
Explainability Applied to a Deep-Learning Based Algorithm for Lung
Nodule Segmentation
Arman Zafaranchi
1,2
, Francesca Lizzi
1
, Alessandra Retico
1
, Camilla Scapicchio
1,2
and
Maria Evelina Fantacci
1,2
1
National Institute for Nuclear Physics (INFN), Pisa, Italy
2
Department of Physics, University of Pisa, Pisa, Italy
Keywords: Lung Nodule Detection, Lung Segmentation, Deep Learning, Segmentation, Lung Cancer.
Abstract: Deep learning and computer-aided detection (CAD) methods play a pivotal role in the early detection and
diagnosis of various cancer types. The significance of AI in the medical field has become particularly
pronounced during the coronavirus pandemic. This study aims to develop a deep learning-based system for
segmenting and detecting nodules in the lung parenchyma, utilizing the Luna-16 challenge dataset. The
algorithm is divided into two phases: the first phase involves lung segmentation using the previously
developed LungQuant algorithm to identify the region of interest (ROI), and the second phase employs a
specifically designed and fine-tuned Attention Res-UNet for nodule segmentation. Additionally, the
explainable AI (XAI) technique, Grad-CAM, was used to demonstrate the reliability of the proposed
algorithm for clinical application. In the initial phase, the LungQuant algorithm achieved an average Dice
Similarity Coefficient (DSC) of 90%. For nodule segmentation, the DSC scores were 81% test sets. The model
also achieved average sensitivity and specificity metrics of 0.86 and 0.92.
1 INTRODUCTION
Lung cancer imposes a significant global health
burden, with an alarming annual incidence of over 1.6
million new cases worldwide. As the second most
common form of cancer, it surpassed breast cancer in
incidence among women in developed nations.
Despite advances in medical technology, the
prognosis for lung cancer remains challenging
(Houda et al., 2024).
Early detection of lung cancer is crucial for
effective treatment and improved survival rates
(Mohamed et al., 2024). Despite physical symptoms
(Durstenfeld et al., 2022), more accurate diagnostic
methods are necessary to initiate treatment. Computed
Tomography (CT) is a highly sensitive imaging
modality. However, frequent CT scans, as required by
possible screening programs, can lead to overexposure
to ionizing radiation. To mitigate this risk, Low Dose
CT (LDCT) scans are now employed for high-risk
patients, allowing the reduction of radiation exposure
through advanced reconstruction and analysis software
(Barca et al., 2018). LDCT is effective in detecting
early-stage lung cancer and has been shown to reduce
mortality rates by 20% (Silva et al., 2022).
Medical image analysis is a challenging task that
requires a high degree of concentration and substantial
expertise, with significant variability among
specialists. This is particularly true in the context of
lung cancer, where small nodules indicate positive
cases, yet these nodules frequently lack uniform size,
volume or location which make them difficult to
detect. This variability is crucial during the early stages
of treatment and can greatly affect a patient's long-term
survival prospects (Peters et al., 2021).
The significance of AI in medical imaging has been
further underscored during the COVID-19 pandemic,
where researchers have developed CAD systems to aid
in detecting infected lesions in lung CT scans. These
AI-powered tools serve as invaluable aids to
radiologists, enhancing diagnostic accuracy and
expediting patient care processes (Greenspan et al.,
2020).
During the COVID-19 pandemic, researchers
developed several CAD systems (Karimkhani et al.,
2022; Lizzi et al., 2023) to assist physicians in
detecting infected lesions in lung CT scans. AI-based
software has proven to be a supportive tool for
radiologists, capable of highlighting potential
abnormalities in CT scans that might be overlooked,
132
Zafaranchi, A., Lizzi, F., Retico, A., Scapicchio, C. and Fantacci, M. E.
Explainability Applied to a Deep-Learning Based Algorithm for Lung Nodule Segmentation.
