
Medical Image Classification Based on Transformer Model
and Ordinal Loss
Yan Liu
a
School of Electronics Engineering and Computer Science, Peking University, Beijing, China
Keywords: Medical Image Classification, ViT, Loss Function, Ordinal Regression.
Abstract: This study centers on the application of transformer models for general medical image classification, a crucial
step towards automating medical diagnostics. By comparing transformer models with classical methods
across diverse medical image datasets, this research aims to enhance performance on specific tasks within
these datasets. The core model, Medical Vision Transformer (MedViT), effectively learns multi-scale features
by integrating convolutional layers with specialized transformer modules, thereby catering to various medical
image classification tasks across different categories. Moreover, this study introduces Ordinal Loss to
augment the model's performance on ordinal regression subtasks. Unlike conventional cross-entropy loss,
Ordinal Loss facilitates improved learning of sequential relationships between categories. Experiments
conducted on MedMNIST validate that MedViT surpasses classical methods on most datasets, with Ordinal
Loss further enhancing performance on ordinal regression subtasks. Visual analysis also confirms that the
new loss function aids the model in effectively discerning key differences between adjacent categories. This
research demonstrates the feasibility of employing a general-purpose transformer model to address medical
image classification challenges across multiple domains. Additionally, plug-and-play modules can be
leveraged to optimize the model for specific tasks, underscoring its versatility and potential for broader
application in medical diagnostics.
1 INTRODUCTION
Medical imaging classification is a crucial process in
healthcare as it involves the automated analysis of
medical images to identify and categorize various
medical conditions. This process assists in diagnosing
diseases, monitoring treatment progress, and
facilitating early detection of abnormalities, which
can significantly improve patient outcomes. By
leveraging advanced algorithms, it enhances the
precision and speed of interpretation, reducing the
workload on radiologists and potentially lowering the
rate of misdiagnosis. Overall, medical imaging
classification serves as an essential tool in modern
medicine, enabling more accurate and efficient
patient care. In recent years, the development of
convolutional neural networks has greatly aided the
progress of computer-aided medical imaging
classification (Lo, 2022; Hu, 2022; Yang, 2021).
Convolutional Neural Network (CNN) can learn
key features from images effectively and represent a
a
https://orcid.org/0009-0000-9961-3162
crucial model architecture for image classification in
computer vision. VGGNet demonstrated the
importance of depth in CNN architectures, featuring
up to 19 layers and achieving remarkable success in
the ImageNet challenge (Simonyan, 2014). ResNet
addresses the problem of degradation in network
depth by introducing residual connections, winning
the ImageNet challenge and profoundly influencing
future research on deep learning architectures (He,
2016).
Transformer is a model originating from the field
of natural language processing. Vision Transformer
transforms images into tokens for classification,
pioneering the use of transformers in computer vision
(Dosovitskiy, 2020). Pooling-based Vision
Transformer innovates by integrating learnable
pooling operations into the transformer architecture,
enabling it to dynamically adjust the resolution of
feature maps and improve efficiency and
performance across various vision tasks (Heo, 2021).
Medical Vision Transformer (MedViT) is a kind of
708
Liu, Y.
Medical Image Classification Based on Transformer Model and Ordinal Loss.
DOI: 10.5220/0012969200004508
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 1st International Conference on Engineering Management, Information Technology and Intelligence (EMITI 2024), pages 708-713
ISBN: 978-989-758-713-9
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

Vision Transformer model specifically designed for
medical imaging tasks. It leverages the transformer's
ability to capture global dependencies and complex
patterns within images to improve the accuracy and
efficiency of diagnosing and analyzing medical
images (Manzari, 2023).
This study focuses primarily on utilizing MedViT
for medical image classification tasks, with a
comprehensive analysis of factors influencing its
performance and subsequent enhancements.
Addressing the challenge of limited dataset sizes, data
augmentation techniques are deployed to augment the
model's generalization capability. Moreover, to
mitigate overfitting concerns, various model depths
are explored through comparative experiments. In
response to the performance limitations in ordered
regression subtasks, a novel loss function termed
ordinal loss is developed, directly applicable to the
model. Comparative experiments between ordinal
loss and the original model are conducted, with
results visualized using interpretable models. The
findings indicate that MedViT offers a significant
advantage over classical methods, and the newly
designed loss function effectively enhances the
model's performance in ordered regression subtasks.
