
Predicting Post Myocardial Infarction Complication: A Study Using
Dual-Modality and Imbalanced Flow Cytometry Data
Nada ALdausari
1 a
, Frans Coenen
1 b
, Anh Nguyen
1 c
and Eduard Shantsila
2 d
1
Department of Computer Science, The University of Liverpool, Liverpool, U.K.
2
Institute of Population Health, The University of Liverpool, Liverpool, U.K.
{n.al-dausari, coenen, anh.nguyen}@liverpool.ac.uk, eduard.shantsila@liverpool.ac.uk
Keywords:
Post-Myocardial Infarction Complications, Flow Cytometry, Artificial Neural Networks, Data Normalisation,
Imbalanced Data Handling, Binary Classification.
Abstract:
Previous research indicated that white blood cell counts and phenotypes can predict complications after My-
ocardial Infarction (MI). However, progress is hindered by the need to consider complex interactions among
different cell types and their characteristics and manual adjustments of flow cytometry data. This study aims
to improve MI complication prediction by applying deep learning techniques to white blood cell test data ob-
tained via flow cytometry. Using data from a cohort study of 246 patients with acute MI, we focused on Major
Adverse Cardiovascular Events as the primary outcome. Flow cytometry data, available in tabular and image
formats, underwent data normalisation and class imbalance adjustments. We built two classification models:
a neural network for tabular data and a convolutional neural network for image data. Combining outputs from
these models using a voting mechanism enhanced the detection of post-MI complications, improving the av-
erage F1 score to 51 compared to individual models. These findings demonstrate the potential of integrating
diverse data handling and analytical methods to advance medical diagnostics and patient care.
1 INTRODUCTION
Cardiovascular Disease (CVD) remains one of the
leading causes of mortality (Bhatnagar et al., 2015;
Centers for Disease Control and Prevention, 2022),
significantly impacting global health trends. Reports
from the National Center for Health Statistics high-
light that between 2019 and 2021, CVD was a major
cause of death in the US (Murphy et al., 2021). Sim-
ilarly, the British Heart Foundation identifies CVD
as more prevalent than cancer in the UK (Bhatnagar
et al., 2015), underscoring its severity as a health con-
cern. Among the various types of CVD, myocardial
infarction (MI), commonly known as a heart attack,
presents particularly complex challenges. It occurs
when blood flow to part of the heart is obstructed,
resulting in heart muscle damage (Thygesen et al.,
2012). Post-MI, patients face significant risks, includ-
ing heart failure and increased mortality; about 20%
of those suffering an acute MI die within the first year,
a
https://orcid.org/0009-0003-3014-059X
b
https://orcid.org/0000-0003-1026-6649
c
https://orcid.org/0000-0002-1449-211X
d
https://orcid.org/0000-0002-2429-6980
with a substantial portion of these deaths occurring af-
ter the initial 30 days (Qing Ye, 2020). This array of
adverse outcomes after a MI is collectively referred
to as Major Adverse Cardiac Events (MACE) (Clinic,
2022).
Recent medical studies have explored potential
predictors for post-MI complications (Boidin et al.,
2023; Shantsila et al., 2013; Shantsila et al., 2019), in-
cluding dynamic changes in specific subsets of white
blood cells, particularly those expressing CD14 and
CD16 markers. High levels of CD14 and CD16 white
blood cells are associated with higher occurrences
of MACE, making these counts useful for predict-
ing post-MI complications and managing patient re-
covery. However, analysing these cells is challenging
due to the complexity of interactions and the need for
manual calibration in flow cytometry. Additionally,
small sample sizes limit the generalisation of find-
ings and focusing solely on cell subsets may overlook
other critical factors. These challenges underscore the
need for further research and improved methodolo-
gies to enhance predictive accuracy and improve pa-
tient outcomes.
Recent deep learning efforts have focused on us-
ing patient data such as age, gender, lifestyle, and
ALdausari, N., Coenen, F., Nguyen, A. and Shantsila, E.
Predicting Post Myocardial Infarction Complication: A Study Using Dual-Modality and Imbalanced Flow Cytometry Data.
DOI: 10.5220/0012998300003838
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2024) - Volume 1: KDIR, pages 81-90
ISBN: 978-989-758-716-0; ISSN: 2184-3228
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
81

isolated biomarker data, typically reflecting the de-
gree of myocardial damage (e.g., troponins) (Moham-
mad et al., 2022; Khera et al., 2021; Li et al., 2023;
Oliveira et al., 2023; Piros et al., 2019; Ghafari et al.,
2023; Newaz et al., 2023; Saxena et al., 2022). These
studies have incorporated a broad range of features,
not solely blood cells, and none have applied convo-
lutional neural networks (CNNs). This paper aims to
bridge the gap between traditional medical research
and the deep learning community by incorporating
white blood cell data into predictive models. We
present a deep learning approach to analysing flow
cytometry data to predict post-MI complications, ad-
dressing two significant technical challenges: the dual
modality of the data and the imbalanced nature of
the available flow cytometry data. Overcoming these
challenges is crucial for enhancing the accuracy of
predictions and improving patient outcomes after MI.
The main contributions of this paper are:
1. Developing preprocessing techniques to explore
and identify data representations that significantly
enhance performance outcomes.
2. Designing and implementing two neural network
models to effectively manage the multi-modality
inherent in the dataset.
3. Investigating and assessing various balancing
techniques to achieve an equitable distribution of
samples across different classes.
4. Employing diverse evaluation methodologies to
identify the most effective balancing technique,
ensuring robust model performance.
2 RELATED WORK
Previous studies have focused on applying machine
and deep learning techniques to predict MI mortal-
ity and hospital admissions due to complications.
These studies, detailed in various research papers,
have utilised a range of machine learning algorithms,
dataset sizes, and features (Mohammad et al., 2022;
Khera et al., 2021; Li et al., 2023; Oliveira et al.,
2023; Piros et al., 2019; Ghafari et al., 2023; Newaz
et al., 2023; Saxena et al., 2022).
