervention. This introduction
sets the stage for a comprehensive exploration of
disease prediction methodologies and their potential
impact on healthcare outcomes.
2 METHODS
2.1 Data Source
These data are sourced from the UC Irvine Machine
Learning Repository. The data collection involved
direct questionnaire surveys with patients from Sylhet
Diabetes Hospital in Bangladesh, and it has received
approval from a doctor.
2.2 Indicator Selection and Description
Table 1 displays various symptoms and health
characteristics related to diabetes in the dataset used
for the study.
Table 1: Variable information.
Sympto
m
Introduction
Gende
r
1=Male, 0=Female
Pol
y
uria 1=Yes, 0=No
Pol
y
di
p
sia 1=Yes, 0=No
Sudden Weight Loss 1=Yes, 0=No
Weakness 1=Yes, 0=No
Polyphagia 1=Yes, 0=No
Genital Thrush 1=Yes, 0=No
Visual Blurrin 1=Yes, 0=No
Itchin
g
1=Yes, 0=No
Irritabilit
y
1=Yes, 0=No
Dela
y
ed Healin
g
1=Yes, 0=No
Partial Paresis 1=Yes, 0=No
Muscle Stiffness 1=Yes, 0=No
Obesity 1=Yes, 0=No
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
698

2.3 Method Introduction
2.3.1 Data Processing
In this experiment, Python is used to systematically
process the data, employing the Pandas library to read
the dataset related to diabetes. The initial step
involves feature encoding, where the Label Encoder
is utilized to convert binary categorical features (such
as gender, presence of polyuria, etc.) into numerical
representations. For multi-categorical features, the
One Hot Encoder is used for independent encoding.
Subsequently, label encoding is performed using the
Label Encoder to convert categorical labels into
numerical format.
After that, the data set is separated into a training
and a test set, typically with an 80-20 ratio. Finally,
data standardization is conducted to normalize the
features, ensuring consistent scales across different
features, and enhancing the stability of the model
during training. This standardization is achieved
using the Standard Scaler function.
2.3.2 Support Vector Machine Model
Support Vector Machine is a supervised learning
algorithm suitable for binary classification problems.
Its main idea is to find an optimal hyperplane that
separates data points belonging to different types.
And then a linear kernel function is used to map the
data into a high-dimensional space, making it easier
to separate in the new space.
In the SVM model, the data was initially divided
into feature variables (X) and target variables (y).
Categorical features were processed using one-hot
encoding, while Label Encoder was employed to
convert the target variable into numerical form. The
data was divided into test and training sets in a two to
eight ratio. An SVM model was constructed with a
linear kernel function and a regularization parameter
C=1. Model performance was verified using 5-fold
cross-validation. The learning_curve function was
utilized to visualize the accuracy trends of the model
with different numbers of training samples.
2.3.3 Random Forest Model
The quarterly differential autoregressive moving
equilibrium model (SARIMA model) is an
improvement on the (ARIMA model), which is used
to transform non-smooth time series into smooth
periodic series. The model achieves the difference
and autoregressive moving average of the time series
by regression of the current and lagging values of the
dependent variable, as well as taking into account the
random error term. This transformation makes the
time series smoother and better able to capture
periodic features.
In the random forest model, categorical variables
were initially mapped to numerical representations.
Subsequently, the dataset was partitioned into feature
variables (X) and target variables (Y), and a random
forest classifier was employed for binary
classification. Model performance was assessed
through 5-fold cross-validation, followed by an
analysis of learning curves to examine the model's
ability at various training set sizes. The accuracy from
cross-validation was ultimately derived, and the
swerve of the model's training and validation
performance with increasing training data was
visualized using learning curve plots.
2.3.4 Multilayer Perceptron Model
A Multilayer Perceptron model is an artificial neural
network composed of multiple layers of neurons,
such as input, hidden and an output layer. Every
neuron has a connection with all neurons in the
preceding layer and is associated with weights and
biases. MLP captures complex relationships between
input features by learning these weights and biases.
In the MLP model, categorical variables were
initially converted into numerical representations
using Label Encoder. Subsequently, feature variables
(X) and target variables (y) were extracted. Following
this, Standard Scaler was applied to standardize the
features, ensuring uniform scales. The MLP model
was defined, comprising two hidden layers with 100
and 50 neurons, respectively, and a maximum
iteration limit set to 500. 5-fold cross-validation was
performed using Stratified K Fold, recording training,
and testing accuracies, as well as losses for each fold.
Finally, charts depicting the variation of training loss
and training/testing accuracies with the number of
iterations were generated to analyze the model's
performance.
2.3.5 Measurement
The Logistic Regression is a linear model which is the
main tool to solve binary classification problems. It
achieves the result by passing the weighted sum of
input features through a sigmoid function, mapping
the input to a probability range between 0 and 1,
facilitating classification.