This study marks a notable progress in lightweight
medical image classification, especially in tackling
ordinal regression subtasks.
2 METHODOLOGIES
2.1 Dataset Description and
Preprocessing
MedMNIST (Yang, 2023) is a lightweight, cross-
domain medical image classification dataset
structured similarly to MNIST. It comprises 12 types
of 2D data and 6 types of 3D data. This study
primarily experiments on the 12 types of 2D data, and
their image types and labels are as follows:
PathMNIST consists of histopathological slices of
colorectal cancer tissue, with dimensions of 3x28x28,
and includes labels for 9 types of tissue.
ChestMNIST consists of frontal-view X-ray
images of the chest, with dimensions of 1x28x28, and
includes 14 disease labels, constituting a multi-label
binary classification task.
DermaMNIST consists of dermatoscopic images,
with dimensions of 3x28x28, and includes labels for
7 types of pigmented skin diseases.
OCTMNIST consists of optical coherence
tomography (OCT) images of the retina, with
dimensions of 1x28x28, and includes labels for 4
diagnosis categories.
PneumoniaMNIST comprises pediatric chest X-
ray images, with dimensions of 1x28x28, constituting
a binary classification task for pneumonia and healthy
cases.
RetinaMNIST consists of retinal fundus images,
with dimensions of 3x28x28, forming a 5-level
ordinal regression for diabetic retinopathy severity.
BreastMNIST consists of breast ultrasound
images, with dimensions of 1x28x28, forming a
binary classification task for benign and malignant
breast cancer.
BloodMNIST consists of microscope images of
blood cells, with dimensions of 3x28x28, comprising
8 labels.
TissueMNIST comprises microscope slice images
of human kidney cortex cells, with dimensions of
1x28x28, containing 8 different category labels.
OrganMNIST consists of CT images of human
body organs from different orientations, with
dimensions of 1x28x28, and includes labels for 11
organ categories.
These datasets vary in size and encompass various
subtasks of image classification. Random scaling,
rotation, cropping, and horizontal flipping are
employed as data augmentation techniques.
2.2 Proposed Approach
The original dataset provides benchmarking for some
classical CNNs and AutoML methods on the 12 2D
datasets, using accuracy (ACC) and area under the
receiver operating characteristic (ROC) curve (AUC)
as evaluation metrics. To compare the performance
differences between MedViT and classical
convolutional networks, a MedViT model is trained
on the same datasets and evaluated using the same
metrics. One of these datasets is RetinaMNIST, used
for classifying retinal image lesion severity, which
falls under ordinal regression tasks. Most models,
including MedViT, generally get low ACC on this
task. To address this, a new loss function (ordinal
loss) with hyperparameters is designed to replace the
original cross-entropy loss function and train it under
different hyperparameters to compare its
performance. Subsequently, the researcher selects the
model with the greatest performance improvement
and uses GradCAM for visual monitoring to visualize
and analyze the effect of the new loss function on
enhancing ordered regression tasks, validating the
design concept. The entire experimental process is
illustrated in Figure 1.