DOI: 10.5220/0013014600003886
In Proceedings of the 1st International Conference on Explainable AI for Neural and Symbolic Methods (EXPLAINS 2024), pages 132-138
ISBN: 978-989-758-720-7
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

thereby prompting further review or additional tests
by human experts. (Gozes et al., n.d.) developed a
deep learning-based CT image analysis system that
could accurately differentiate between COVID-19
positive and negative patients. This system localized
lung abnormalities and provided quantitative
measurements, supporting radiologists' diagnostic
and prognostic assessments.
The AI system consisted of multiple components,
analysing CT cases at two levels: 3D analysis for
nodules and focal opacities using existing algorithms,
and 2D analysis of each slice to detect larger diffuse
opacities, such as ground-glass infiltrates.
Additionally, (Fang et al., 2021) designed an AI-
powered framework to assess disease severity and
predict outcomes for COVID-19 patients. This
framework was evaluated using datasets from two
hospitals and compared against manual assessments
by radiologists, demonstrating superior accuracy in
predicting ICU admissions and mortality. The study
highlighted the potential of AI-based methodologies
to enhance the management of COVID-19 patients
(Scapicchio et al., n.d.).
The AI system's performance was compared to eight
human observers and the clinical assessments of
patients, including RT-PCR testing. The findings
revealed that CORADS-AI successfully automated
the scoring of chest CT scans, aligning with the CO-
RADS and CT severity score metrics, and performed
comparably to human observers in terms of CT
severity scores, with equal or superior proficiency in
identifying COVID-19 positive patients.
In recent years, deep learning (DL) methods have
emerged as powerful tools for medical image
analysis, offering significant improvements in the
segmentation of lung nodules. These methods
leverage large datasets and complex algorithms to
identify and delineate nodules with high precision.
One such algorithm, adapted from the LungQuant
approach, forms the foundation of our method’s
initial phase in finding the ROI.
Despite their potential, the "black-box" nature of DL
models raises concerns about their transparency and
interpretability, which are crucial for clinical
adoption. Therefore, incorporating XAI techniques is
imperative to ensure the transparency and reliability
of these models, thereby fostering trust among
medical professionals. We will present our approach
to lung nodule segmentation using DL methods,
supplemented by XAI results, to demonstrate the
accuracy and interpretability of our models. By doing
so, we aim to highlight the transformative potential of
DL in lung cancer diagnosis and advocate for the
integration of XAI in clinical practice.
2 MATERIAL AND METHODS
The main goal of our project is to create a reliable and
robust CAD for lung cancer detection utilizing deep
learning methods. In the first step of our paper we
implemented a two-step algorithm using the Luna-16
dataset alongside with explainable AI techniques to
demonstrate the reliability of the model. Fig.1
illustrates the schematic representation of the
proposed algorithm.
2.1 Dataset
A noteworthy dataset used in our study is the Lung
Nodule Analysis 2016 challenge (Luna-16) (Murphy
et al., 2009), renowned for its application in lung
cancer detection. Comprising CT scans from 888
patients, Luna-16 provides ground truth information
for ROI segmentation, along with the coordinates of
nodules in a 3D scale. Luna-16 is derived from the
LIDC-IDRI dataset, featuring specific nodule
volumes and low-dose CT screening. For the first
phase obviously, we used original CTs with ground
truth of lung parenchyma for training. Then, for the
second phase of the algorithm, we generated a 3D
cube with nodules in the determined coordinates, so
during the training process each slice of segmented
ROI can match with the generated mask. Before
segmentation, we applied initial preprocessing to the
CT scans, which included normalizing the image
intensities and the Hounsfield Unit of CTs.
2.2 Phase 1: Lung Segmentation
Lung nodule segmentation is a challenging task for
AI due to factors such as image noise, imbalanced
data, and the complex structure of lung tissues. To
address these challenges, we implemented several
techniques. In the initial step, identifying the ROI
helps to reduce the complexity of the image structure.