In the logistic model, the data was initially
preprocessed and features were standardized.
Subsequently, the data was split into a training and a
testing set. This model was defined as a logistic
regression model with a linear layer, a Sigmoid
The Comparison of Diabetes Risk Prediction Accuracy Across Different Models
699
activation function, BCELoss loss function, and
trained for 500 epochs using the SGD optimizer.
Learning curves were employed to illustrate the
trends in training and testing accuracies. Ultimately,
the data of the model accuracy on the testing set was
reported.
3 RESULTS AND DISCUSSION
3.1 Analysis of Results
This study utilizes the pre-defined training set and
employs the Python language for visualizing the
output.
3.1.1 SVM Model Results
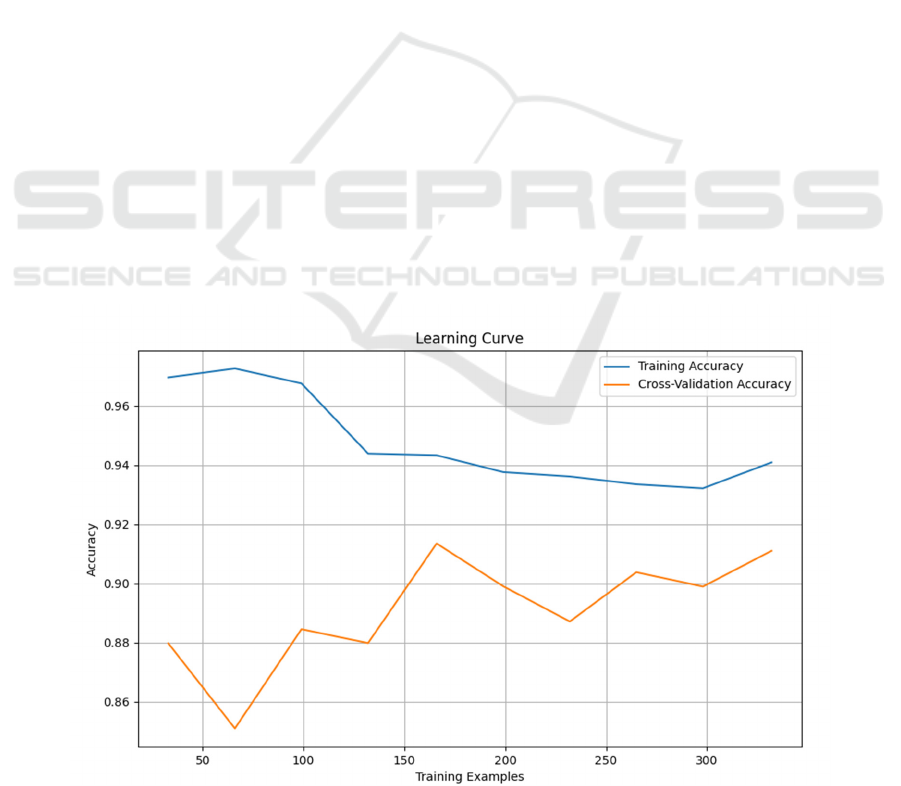
From Figure 1, with the increase in the number of
training samples, the Training Accuracy initially
experiences a slight decline before stabilizing. The
initial decrease may be attributed to overfitting noise
or local features when training on a small amount of
data. As the dataset grows, the model generalizes
better to overall features.
Test Accuracy exhibits fluctuations, but the
overall trend is a slow increase. The initial
fluctuations may arise from suboptimal performance
on specific validation sets during cross-validation
with a small dataset. With an increase in data, the
model's performance averages across different
validation sets, leading to a gradual rise in Test
Accuracy.
The trend of accuracy shows that the SVM has a
good performance on the training set without
significant overfitting. The gradual increase in Cross-
Validation Accuracy suggests that with more data, the
model performs better over a larger range and is likely
to generalize to unseen data. The model might benefit
from additional data to further enhance performance,
as the current sample size results in relatively high
fluctuations in Test Accuracy.
3.1.2 Random Forest Model Results
From Figure 2, Random Forest demonstrates strong
fitting capability, easily achieving perfect accuracy
on smaller training sets, resulting in a Training
Accuracy that remains stable at 1. The Test Accuracy
stabilizes initially, then rapidly increases with an
increase in training samples, eventually levelling off
at 97.3%. This suggests that the model starts
generalizing on larger samples and performs well on
the test set.
The strength of Random Forest lies in its fitting
ability to the data, but it is also prone to overfitting on
small datasets. Therefore, when using Random Forest
for predictive classification tasks, efforts should be
made to avoid overfitting, ensuring the model
maintains stable generalization on unseen data.
Figure 1: Support Vector Machine.