CVD Datasets. Studies on predicting CVD com-
plications have employed many datasets and fea-
tures to enhance model accuracy. These datasets
vary significantly in size, with some studies using
smaller datasets of approximately 1,000 to 1,700 pa-
tients (Oliveira et al., 2023; Ghafari et al., 2023;
Newaz et al., 2023; Saxena et al., 2022). In compar-
ison, others utilised much larger datasets, including
those exceeding 100,000 patients (Mohammad et al.,
2022; Khera et al., 2021; Li et al., 2023; Piros et al.,
2019). Common features across these studies encom-
pass patient demographics such as age and gender,
medical history, lifestyle factors, clinical markers like
troponin levels, and diagnostic test data such as ECG
results (Newaz et al., 2023). Larger datasets typi-
cally include more detailed and diverse features, such
as in-hospital treatment details and discharge medi-
cations. The variety of features used underscores the
importance of diverse data in improving the predictive
power of machine learning models for CVD compli-
cations.
Machine Learning for Post-MI Complications
Analysis. Various machine learning algorithms
have been employed in these studies, yielding no-
table successes. Commonly used algorithms include
Logistic Regression, Support Vector Machine, Ran-
dom Forest, XGBoost, and Artificial Neural Net-
works. Smaller datasets, ranging from 1,000 to 1,700
patients (Oliveira et al., 2023; Ghafari et al., 2023;
Newaz et al., 2023; Saxena et al., 2022), often utilised
combinations of Support Vector Machine, Logistic
Regression, k-nearest neighbours, and Naive Bayes,
achieving high accuracy and robust performance met-
rics. Larger datasets, such as those with over 100,000
patients (Mohammad et al., 2022; Khera et al., 2021;
Li et al., 2023; Piros et al., 2019), typically employed
more sophisticated algorithms like XGBoost and Ar-
tificial Neural Networks, demonstrating their effec-
tiveness with high accuracy and strong performance
scores. Overall, XGBoost and Artificial Neural Net-
works consistently emerged as top-performing mod-
els across various studies, highlighting their capabil-
ity to handle diverse and complex datasets to pre-
dict cardiovascular disease complications accurately.
These studies emphasise the importance of selecting
appropriate algorithms tailored to the dataset size and
feature complexity to optimise prediction.
While previous studies have concentrated on em-
ploying machine and deep learning models trained on
general patient data, this paper diverges by explic-
itly focusing on blood cell data, mainly white blood
cells. White blood cells are pivotal in the context
of cardiovascular damage and repair. This focus not
only introduces a novel dataset for machine learning
applications but also aligns with medical research,
as highlighted in previous studies (Shantsila et al.,
2011; Shantsila et al., 2019), underscoring the criti-
cal role of white blood cells in cardiovascular health.
This approach bridges a gap between machine learn-
ing methodologies and medical insights, providing a
unique perspective on predicting post-MI complica-
tion.
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
82
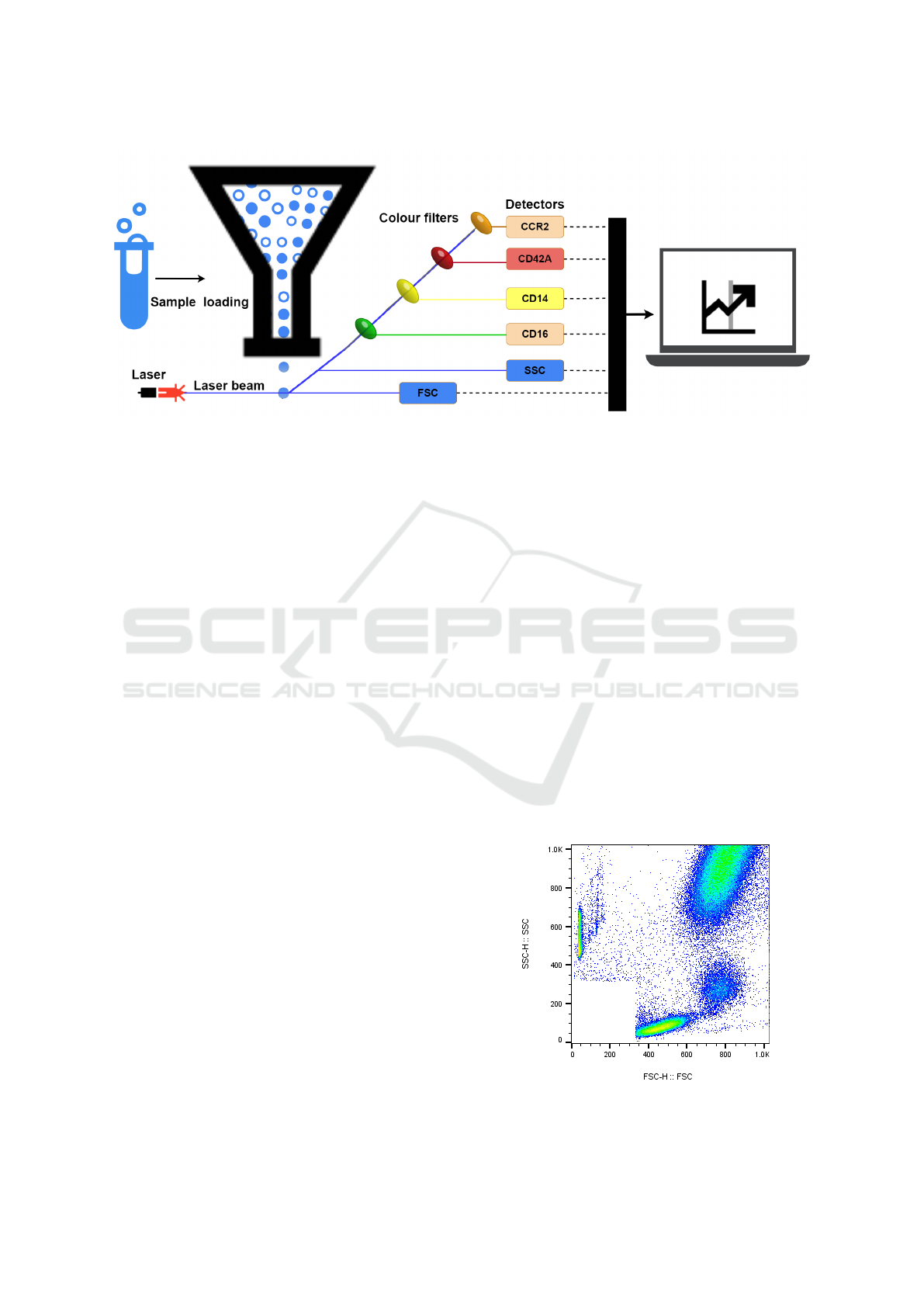
Figure 1: Collect Data by flow cytometry.