For this purpose, we utilized first part of the
LungQuant algorithm to segment the lung region
from body organs in CT scans.
LungQuant is a fully automated deep learning-
based system designed to assist radiologists in
detecting lung lesions indicative of COVID-19
infection (Lizzi et al., 2022). The initial version,
introduced in 2023, demonstrated significant
promise. A subsequent version was released with a
refined structure to enhance the segmentation
accuracy of lung parenchyma and COVID-19
pneumonia in CT scans (Lizzi et al., 2023). This
section will explore the details of the LungQuant
methodology.
Explainability Applied to a Deep-Learning Based Algorithm for Lung Nodule Segmentation
133
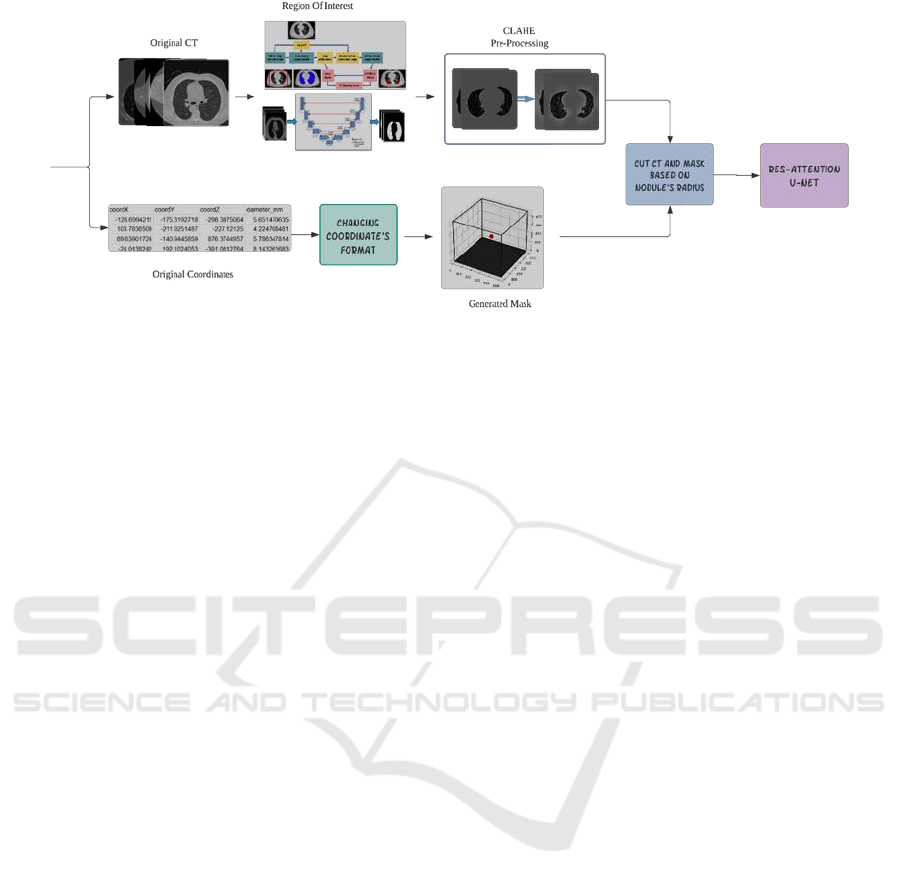
Figure 1: Diagram of proposed algorithm.
LungQuant was developed using deep learning
algorithms in multiple steps and have been evaluated
to asses with various datasets (Scapicchio et al.,
2023). Initially, an AlexNet-based DNN predicts two
points to define a bounding box around the 3D voxel
data of the lungs, aiding in the localization of the lung
parenchyma for further analysis. The next phase
employs two U-nets: the first segments the lung
parenchyma, which we have utilized in this paper,
and the second uses these results to accurately
identify and delineate COVID-19 lesions. Pre-
processing and data augmentation were applied to
prevent overfitting and improve model performance.