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
700
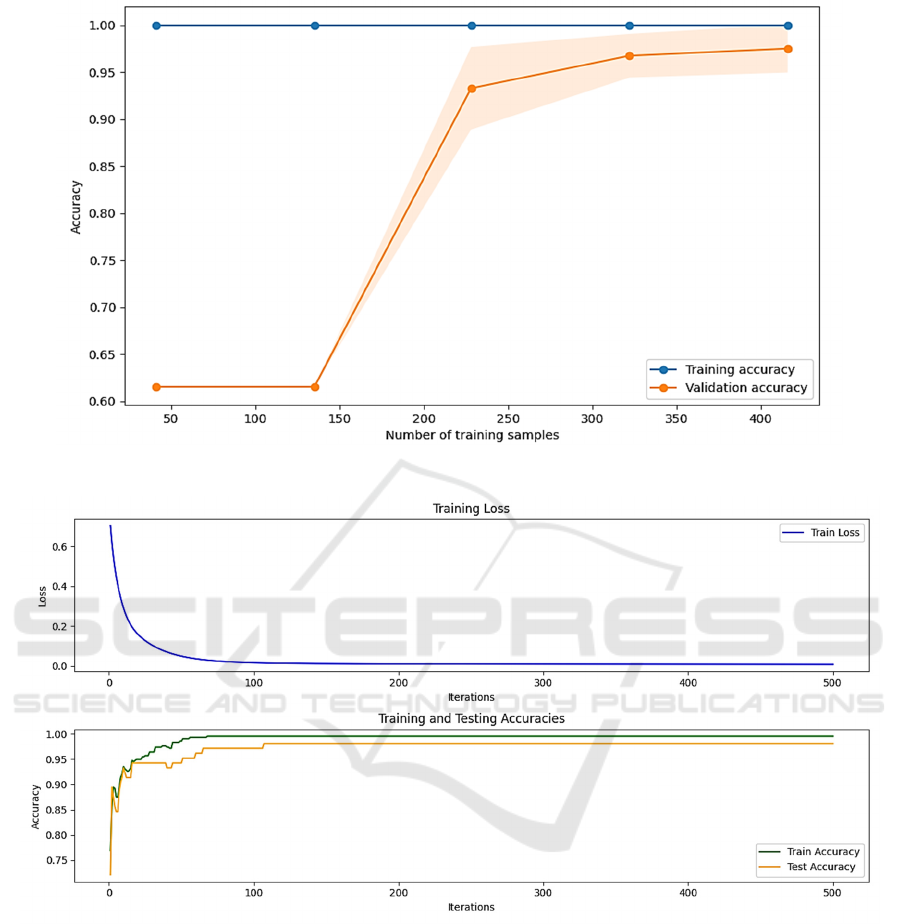
Figure 2: Random Forest.
Figure 3: Multilayer Perceptron.
3.1.3 Multilayer Perceptron Model
From Figure 3, the training loss curve of the MLP
rapidly decreases during the initial stages of training
iterations before stabilizing, indicating that the model
has known the features of the data successfully. Both
training accuracy and test accuracy gradually increase
with an increase in the number of model iterations.
The test accuracy is a little lower than the training’s,
suggesting that the MLP performs well on both two
sets without exhibiting overfitting. The final model
achieves a test accuracy of 99.2%.
3.1.4 Logistic Regression Model Result
From Figure 4, the test accuracy figure is a little lower
than the trainings, and their accuracy curves exhibit a
similar upward trend with the number increase of
training epochs, eventually converging around a
stable value. This indicates that with an increase of
the model iterations, the accuracy of predicting
disease status significantly improves, ultimately
reaching 83.7%.