3 FLOWCYTO-MI: THE FLOW
CYTOMETRY POST-MI
COMPLICATION DATASET
3.1 Data Collection
There are multiple risk factors predictive of mortal-
ity after the diagnosis of MI, including those based
on imaging (echocardiography) and blood tests (tro-
ponin levels) (Reddy et al., 2015). This paper focuses
on predicting MI complications using blood pathol-
ogy data from flow cytometry. This technology uses
lasers at a sequence of white blood cells moving in a
directed fluid stream to generate light signals, causing
them to emit light at different wavelengths. Colour
filters play a crucial role in this process. They sepa-
rate the emitted fluorescence light into distinct wave-
length bands, allowing only specific wavelengths of
light to pass through while blocking others. For ex-
ample, a filter might permit green light to pass while
blocking light of other wavelengths (such as blue or
red). Following filtration, the light reaches the de-
tectors, which measure the intensity of the filtered
light. Detectors measure the scatter of light and flu-
orescence emission concerning each cell. Scatter is
measured along the laser signal path Forward Scatter
(FSC) and at a 90-degree angle to the path Side Scat-
ter (SSC). FSC measures cell size, while SSC mea-
sures cell complexity or granularity. The fluorescence
helps identify the surface expression (density) of vari-
ous types of molecules found on the surface of a blood
cell. These surface expressions indicate multiple cell
functions, labelled by the Cluster of Differentiation
(CD) protocol. The data collected by the detectors
is then processed and quantified using sophisticated
software, converting the raw light intensity measure-
ments into meaningful numerical values. Figure 1 il-
lustrates this process.
The collective effect of these measures is that they
allow the separation of individual cells by plotting
pairs of features. Figure 2 provides an example of
such a plot, generated using FlowJo (FlowJo, 2024),
a software system that supports analysing data ob-
tained through flow cytometry. The figure plots FSC
density on the x-axis and SSC density on the y-axis.
The colours used in the figure indicate cell density:
blue and green for low density, red and orange for
high density, and yellow for medium density (FlowJo,
2024). The white area in the bottom left corner, which
does not have any data, shows electronic noise and
tiny particles smaller than cells, thus it is not included
in the data collection. The image data is characterised
by dimensions of 611 × 620 × 4, denoting the height
and width (in pixels) and the RGBA (Red, Green,
Blue, and Alpha) values.
Figure 2: Example density plot (FSC against SSC) gener-
ated using Flowjo(FlowJo, 2024).
Predicting Post Myocardial Infarction Complication: A Study Using Dual-Modality and Imbalanced Flow Cytometry Data
83
This paper collected flow cytometry data for 246
patients from several hospitals in Birmingham, UK,
including City Hospital, Sandwell General Hospital,
Heartlands Hospital, and Queen Elizabeth Hospital,
from November 2009 to November 2012. For each
patient, the data was provided in two formats: (i) a
tabular data file (one line per cell) and (ii) a Portable
Network Graphics (PNG) image.
3.2 Data Statistics
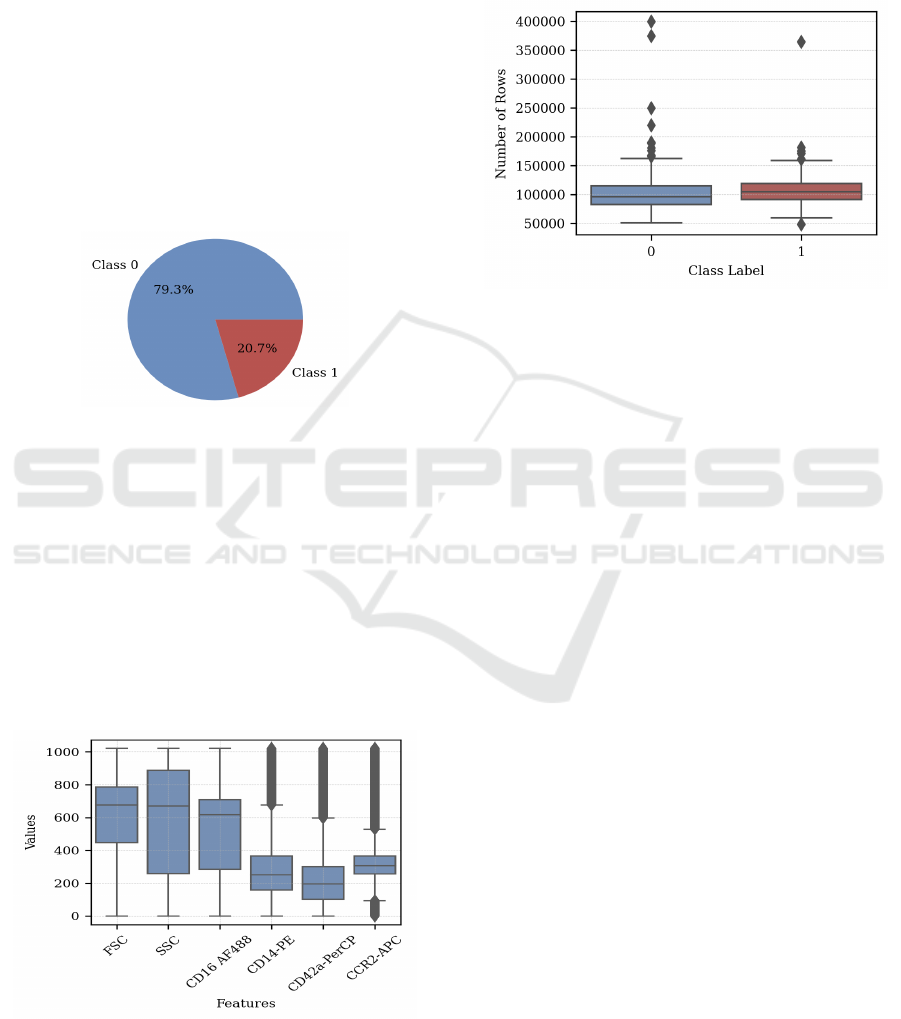
In Figure 3, we illustrate the dataset distribution,
which includes 195 instances from class 0 (patients
without post-MI complications) and 51 instances
from class 1 (patients with post-MI complications,
heart failure, or death).