2.3 CLAHE Preprocessing
After segmenting the lung region using LungQuant,
we applied Contrast Limited Adaptive Histogram
Equalization (CLAHE) to the segmented lung images
(Kyriakopoulou, 2020). CLAHE is an advanced
image preprocessing technique used to enhance the
contrast of images, particularly in medical imaging
for improving the visibility of features within an
image. This technique improves the contrast of an
image in a localized manner, making it easier to
detect features like lung nodules in medical images.
By limiting the contrast enhancement, CLAHE
reduces the risk of noise amplification while
preserving fine details and edges in the image, which
is crucial for accurate diagnosis and analysis in
medical imaging.
2.4 Phase 2: Nodule Segmentation
In the second phase of our methodology, we focus on
the segmentation of lung nodules using an advanced
deep learning model. This phase builds upon the
output of the first phase, where the lung region was
isolated using the LungQuant algorithm.
To achieve accurate nodule segmentation, we
employed an Attention Res-UNet architecture. This
model is designed to enhance the focus on relevant
features while maintaining the spatial details crucial
for precise nodule detection. The Attention Res-UNet
incorporates attention blocks that selectively
highlight important features in the image, reducing
the impact of irrelevant background information. This
mechanism improves the model’s ability to detect
small and subtle nodules amidst the lung parenchyma.
Moreover, the architecture utilizes residual
connections, allowing the model to learn more
effectively by mitigating the vanishing gradient
problem. This enhancement helps in preserving the
gradient flow through deep layers, ensuring better
learning of complex patterns. For the training process,
we generated 3D cubes with nodules at the specified
coordinates provided by the LUNA-16 dataset. Using
the nodule coordinates from the dataset, we created
binary masks for each nodule. These masks are
essential for training the model, providing the ground
truth for the nodule locations. We used the Dice Loss
function (Sudre, C.H., Li, W., Vercauteren, T.,
Ourselin, S., Jorge Cardoso, 2017), which is
particularly effective for imbalanced data, where
background voxels are more than nodules one. The
fine-tuning process involved training on the
generated data and refining the model’s architecture
to enhance its ability to distinguish nodules from
surrounding tissue, thereby yielding promising results
in lung nodule segmentation This pre-processing
ensures that each slice of the segmented ROI can be
matched with the corresponding mask.
EXPLAINS 2024 - 1st International Conference on Explainable AI for Neural and Symbolic Methods
134

Figure 2: Results of Lung segmentation with LungQuant’s first phase.
2.5 Model Explanation and
Performance Evaluation
Explainable AI is crucial in various applications,
especially in high-stakes fields like healthcare, for
building trust and transparency in order to demystify
the “black box” nature of deep learning models to
make their decision transparent. Moreover, In
healthcare, decisions based on AI can have significant
consequences. XAI ensures that AI models can be
held accountable for their decisions, providing
explanations that can be analysed.
To ensure the interpretability of our model, we
applied the Grad-CAM (Gradient-weighted Class
Activation Mapping) technique. Grad-CAM
(Selvaraju et al., 2016) is a powerful visualization
tool that helps in understanding and interpreting the
decisions made by deep learning models. It highlights
the regions in the input image that contribute most
significantly to the model's predictions, thereby
providing a visual explanation of the model's focus
and attention. For each CT scan slice processed by the
Attention Res-UNet, we generated Grad-CAM
heatmaps.
These heatmaps were overlaid on the original CT
images to highlight the regions where the model
focused its attention while identifying nodules. The
visual explanations provided by Grad-CAM helped in
validating the model’s predictions by confirming
whether the identified regions correspond to actual
nodules. This step is crucial for gaining the trust of
medical professionals and ensuring the reliability of
the AI system. By analysing the Grad-CAM
heatmaps, we could identify any potential areas
where the model might be making incorrect
predictions or missing nodules. This feedback loop
allowed us to fine-tune the model and improve its
performance iteratively.