Medical Image Classification Based on Transformer Model and Ordinal Loss
709
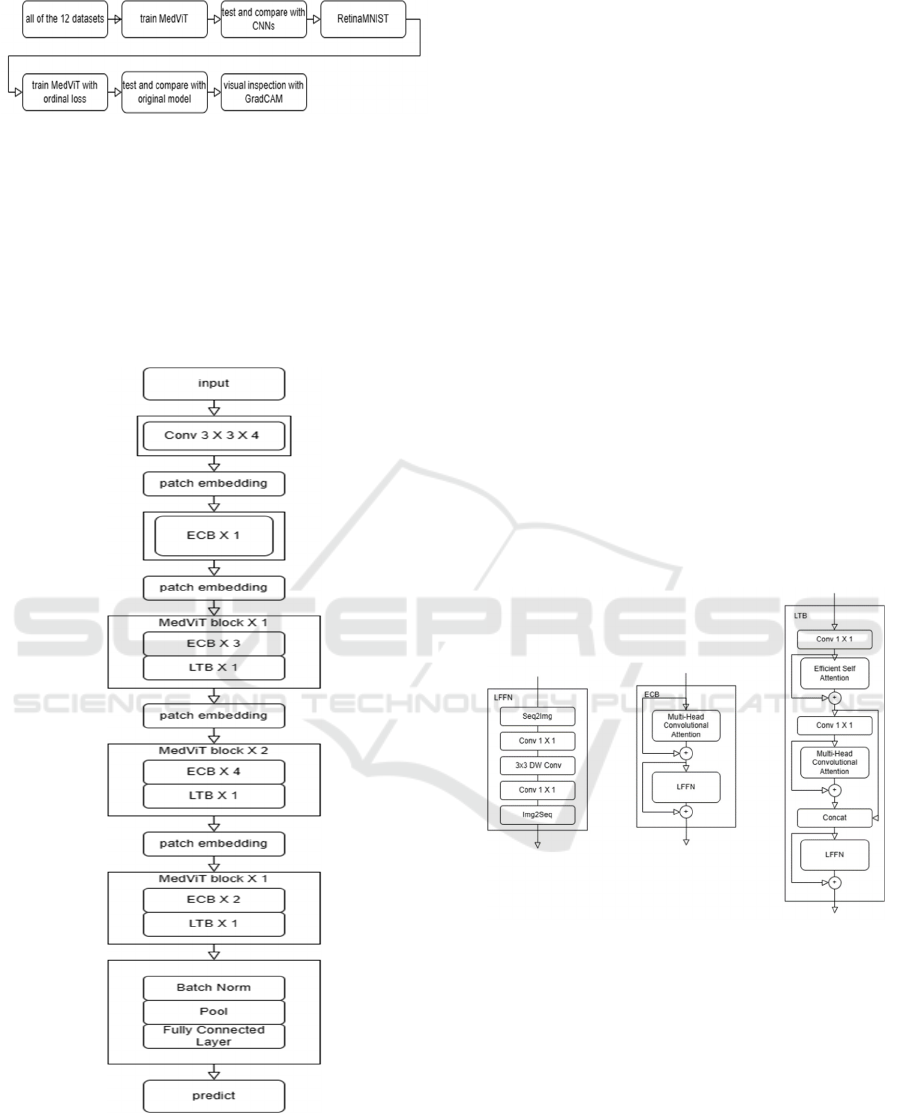
Figure 1: The pipeline of the research
(Photo/Picture credit:
Original).
2.2.1 MedViT
MedViT is a hybrid architecture of CNN and
transformer. It contains two main modules, Local
Transformer Block (LTB) and Efficient Convolution
Block (ECB). Both modules contain a Locally Feed
Forward Network (LFFN).
Figure 2: The architecture of MedViT
(Photo/Picture credit:
Original).
LFFN rearranges the token sequence into a 2D
grid and performs convolution, then rearranges it
back into a token sequence. In this way, it can capture
the locality information in the data. An ECB block is
made by a multi-head convolution attention block and
LFFN connected together. It is used to learn the long-
range dependencies between pixels corresponding to
the background. In an LTB, an improved version of
the self-attention block is used to capture low-
frequency signals, while the multi-head convolution
attention block is used to capture high-frequency
signals in different parallel representation subspaces.
Their outputs are then concatenated to achieve a mix
of high and low-frequency signals. The architecture
of MedViT is shown in Figure 2 and the specific
structure of LFFN, LTB and ECB are shown in Figure
3. A MedViT block consists of one or more ECB and
LTB blocks stacked together. The image input passes
through an initial convolutional layer and an ECB
block. Then, it traverses through multiple sets of
MedViT blocks of varying scales to comprehensively
learn features at different scales. Finally, it goes
through pooling and fully connected layers to obtain
classification predictions. Given the smaller size of
the data images, shallower model depths are
employed to prevent overfitting. However, in other
application scenarios, stacking more MedViT blocks
can lead to improved performance.
Figure 3: The structure of LFFN, LTB and ECB
(Photo/
Picture credit: Original).