Figure 3: Distribution of dataset.
The tabular data comprised six attributes
(columns). The first two were the FSC and SSC
values (see section 3.1), and the remaining four were
counts of particular surface molecules indicating pro-
teins of various kinds labelled using the CD protocol
(CD16, CD14, CD42a, and CCR2)(FlowJo, 2024).
Figure 4 shows the range, median, and variability of
each feature in the dataset. Features like FSC, SSC,
and CD16 AF488 have higher medians and broader
distributions, while CD14-PE, CD42a-PerCP, and
CCR2-APC show lower medians with significant
variability, highlighted by numerous outliers.
Figure 4: Features Distribution.
The tabular files also varied in length (number of
records/rows) because flow cytometry does not al-
ways process the same number of cells. Figure 5
shows the median number of rows per patient is sim-
ilar for both classes, around 100,000, with a slightly
larger interquartile range for Class 0. Additionally,
there are significant outliers in both classes, with
some patients having up to 400,000 rows.
Figure 5: Number of Rows.
4 POST MYOCARDIAL
INFARCTION COMPLICATION
PREDICTION
The work presented in this paper is directed at us-
ing neural networks to predict post-MI complications.
The use of neural networks was influenced by the ob-
servation that previous research has demonstrated that
neural networks are robust and practical techniques
for classification (Zhang, 2000). In addition, numer-
ous medical diagnosis applications have shown signif-
icant success by utilising neural networks (Zhou and
Jiang, 2003).
To build a machine learning model that would
work with such dual-modality data, there were two
options: (i) use some form of unifying representation
and build a single model, or (ii) build individual mod-
els, one for each modality and combine the result (for
example, by voting). The first was used in the case of
Aldosari et al.(2022) in the context of electrocardio-
gram (ECG) and patient data to predict the likelihood
of CVD. This required features to be extracted from
each data format to unify the data representation that
could be constructed. The disadvantage was that the
feature extraction process could result in information
loss. When building separate models for each modal-
ity, the disadvantage is that it is assumed that each
modality is entirely independent of the others when
this might not be the case. Given the challenge of ex-
tracting features from the blood cell data, the second
option was to construct two models. However, in this
paper, we tried to avoid the disadvantage of informa-
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
84

tion separation by combining the two models, one for
the tabular comma-separated values (CSV) data and
one for the image data, with a voting method Algo-
rithm 1.
Data: Tabular dataset T , Image dataset I, Number
of folds K = 5, Tabular model M
T
, Image
model M
I
.
Result: Comprehensive average results with
statistical significance analysis
Initialise results list R;
for fold f from 1 to K do
Split T and I into stratified training,
validation, and test sets;
• Normalise T features using RobustScaler;
• Normalise I images using ToTensor();
• Apply data augmentation in I;
Apply data balancing methods to Train T and
Train I:
• Use Random Over-Sampling, Random
Under-Sampling and SMOTE;
• Use Geometric Transformation, Focal
Loss and Random Under-Sampling;
Train Models:
• Perform hyperparameter tuning for both
M
T
on Train T and M
I
on Train I, tuning
parameters such as learning rate, batch
size, and number of epochs, with cross-
validation ;
• Train both M
T
on Train T and M
I
on Train I
using their respective best hyperparameters;
• Evaluate M
T
on Test T and M
I
on Test I,
saving detailed metrics (precision, recall,
F1-score) to R
T
and R
I
respectively;
Combine results R
T
and R
I
:
• Compute the weighted average of
predictions based on validation
performance;
• If biased, default to class 1;
• Save combined results R
f
with all
detailed metrics;
Append R
f
to R;
end
Calculate comprehensive average results R:
• Average of all metrics (precision, recall, F1-score);
return Comprehensive average results with
statistical significance analysis.;
Algorithm 1: Cross-Validation with Dual Models for Tabu-
lar and Image Data.
Data Transformation. Each patient’s dataset has a
varying number of rows for tabular data but consis-
tently includes six specific columns. We convert the
data into a unified tensor format to prepare for neu-
ral network processing with PyTorch. This involves:
identifying the maximum number of rows (399078),
padding shorter sequences with zeros to match this
length, converting each DataFrame into a PyTorch
tensor, and concatenating these tensors into a master
tensor. This results in a tensor format that includes
the number of datasets, rows, and columns. For im-
age data, data augmentation techniques enhance the
size and quality of training datasets, improving deep-
learning models (Yang et al., 2022). The applied
transformations include converting to tensors, resiz-
ing images to 256x256 pixels, randomly rotating them
by up to 20 degrees, and flipping them vertically with
a 0.4 probability and horizontally with a 0.5 probabil-
ity.