2.6 Metrics
To assess the performance of each phase, we applied
appropriate metrics for thorough evaluation and
comparison. For the first phase, lung segmentation
performance validation, we used the DSC to measure
the overlap between prediction and ground truth. For
the second phase of nodule segmentation, we utilized
sensitivity, specificity, and the average False Positive
Rate (FPR) per scan. These metrics provide a
comprehensive evaluation of the algorithm's accuracy
and reliability in both lung region segmentation and
nodule detection.
3 RESULTS
In this section, we present the outcomes of our study
on lung nodule segmentation using DL methods,
supported by XAI. The results are organized to
demonstrate the efficacy of our approach, the
performance of the model, and the interpretability of
its decisions.
Up to this point, we have elaborated on the details
of the proposed algorithm. Broadly speaking, we have
three distinct objectives in this paper. The first
objective is to use and evaluate the performance of
LungQuant for lung segmentation purposes. By
achieving this, we aim to obtain a more precise ROI
and demonstrate the robustness of our deep learning-
based algorithm. This step is crucial in ensuring the
accuracy of subsequent phases and in showcasing the
efficacy of LungQuant in clinical applications. In the
original LungQuant paper, a 96% DSC was achieved
on the COVID-19-CT-Seg dataset. For our first
objective, we evaluated the lung segmentation task
using DSC and obtained an average score of 90%
based on the provided ground truth. Fig. 2
demonstrates the algorithm's robustness across
different datasets and highlights LungQuant's
exceptional performance in the more challenging
regions of the lung, i.e. the bases.
In the second phase, we developed an Attention
Res-UNet architecture specifically for the nodule
segmentation task. To enhance the clarity of lung
tissue and reduce noise, we applied CLAHE to the
outputs from the first step. This preprocessing step
Explainability Applied to a Deep-Learning Based Algorithm for Lung Nodule Segmentation
135
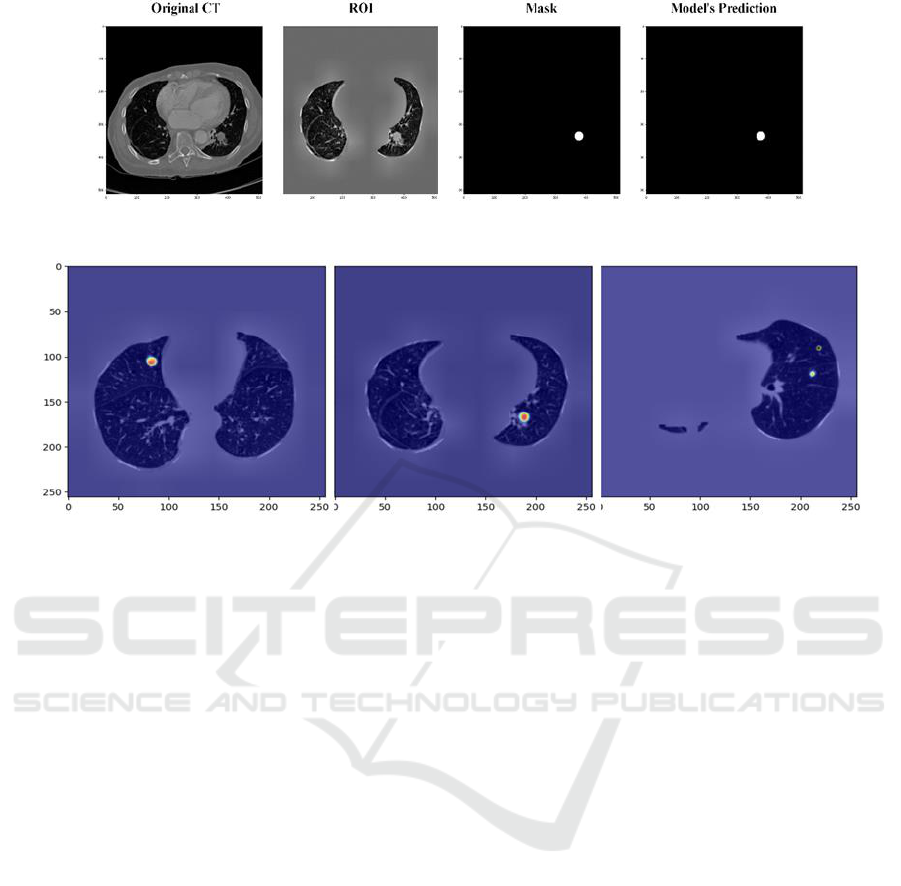
Figure 3: Prediction of Attention Res-Unet.