2.2.2 GradCAM
Gradient-weighted Class Activation Mapping
(GradCAM) is a technique utilized to visualize the
significance of specific regions within an image for a
neural network's predictions. It provides insights into
the prediction process of the model by highlighting
the portions of the input image that contribute most
significantly to its prediction. Although GradCAM
was initially developed for analyzing CNNs, it can
also be applied to transformer models. Unlike when
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
710

applied to CNNs, where GradCAM typically utilizes
the output and gradients from the last convolutional
layer, when applied to transformer models, it
generally leverages the output and gradients from the
final transformer block. These gradients represent the
importance of each feature map for the final
prediction. Using these gradients, the contribution of
each token to the classification result is calculated.
These contributions are subsequently correlated with
the spatial positions on the original image to generate
a heatmap. Elevated values in the heatmap denote
areas where the model concentrates its attention
during predictions.
By overlaying this heatmap onto the input image,
it can be visually interpreted which parts of the image
are most important for the neural network's
prediction. This can visually represent the process
and basis of the model’s classification.
2.2.3 Loss Function
Cross-entropy loss function measures the difference
between two probability distributions: the predicted
probability distribution output by the model and the
true probability distribution of the labels. This loss
function penalizes incorrect predictions more
severely as their confidence increases, leading to
more effective training of classification models.
Cross-entropy loss function is commonly expressed
as:
𝐿
= −
∑
𝑦
𝑙𝑜𝑔𝑦
(1)
where N is the number of labels, 𝑦
is the true
probability (0 or 1) of the sample belonging to class i,
and
𝑦
is the predicted.
This research designs a new loss function, Ordinal
Loss, for addressing ordinal regression tasks. The
motivation behind this design is that the loss function
should optimize the model's output towards a
unimodal distribution closer to the true probability
distribution, while simultaneously enhancing the
sensitivity between adjacent categories as much as
possible. Ordinal Loss is calculated as:
𝑀𝑎𝑟𝑔𝑖𝑛 𝑅𝑎𝑛𝑘
𝑦
, 𝑦
, 𝑡
=max
0, −𝑡
𝑦
−𝑦
+
𝑚𝑎𝑟𝑔𝑖𝑛
(2)
𝑅𝑎𝑛𝑘 𝐿𝑜𝑠𝑠=
∑∑
𝑀𝑎𝑟𝑔𝑖𝑛 𝑅𝑎𝑛𝑘𝑦
, 𝑦
, 𝑡
,
(3)
𝑡
=
1, 𝑖𝑓 𝑙𝑎𝑏𝑒𝑙>
−1, 𝑜𝑡ℎ𝑒𝑟𝑤𝑖𝑠𝑒
(4)
𝑂𝑟𝑑𝑖𝑛𝑎𝑙 𝐿𝑜𝑠𝑠= 𝛼𝑅𝑎𝑛𝑘 𝐿𝑜𝑠𝑠+
1 −𝛼
𝐿
(5)
where α is a hyperparameter, label is the true class of
the sample, and margin is a parameter fixed at 0.1. It
can be seen that Ordinal Loss is a combination of rank
loss and traditional cross-entropy loss. The rank loss
encourages the model's predicted probability
distribution to approach a more realistic unimodal
distribution, while also enhancing the sensitivity of
the model to adjacent categories. As α increases, the
model's optimization direction becomes more
influenced by the rank loss.
2.3 Implementation Details
The training is conducted on an Nvidia A800 GPU.
The model is independently trained for 50 epochs on
12 datasets, with a batch size of 128. The learning rate
is set to 0.001 at the beginning, adopting a cosine
decay strategy over a cycle of 50 epochs. In the
experimental section of ordinal loss, keeping other
settings unchanged, training is conducted on
RetinaMNIST, and is set to 0, 0.1, 0.2, 0.3, 0.4, and
0.5, respectively. All training is performed using the
AdamW optimizer.
3 RESULTS AND DISCUSSION
The experimental results include the testing
performance of MedViT on 12 datasets and the
accuracy of MedViT with ordinal loss on
RetinaMNIST. The comparison between MedViT's
ACC and AUC on all 12 datasets and classical
methods is illustrated in Table 1. MedViT performs
exceptionally well on the PathMNIST dataset, with
the highest AUC of 0.992 and a very high ACC of
0.909 compared to the other methods listed. This
suggests MedViT is very effective at distinguishing
between the different classes in this particular dataset.