Data Splitting. The dataset was divided into a 06%
training set, a 20 % validation set and a 20% test-
ing set, following standard practice (Mpanya et al.,
2021). Five-fold cross-validation was used for evalu-
ation, partitioning the dataset into five folds and run-
ning training and testing five times. Stratified sam-
pling ensured equal class distribution across folds us-
ing Python’s StratifiedKFold with five splits. This
maintains a 60-20-20 split, with about 10 or 11 in-
stances of Class 1 in the test set. Reducing to three
folds increases the test set to 17 instances for Class
1 while increasing to seven folds reduces it to five
instances. This affects data balance for training and
testing, though variations are minor. The data distri-
bution is shown in Table 1.
Table 1: Data distribution in training, validation, and testing
sets.
Fold
Training data Valid. data Test data
Total
0 1 0 1 0 1
1 156 40 39 10 39 11 246
2,3,4,5 156 41 39 11 39 10 246
Data Normalisation. Data normalisation ensures
that each attribute contributes equally numeri-
cally (Garc
´
ıa et al., 2015), which enhances classifi-
cation performance, especially in medical data clas-
sification (Jayalakshmi and Santhakumaran, 2011;
Singh and Singh, 2020). In tabular flow cytometry
data, varying feature ranges required normalisation.
The RobustScaler method was applied (Izonin et al.,
2022), which uses the median and Interquartile Range
(IQR) for scaling, as shown in Equation 1:
X
′
=
X −X
med
IQR
(1)
where X
′
is the normalised attribute, X
med
is the me-
Predicting Post Myocardial Infarction Complication: A Study Using Dual-Modality and Imbalanced Flow Cytometry Data
85
dian, and IQR is the Interquartile Range. In PyTorch,
transforms.ToTensor() normalizes RGBA values
from [0, 255] to [0, 1] by dividing by 255 and storing
the data as a tensor.
Data Balancing. The available data predominantly
consists of myocardial infarction (MI) patients with-
out post-MI complications, resulting in an imbalance,
with a higher prevalence of patients without compli-
cations. The dataset comprises 195 instances from
class 0 (no MI complications) and 51 instances from
class 1 (MI complications), creating a 4:1 ratio. Var-
ious techniques were employed on both tabular and
image data to address this imbalance. For tabular
data (Zhang et al., 2023; Khushi et al., 2021), random
over-sampling generated additional records for the
minority class, random under-sampling reduced the
majority class records, and SMOTE (Synthetic Mi-
nority Oversampling Technique) augmented the mi-
nority class using synthetic data created through in-
terpolation. This process involves the existing minor-
ity class samples and their nearest neighbours, with K
set to 5, to ensure that the number of records in the
minority class matches those in the majority class.For
image data, balancing strategies involved geometric
transformation, random under-sampling similar to the
tabular data approach, and focal loss, which modu-
lates cross-entropy loss to focus on minority examples
by down-weighting easy examples and emphasising
hard-to-classify ones. The weighting factor α is set
to 0.80, calculated by the ratio of majority class sam-
ples to total samples on the training set, emphasising
the minority class. The focusing parameter γ is set to
2, ensuring the model focuses on hard-to-classify ex-
amples (Lin et al., 2017). Augmentation conducted
through horizontal and vertical axis flipping improves
the model’s ability to recognise patterns regardless
of position. Consequently, two additional images for
each image in the minority class could be generated
this way.The use of random under-sampling for both
tabular and image data effectively reduced the major-
ity class without impacting the minority class, ensur-
ing that all information from Class 1 was preserved,
which is critical for accurate modeling. Additionally,
with each patient contributing approximately 100,000
rows (see Figure 5), there remained ample data to
train the model effectively despite the reduction in the
majority class.
Model Generation. This paper addresses the chal-
lenge of data’s dual-modality by employing two neu-
ral network models, each characterised by distinct
architectures and design patterns tailored to its data
type, with the predictions from both models combined
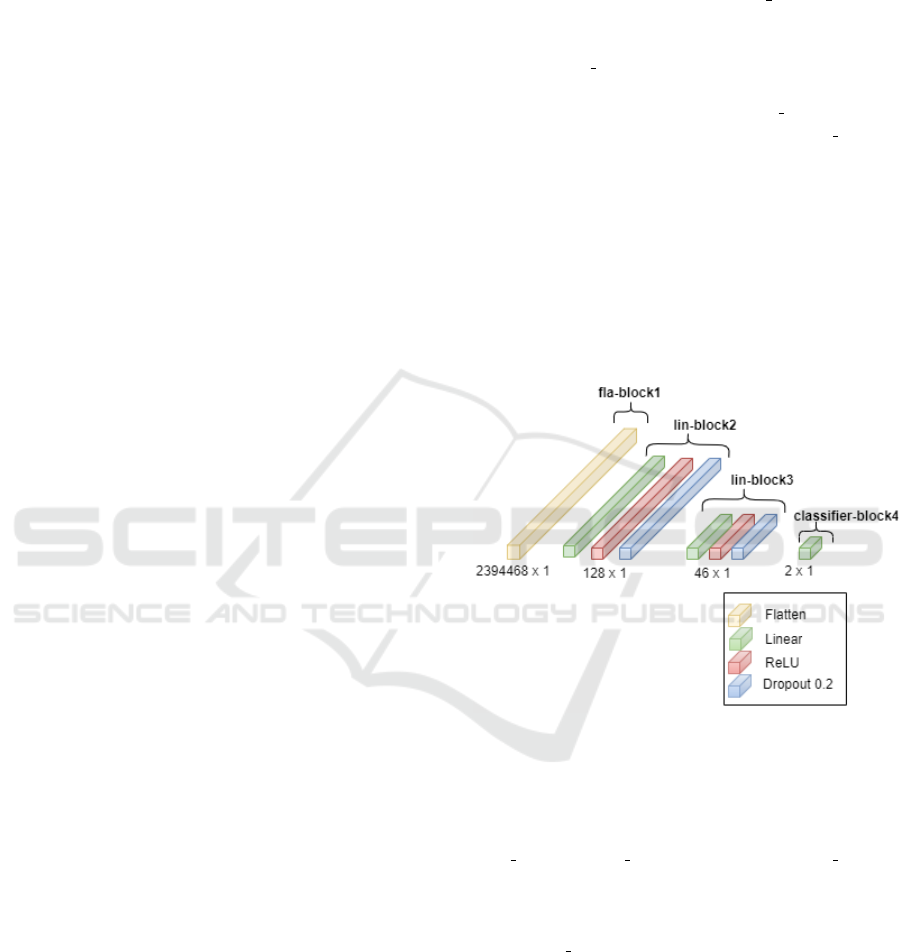
to improve overall performance. The tabular data neu-
ral network comprises a standard feed-forward neural
network consisting of a sequence of layers organised
into four blocks. The first block, fla block1, the flat-
ten layer, transforms the input to 2394468, which is
the product of 399078 and 6, the input size. The sec-
ond block, lin block2, includes linear, batch normal-
isation, Rectified Linear Unit (ReLU) activation, and
dropout layers. The next block, lin block3, is the
same as block 1. The last block, classifier block4,
includes a linear layer, as shown in Figure 6. Imple-
mentation was conducted using the Python PyTorch
library. This model employed cross-entropy loss to
measure the disparity between predicted class prob-
abilities and the actual class labels. The loss, which
falls between 0 and 1, indicates the model’s accuracy
and aims to minimise it as much as possible (PyTorch,
2024). The model’s parameters were also updated
during training using the Adam Optimiser, which has
a learning rate of 1e-3 and a batch size of 8.