The Comparison of Diabetes Risk Prediction Accuracy Across Different Models
701
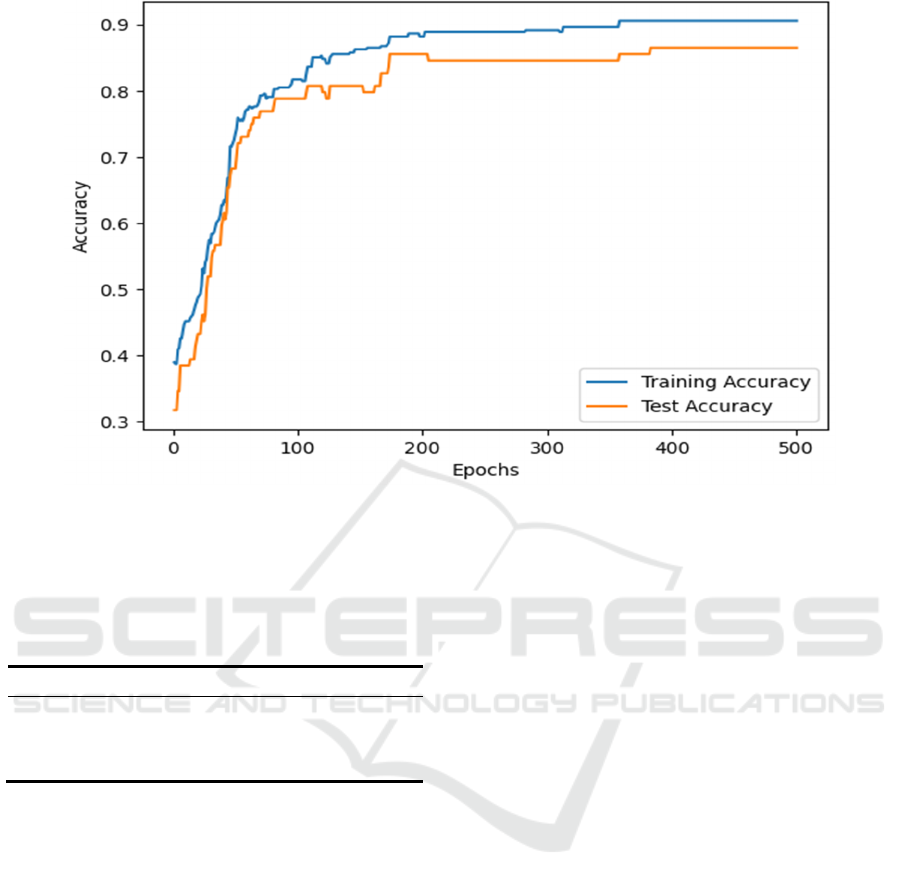
Figure 4: Logistic Regression.
3.2 The Comparison of Results
Table 2 shows the results of the four models.
Table 2: The Comparison of results.
Types of Models Test Accuracy
Support Vecto
r
Machine 0.911
Random Forest 0.973
Multilaye
r
Perceptron 0.992
Lo
g
istic Re
g
ression 0.837
Through comparing the performance of multiple
models, it was found that the Multilayer Perceptron
(MLP) excelled in this task, achieving an impressive
accuracy of 99.2%.
4 CONCLUSION
This study delved into the early diabetes prediction
domain, comparing the performance of various
models. It was observed that the Multilayer
Perceptron (MLP) exhibited outstanding performance
in this task, achieving 99.2% as the accuracy. Future
research could consider expanding the dataset to
validate the model's ability on a larger scale or
explore more sophisticated feature engineering to
improve the model's sensitivity to early diabetes
indicators or experiment with combinations of
different algorithms, potentially further improving
model performance.
AUTHORS CONTRIBUTION
All the authors contributed equally and their names
were listed in alphabetical order.
REFERENCES
Aslam, S., Rasool, A., Wu, H., Li, X., 2024. CEL: A
Continual Learning Model for Disease Outbreak
Prediction by Leveraging Domain Adaptation via
Elastic Weight Consolidation. Working paper.
Dahiwade, D., Patle, G., Meshram, E., 2019. March.
Designing disease prediction model using machine
learning approach. In 2019 3rd International
Conference on Computing Methodologies and
Communication (ICCMC). IEEE.
Dyoub, A., Letteri, I., 2023. Dataset Optimization for
Chronic Disease Prediction with Bio-Inspired Feature
Selection. Working paper.
Frasca, M., La Torre, D., Cutica, I., 2023. Combining
Convolution Neural Networks with Long-Short Time
Memory Layers to Predict Parkinson's Disease
Progression. Working paper.
Hosseininian, S. Z. S., Tajari, A., Ghalehnoie, M., Alfi, A.,
2024. Evaluating Echo State Network for Parkinson's
Disease Prediction using Voice Features. Working
paper.
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
702

Kanchan, B. D., & Kishor, M. M., 2016. December. Study
of machine learning algorithms for special disease
prediction using principal of component analysis.
In 2016 international conference on global trends in
signal processing, information computing and
communication (ICGTSPICC). IEEE.
Niu, S., Zhou, Y., Li, Z., Huang, S., & Zhou, Y., 2024. An
Improved Grey Wolf Optimization Algorithm for Heart
Disease Prediction. Working paper.
Patro, S. P., Nayak, G. S., & Padhy, N., 2021. Heart disease
prediction by using novel optimization algorithm: A
supervised learning prospective. Informatics in
Medicine Unlocked, 26, 100696.
Uddin, S., Khan, A., Hossain, M. E., & Moni, M. A., 2019.
Comparing different supervised machine learning
algorithms for disease prediction. BMC medical
informatics and decision making, 19(1), 1-16.
Wang, C., Lei, Y., Chen, T., Zhang, J., Li, Y., Shan, H.,
2024. HOPE: Hybrid-Granularity Ordinal Prototype
Learning for Progression Prediction of Mild Cognitive
Impairment. Journal of Biomedical and Health
Informatics.
The Comparison of Diabetes Risk Prediction Accuracy Across Different Models
703