For the PneumoniaMNIST dataset, MedViT again
has an impressive AUC of 0.978 and ACC of 0.939,
outperforming all other methods by a notable margin
in AUC, indicating strong performance in terms of the
model's ability to rank predictions correctly. In the
OCTMNIST, MedViT has good AUC and ACC
scores, but not the highest. Its AUC of 0.960 and ACC
of 0.783 are strong, but Google AutoML Vision has
slightly better performance with an AUC of 0.963 and
ACC of 0.771. MedViT's performance on the other
datasets is also generally strong, often within the top
three methods. For example, it performs very well on
the BloodMNIST with an AUC of 0.997 and an ACC
of 0.968, suggesting a high capability of
distinguishing between classes accurately.
Medical Image Classification Based on Transformer Model and Ordinal Loss
711
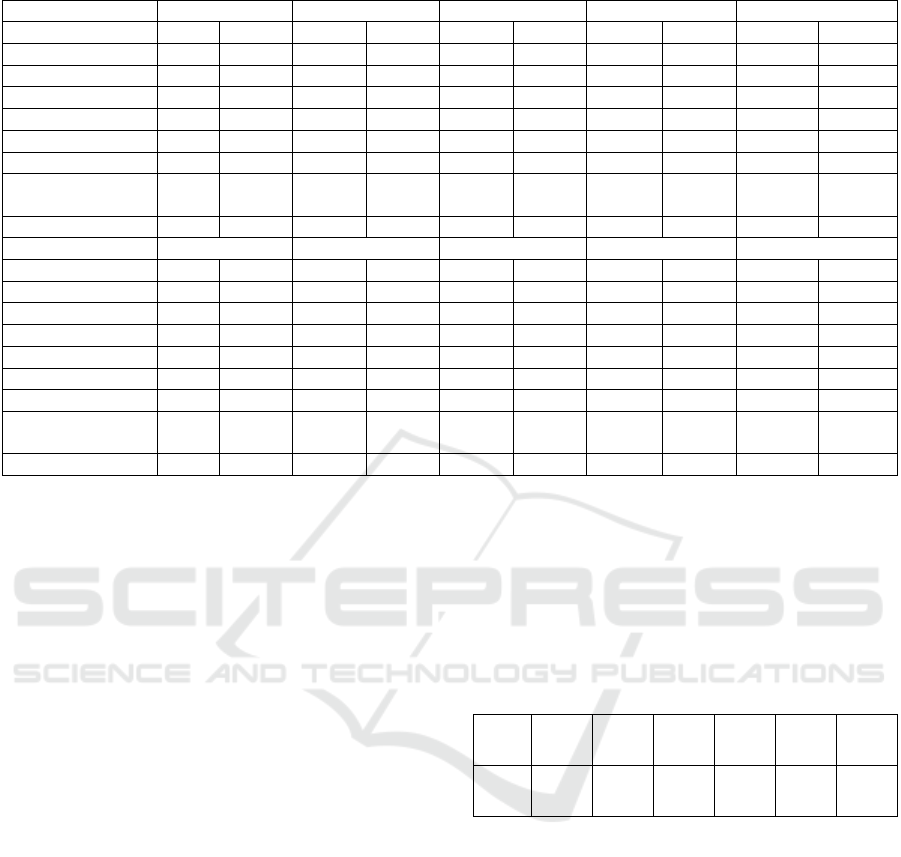
Table 1: Performance of MedViT and other classical methods on 12 datasets.