Figure 6: Architecture for the Tabular Data Feed Forward
Neural Network.
The image data neural network model was a
Convolutional Neural Network (CNN). This model
was organised into three components, referred to as
conv block1, conv block2, and classifier block3.
The first two convolutional blocks comprised sev-
eral layers, including convolutional layers, batch nor-
malisation, and pooling layers. The role of the
classifier block3 is to take the output from the con-
volutional layers, flatten it, and then pass it through
a fully connected (linear) layer for making classifica-
tion predictions. The architecture of the CNN model
is illustrated in Figure 7. All input images were re-
sized to (256, 256). The model employs binary cross-
entropy loss, typically utilised for binary classifica-
tion tasks. This loss function compares the predicted
logits and target labels. Similar to the previous model,
it employs the Adam optimiser.
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
86

Figure 7: Architecture for the Image Data CNN.
The classifications from these two models were
combined using voting (Bin Habib and Tasnim, 2020;
G
´
eron, 2017). A straightforward method to enhance
the classifier performance is combining the predic-
tions from multiple classifiers and selecting the class
with the highest number of votes. In case of a tie-
break situation, class 1 was selected, as in medical
diagnosis, it is considered more critical to avoid miss-
ing an actual illness than to diagnose someone as ill
incorrectly. This form of ensemble classifier is known
as a complex voting classifier.
5 RESULTS
This section evaluates the proposed process using
FlowCyto-MI data. The evaluation metrics adopted
were as follows (Zhou, 2020):
Precision =
True Positives
True Positives +False Positives
(2)
Recall =
True Positives
True Positives +False Negatives
(3)
F1 =
2 · Precision · Recall
Precision + Recall
(4)
The same data splitting for training, validation,
and testing was used in both models, which handle
tabular and image data, respectively. The number of
data points used in each epoch is shown in Table 1.
For each fold, only the test set was used for eval-
uation, without using any data from the training or
validation sets. After experimenting with different
epochs, the proposed model’s results are presented in
Tables 2 and 3.
Note that results are presented using each of the
imbalanced data techniques and no technique (Base-
line).
Evaluation Data Balancing. Considering the tabu-
lar data, an inspection of Table 2 indicates that with-
out data balancing, the tabular model performed ex-
ceptionally well for class 0 (patients without post-MI
complications), achieving an F1 score of 87. How-
ever, class 1 (patients with post-MI complications)
recorded an F1 score of 0 for epochs 10 and 3 for
epoch 15. All methods, including random over-
sampling, random under-sampling, and SMOTE, per-
formed better than the baseline for class 1. For epoch
10, the highest average F1 score, 49, was obtained
with SMOTE. For epoch 15, the best average F1
score, 50, was achieved using random over-sampling.
This is the best result for this model.
Table 2: Tabular data Feed Forward Neural Network Re-
sults, using 10 and 15 Epochs.
Method Class 10 Epochs 15 Epochs
(5 folds) Prec. Rec. F1 Prec. Rec. F1
Baseline 0 78 97 87 79 97 87
1 0 0 0 20 2 3
Random 0 79 95 86 79 91 85
over-sampling 1 21 8 11 31 21 15
Random 0 79 60 62 78 62 61
under-sampling 1 18 37 19 27 35 18
SMOTE 0 79 91 85 79 94 86
1 42 10 13 13 6 8
Abbreviations: Prec.= Precision, and Rec.= Recall
Regarding the image data, the examination of Ta-
ble 3 reveals that the baseline performance for class 1
was superior compared to that observed with tabular
data. At 200 epochs, the highest average F1 scores
recorded were 50.5. Methods such as augmentation,
random under-sampling, and focal loss demonstrated
improvements in the F1 score for class 1 beyond the
baseline. Focal loss, applied at epochs 100 and 200,
achieved an F1 score of 50.5. Because the average F1
scores are equal in the baseline and with focal loss,
we compare based on the recall of class 1. In medical
diagnostics, recall is often prioritised over precision
as it focuses on the proportion of actual positive cases
(patients with the disease) correctly identified by the
model. Focal loss at epochs 100 and 200 was selected
based on the recall score for class 1.
Evaluation of Combined Model. Table 4 presents
the outcomes of model integration, where random
over-sampling with 15 epochs was chosen for the tab-
ular model, and focal loss with 100 and 200 epochs
was selected for the image model. This setup enabled
Predicting Post Myocardial Infarction Complication: A Study Using Dual-Modality and Imbalanced Flow Cytometry Data
87

Table 3: Image data CNN Results, using 100 and 200
Epochs.
Method Class 100 Epochs 200 Epochs
(5 folds) Prec. Rec. F1 Prec. Rec. F1
Baseline 0 78 88 83 80 89 84
1 18 9 12 18 15 17
Augmentation 0 79 64 68 78 74 70
1 24 41 28 25 31 22
Focal Loss 0 80 71 75 79 74 77
1 22 33 26 21 27 24
Random 0 77 48 57 78 55 64
under-sampling 1 22 53 30 19 42 26
Abbreviations: Prec.= Precision, and Rec.= Recall
two voting scenarios between these models. In the
first scenario, the vote was between the final result
from the best tabular model (random over-sampling
with 15 epochs) and the final result from the best im-
age model (focal loss and 100 epochs). In the case
of a tie, class 1 was selected. In the second scenario,
the voting process was identical, except that the im-
age model used focal loss with 200 epochs instead
of 100. The best result was achieved using random
over-sampling and focal loss with 200 epochs, result-
ing in an average F1 score of 51. While this repre-
sents a slight improvement over the best individual
results from the tabular and image models, the dif-
ference in performance compared to the CNN model
with focal loss and 100 epochs is minimal. Specifi-
cally, for Class 0, both models produced nearly iden-
tical results (Precision = 80, Recall = 71, F1 = 75),
and for Class 1, the difference is very slight, with the
CNN model yielding Precision = 22, Recall = 33, and
F1 = 26, while our integrated model achieved Preci-
sion = 22, Recall = 34, and F1 = 27. A review of
Table 4 reveals that the integration strategy achieves
the highest F1 score of 51, combining random over-
sampling and focal loss with 200 epochs. However,
while this integration approach addresses the dual
modality and imbalanced nature of the data, the per-
formance improvements are incremental rather than
significant when compared to the CNN model alone.
Additionally, only 34% of post-MI complications are
correctly identified, with 78% of the diagnosed cases
being false positives. This raises concerns about the
practical applicability of the model in real-world clin-
ical settings, where a high rate of false positives may
lead to unnecessary interventions and increased costs.
While the integration strategy provides a slight perfor-
mance boost, further refinement is required to reduce
the false positive rate and improve the model’s relia-
bility for practical use in diagnosing post-MI compli-
cations.
Table 4: Evaluation of Combined Model
Method Combination Epochs Class Prec. Rec. F1
Random over-sampling 15 0 80 65 71
& Focal Loss 100 1 23 39 28
Random over-sampling 15 0 80 71 75
& Focal Loss 200 1 22 34 27
Abbreviations: Prec.= Precision, and Rec.= Recall
6 CONCLUSIONS
This paper presented a deep learning approach to pre-
dict post-MI complications using dual-modal imbal-
anced flow cytometry data, consisting of both tabular
and image data. Unlike previous studies, which did
not utilise blood test data at the individual cell level,
our focus was on leveraging this detailed blood cell
data for more accurate predictions.
To address the dual-modality issue, we developed
two models: one for tabular data and one for image
data. The predictions from these models were then
combined to produce a final prediction. The best re-
sults were achieved using random over-sampling for
the tabular data and focal loss for the image data.
Our evaluation indicates that the image-based model
outperforms the tabular model in predicting post-MI
complications. These findings underscore the poten-
tial of using detailed blood cell data and advanced
modelling techniques to improve prediction accuracy
in medical diagnostics.
REFERENCES
Bhatnagar, P., Wickramasinghe, K., Williams, J., Rayner,
M., and Townsend, N. (2015). The epidemiology of
cardiovascular disease in the uk 2014. 101(15):1182–
1189.
Bin Habib, A.-Z. S. and Tasnim, T. (2020). An ensemble
hard voting model for cardiovascular disease predic-
tion. In 2020 2nd International Conference on Sus-
tainable Technologies for Industry 4.0 (STI), pages 1–
6.
Boidin, M., Lip, G. Y., Shantsila, A., Thijssen, D., and
Shantsila, E. (2023). Dynamic changes of monocytes
subsets predict major adverse cardiovascular events
and left ventricular function after stemi. Scientific re-
ports, 13(1):48.
Centers for Disease Control and Prevention (2022). Prod-
ucts - data briefs - number 456 - september 2022. Na-
tional Center for Health Statistics. Accessed: 2024-
03-30.
Clinic, C. (2022). Congestive heart failure. https://my.cle
velandclinic.org/health/diseases/17069-heart-failure
-understanding-heart-failure.
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
88

FlowJo (2024). Flowjo data analysis software. https://ww
w.flowjo.com/solutions/flowjo. February 21, 2024.
Garc
´
ıa, S., Luengo, J., and Herrera, F. (2015). Data prepro-
cessing in data mining, volume 72. Springer.
G
´
eron, A. (2017). Hands-On Machine Learning with Scikit-
Learn and TensorFlow: Concepts, Tools, and Tech-
niques to Build Intelligent Systems. O’Reilly Media,
Sebastopol, CA.
Ghafari, R., Azar, A. S., Ghafari, A., Aghdam, F. M.,
Valizadeh, M., Khalili, N., and Hatamkhani, S.
(2023). Prediction of the fatal acute complications
of myocardial infarction via machine learning algo-
rithms. The Journal of Tehran University Heart Cen-
ter, 18(4):278–287.
Izonin, I., Ilchyshyn, B., Tkachenko, R., Gregu
ˇ
s, M.,
Shakhovska, N., and Strauss, C. (2022). Towards data
normalization task for the efficient mining of medical
data. In 2022 12th International Conference on Ad-
vanced Computer Information Technologies (ACIT),
pages 480–484.
Jayalakshmi, T. and Santhakumaran, A. (2011). Statistical
normalization and back propagation for classification.
International Journal of Computer Theory and Engi-
neering, 3(1):1793–8201.
Khera, R., Haimovich, J., Hurley, N. C., McNamara, R.,
Spertus, J. A., Desai, N., Rumsfeld, J. S., Masoudi,
F. A., Huang, C., Normand, S.-L., Mortazavi, B. J.,
and Krumholz, H. M. (2021). Use of Machine Learn-
ing Models to Predict Death After Acute Myocardial
Infarction. JAMA Cardiology, 6(6):633–641.