Methods PathMNIST ChestMNIST DermaMNIST OCTMNIST PneumoniaMNIST
AUC ACC AUC ACC AUC ACC AUC ACC AUC ACC
ResNet-18 (28) 0.983 0.907 0.768 0.947 0.917 0.735 0.943 0.743 0.944 0.854
ResNet-18 (224) 0.989 0.909 0.773 0.947 0.92 0.754 0.958 0.763 0.956 0.864
ResNet-50
(
28
)
0.99 0.911 0.769 0.947 0.913 0.735 0.952 0.762 0.948 0.854
ResNet-50
(
224
)
0.989 0.892 0.773 0.948 0.912 0.731 0.958 0.776 0.962 0.884
auto-sklearn 0.934 0.716 0.649 0.779 0.902 0.719 0.887 0.601 0.942 0.855
AutoKeras 0.959 0.834 0.742 0.937 0.915 0.749 0.955 0.763 0.947 0.878
Google AutoML
Vision
0.944 0.728 0.778 0.948 0.914 0.768 0.963 0.771 0.991 0.946
MedViT 0.992 0.909 0.550 0.947 0.924 0.768 0.960 0.783 0.978 0.939
Methods BreastMNIST BloodMNIST TissueMNIST OrganAMNIST OrganCMNIST
AUC ACC AUC ACC AUC ACC AUC ACC AUC ACC
ResNet-18
(
28
)
0.901 0.863 0.998 0.958 0.93 0.676 0.997 0.935 0.992 0.900
ResNet-18
(
224
)
0.891 0.833 0.998 0.963 0.933 0.681 0.998 0.951 0.994 0.920
ResNet-50
(
28
)
0.857 0.812 0.997 0.956 0.931 0.68 0.997 0.935 0.992 0.905
ResNet-50 (224) 0.866 0.842 0.997 0.95 0.932 0.68 0.998 0.947 0.993 0.911
auto-sklearn 0.836 0.803 0.984 0.878 0.828 0.532 0.963 0.762 0.976 0.829
AutoKeras 0.871 0.831 0.998 0.961 0.941 0.703 0.994 0.905 0.99 0.879
Google AutoML
Vision
0.919 0.861 0.998 0.966 0.924 0.673 0.99 0.886 0.988 0.877
MedViT 0.856 0.891 0.997 0.968 0.922 0.672 0.997 0.932 0.993 0.920
For the TissueMNIST dataset, MedViT's
performance is not as strong as on other datasets, with
an AUC of 0.922 and an ACC of 0.672. While the
AUC is relatively high, the ACC is the lowest among
the reported results for this dataset. For OrganMNIST
datasets, MedViT maintains high AUC scores (0.997,
0.992, and 0.975 respectively) and high ACC (ACC
scores of 0.932, 0.920, and 0.796 respectively),
indicating robust overall performance across these
different datasets. Overall, MedViT achieves the top
two highest ACC in 10 out of the 12 data categories
and the top two highest AUC in 9 out of the datasets.
It demonstrates high effectiveness on these medical
imaging datasets, especially for PathMNIST,
PneumoniaMNIST, and BloodMNIST, with
consistently high AUC and ACC scores, indicating
strong predictive performance and reliability. It
would be a good choice for tasks similar to those
datasets where high sensitivity and specificity are
crucial.
Table 2 illustrates the training results of MedViT
on RetinaMNIST with different hyperparameters α
after replacing the loss function with Ordinal Loss.
Here, α=0.0 corresponds to using only the original
cross-entropy loss function. It can be observed that as
α increases, the weight of the Rank Loss increases,
and the training performance shows a trend of
improvement followed by deterioration. At α=0.2, the
model achieves the best performance, with a
significant improvement of 4% compared to the
original, reaching an accuracy close to 60%. This
indicates that Ordinal Loss indeed effectively
enhances MedViT's performance in ordinal
regression tasks. Moreover, the proportion of Rank
Loss should not be maximized; instead, it needs to be
balanced with traditional cross-entropy to achieve
optimal performance.
Table 2: Performance of MedViT with Ordinal Loss on
RetinaMNIST.
𝛼
0.0 0.1 0.2 0.3 0.4 0.5
ACC 0.552 0.570 0.594 0.581 0.557 0.546
Figure 4 shows the attention heatmap of two
models trained using the traditional loss function and
α=0.2 Ordinal Loss, respectively, when identifying
samples from two adjacent classes. The retinal image
above corresponds to a lesion severity level of 1,
while the one below corresponds to level 2. The green
boxes highlight the areas of significant retinal lesions,
which serve as the primary discriminative features.