Khushi, M., Shaukat, K., Alam, T. M., Hameed, I. A., Ud-
din, S., Luo, S., Yang, X., and Reyes, M. C. (2021). A
comparative performance analysis of data resampling
methods on imbalance medical data. IEEE Access,
9:109960–109975.
Li, X., Shang, C., Xu, C., Wang, Y., Xu, J., and Zhou,
Q. (2023). Development and comparison of machine
learning-based models for predicting heart failure af-
ter acute myocardial infarction. BMC Medical Infor-
matics and Decision Making, 23(1):165.
Lin, T.-Y., Goyal, P., Girshick, R., He, K., and Doll
´
ar, P.
(2017). Focal loss for dense object detection. In
Proceedings of the IEEE international conference on
computer vision, pages 2980–2988.
Mohammad, M. A., Olesen, K. K. W., Koul, S., Gale, C. P.,
Rylance, R., Jernberg, T., Baron, T., Spaak, J., James,
S., Lindahl, B., Maeng, M., and Erlinge, D. (2022).
Development and validation of an artificial neural net-
work algorithm to predict mortality and admission to
hospital for heart failure after myocardial infarction: a
nationwide population-based study. The Lancet. Dig-
ital health, 4(1):e37–e45.
Mpanya, D., Celik, T., Klug, E., and Ntsinjana, H. (2021).
Predicting mortality and hospitalization in heart fail-
ure using machine learning: A systematic literature
review. IJC Heart & Vasculature, 34:100773.
Murphy, S. L., Kochanek, K. D., Xu, J., and Arias, E.
(2021). Mortality in the united states, 2020. National
Center for Health Statistics (NCHS), Data Brief Num.
427.
Newaz, A., Mohosheu, M. S., and Al Noman, M. A.
(2023). Predicting complications of myocardial in-
farction within several hours of hospitalization using
data mining techniques. Informatics in Medicine Un-
locked, 42:101361.
Oliveira, M., Seringa, J., Pinto, F. J., Henriques, R., and
Magalh
˜
aes, T. (2023). Machine learning prediction of
mortality in acute myocardial infarction. BMC Medi-
cal Informatics and Decision Making, 23(1):1–16.
Piros, P., Ferenci, T., Fleiner, R., Andr
´
eka, P., Fujita, H.,
F
˝
oz
˝
o, L., Kov
´
acs, L., and J
´
anosi, A. (2019). Compar-
ing machine learning and regression models for mor-
tality prediction based on the hungarian myocardial
infarction registry. Knowledge-Based Systems, 179:1–
7.
PyTorch (2024). torch.nn.CrossEntropyLoss. https://pytorc
h.org/docs/stable/generated/torch.nn.CrossEntropyL
oss.html. Febarary 20, 2024.
Qing Ye, Jie Zhang, L. M. (2020). Predictors of all-
cause 1-year mortality in myocardial infarction pa-
tients. Medicine, 99(23).
Reddy, K., Khaliq, A., and Henning, R. (2015). Recent
advances in the diagnosis and treatment of acute my-
ocardial infarction. World Journal of Cardiology,
7(5):243–276.
Saxena, A., Kumar, M., Tyagi, P., Sikarwar, K., and Pathak,
A. (2022). Machine learning based selection of my-
ocardial complications to predict heart attack. In 2022
IEEE 9th Uttar Pradesh Section International Con-
ference on Electrical, Electronics and Computer En-
gineering (UPCON), pages 1–4. IEEE.
Shantsila, E., Ghattas, A., Griffiths, H., and Lip, G.
(2019). Mon2 predicts poor outcome in st-elevation
myocardial infarction. Journal of internal medicine,
285(3):301–316.
Shantsila, E., Tapp, L. D., Wrigley, B. J., Montoro-Garcia,
S., and Lip, G. Y. (2013). Cxcr4 positive and angio-
genic monocytes in myocardial infarction. Thrombo-
sis and haemostasis, 109(02):255–262.
Shantsila, E., Wrigley, B., Tapp, L., Apostolakis, S.,
Montoro-Garcia, S., Drayson, M., and Lip, G. (2011).
Immunophenotypic characterization of human mono-
cyte subsets: possible implications for cardiovascular
disease pathophysiology. Journal of Thrombosis and
Haemostasis, 9(5):1056–1066.
Singh, D. and Singh, B. (2020). Investigating the impact
of data normalization on classification performance.
Applied Soft Computing, 97:105524.
Thygesen, K., Alpert, J. S., Jaffe, A. S., Simoons, M. L.,
Chaitman, B. R., and White, H. D. (2012). Third uni-
versal definition of myocardial infarction. circulation,
126(16):2020–2035.
Yang, S., Xiao, W., Zhang, M., Guo, S., Zhao, J., and Shen,
F. (2022). Image data augmentation for deep learning:
A survey. arXiv preprint arXiv:2204.08610.
Zhang, G. (2000). Neural networks for classification: a sur-
vey. IEEE Transactions on Systems, Man, and Cyber-
netics, Part C (Applications and Reviews), 30(4):451–
462.
Predicting Post Myocardial Infarction Complication: A Study Using Dual-Modality and Imbalanced Flow Cytometry Data
89

Zhang, Y., Kang, B., Hooi, B., Yan, S., and Feng, J. (2023).
Deep long-tailed learning: A survey. IEEE Transac-
tions on Pattern Analysis and Machine Intelligence,
45(9):10795–10816.
Zhou, V. (2020). Precision, recall, and f score concepts in
detail. https://regenerativetoday.com/precision-recal
l-and-f-score-concepts-in-details/. March 20, 2024.
Zhou, Z.-H. and Jiang, Y. (2003). Medical diagnosis with
c4.5 rule preceded by artificial neural network ensem-
ble. IEEE Transactions on Information Technology in
Biomedicine, 7(1):37–42.
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
90