The deeper red regions in the heatmap indicate areas
that play a more significant role in the model's
classification process. It can be observed that in the
heatmap of the model trained with Ordinal Loss, the
red regions overlap more closely with the green
boxes, indicating that this model better captures the
key features for distinguishing between samples from
two adjacent classes. In contrast, in the heatmap of
EMITI 2024 - International Conference on Engineering Management, Information Technology and Intelligence
712

the model trained with traditional cross-entropy loss,
the red regions are concentrated mainly in the middle
of the image, failing to effectively differentiate
between the two classes of samples. The experimental
results and visual inspection demonstrate that as
expected, Ordinal Loss enables the model to better
distinguish between adjacent classes, thus improving
the performance on ordinal regression tasks.
Figure 4: Visual inspection of models trained with two
different loss functions using GradCAM
(Photo/Picture
credit: Original).
4 CONCLUSIONS
This study concentrates on utilizing transformer
models for image classification tasks on MedMNIST
and enhancing the performance of ordinal regression
subtasks using a novel loss function. The MedViT
model, a hybrid architecture combining CNN and
transformer, is employed to classify all 12 2D datasets
in MedMNIST and compared against classical CNN
models. Experimental findings reveal that MedViT,
adept at capturing multi-scale features, showcases
significant advantages over traditional methods,
yielding superior performance across most of the 12
datasets. The development of Ordinal Loss aims to
address the observed performance limitations across
all models on the ordinal regression subdataset,
RetinaMNIST. This loss function combines
traditional cross-entropy loss with Rank Loss,
emphasizing similarity relationships between ordered
categories during model training. Comparative
experiments with unmodified cross-entropy loss
demonstrate that models trained with Ordinal Loss
achieve higher accuracy on RetinaMNIST for ordinal
regression tasks. Visual inspection using GradCAM
further illustrates that Ordinal Loss enables the model
to better discern key features for distinguishing
adjacent categories. In the realm of fine-grained
recognition, certain methods enhance model
performance by learning pairs of intra-class and inter-
class similar samples. In future research, this
approach could also be considered for integration into
the ordinal regression task to further enhance the
model's ability to discern similar samples effectively.
REFERENCES
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., ... & Houlsby, N. 2020.
An image is worth 16x16 words: Transformers for
image recognition at scale. arXiv:2010.11929.
He, K., Zhang, X., Ren, S., & Sun, J. 2016. Deep residual
learning for image recognition. In Proceedings of the
IEEE conference on computer vision and pattern
recognition. pp: 770-778.
Heo, B., Yun, S., Han, D., Chun, S., Choe, J., & Oh, S. J.
2021. Rethinking spatial dimensions of vision
transformers. In Proceedings of the IEEE/CVF
international conference on computer vision. pp:
11936-11945.
Hu, Q., Chen, C., Kang, S., Sun, Z., Wang, Y., Xiang, M., ...
& Wang, S. 2022. Application of computer-aided
detection (CAD) software to automatically detect
nodules under SDCT and LDCT scans with different
parameters. Computers in Biology and Medicine, vol.
146, p: 105538.
Hu, W., Li, C., Li, X., Rahaman, M. M., Ma, J., Zhang, Y., ...
& Grzegorzek, M. 2022. GasHisSDB: A new gastric
histopathology image dataset for computer aided
diagnosis of gastric cancer. Computers in biology and
medicine, vol. 142, p: 105207.
Lo, C. M., & Hung, P. H. 2022. Computer-aided diagnosis
of ischemic stroke using multi-dimensional image
features in carotid color Doppler. Computers in Biology
and Medicine, vol. 147, p: 105779.
Manzari, O. N., Ahmadabadi, H., Kashiani, H., Shokouhi,
S. B., & Ayatollahi, A. 2023. MedViT: a robust vision
transformer for generalized medical image
classification. Computers in Biology and Medicine, vol.
157, p: 106791.
Simonyan, K., & Zisserman, A. 2014. Very deep
convolutional networks for large-scale image
recognition. arXiv:1409.1556.
Yang, J., Shi, R., Wei, D., Liu, Z., Zhao, L., Ke, B., ... & Ni,
B. 2023. Medmnist v2-a large-scale lightweight
benchmark for 2d and 3d biomedical image
classification. Scientific Data, vol. 10(1), p: 41.
Yang, X., & Stamp, M. 2021. Computer-aided diagnosis of
low grade endometrial stromal sarcoma (LGESS).
Computers in Biology and Medicine, vol. 138, p:
104874.
Medical Image Classification Based on Transformer Model and Ordinal Loss
713