Figure 4: Results of Grad-Cam for Explainability of Nodule segmentation.
was essential for improving the visibility of subtle
features within the lung images. Subsequently, we
fine-tuned the Attention Res-UNet model, optimizing
its parameters to achieve robust performance in
detecting and segmenting lung nodules. The trained
neural network achieved Dice Coefficients of 85%,
83%, and 81% for the training, validation, and test
sets, respectively. Additionally, the model reached
average sensitivity and specificity metrics of 0.86 and
0.92, with an average FPR of 2.25 per scan,
demonstrating its effectiveness and reliability in lung
nodule segmentation. Figure 3 showcases the
accurate segmentation results of our fine-tuned model
for nodule detection. The comparison between the
predicted points and the generated mask highlights
the model’s outstanding performance.
Final objective of this paper is to visualize the
areas where the Attention Res-UNet model focused
during prediction. Grad-CAM generates heatmaps
that highlight important regions in the input image for
predicting lung nodules, providing insights into the
model’s decision-making process. In Fig. 4, the Grad-
CAM visualization shows a focused heatmap around
a small, distinct region within the lung parenchyma.
The highlighted region corresponds to a suspected
nodule, indicating that the model successfully
identified this area as important for nodule detection.
The concentration of the heatmap around the nodule
demonstrates the model's ability to localize the
nodule accurately. Moreover, in the case with
presence of two nodules the high-intensity heatmap
accurately highlights the nodule's location.
4 DISCUSSION
As mentioned before, our project’s goal is to develop
a deep learning-based CAD algorithm for lung cancer
detection. Up to this point, we have designed, fine-
tuned, and tested several complex deep neural
networks to evaluate and compare the performance of
different models, i.e. U-Net, Res U-Net, Attention U-
Net, on the LUNA-16 dataset.
Recent research indicates that attention
mechanisms can perform well with complex data like
medical images. Specifically, in our scenario of
detecting lung nodules with low volume amidst lung
tissues, the attention mechanism can effectively focus
on the target parts. Additionally, residual blocks help
to mitigate the vanishing gradient issue, which is
likely due to the similar structure of the data.
One of the long-term goals of this project is to
implement the developed algorithm in clinical
environments, which necessitates ensuring the
reliability and robustness of the CAD system. The
integration of our proposed DL-based methodology,
particularly the use of LungQuant for lung
segmentation, and an Attention Res-UNet for nodule
EXPLAINS 2024 - 1st International Conference on Explainable AI for Neural and Symbolic Methods
136

segmentation, has the potential to improve diagnostic
workflows in clinical settings. This approach can
assist radiologists by providing accurate and reliable
segmentation, thereby reducing workload and
improving early detection rates of lung cancer.
Incorporating XAI techniques, such as Grad-
CAM, is vital for guaranteeing the transparency and
trustworthiness of AI models in medical imaging.
XAI offers insights into the model’s decision-making
process, thereby enhancing the interpretability and
acceptance of AI-based tools by medical
professionals.
In this process, we encounter several challenges.
One limitation of our study is the relatively small
dataset size, which may impact the generalizability
and robustness of our results. Furthermore, variations
in image quality and the assumptions made during
model training and evaluation could influence the
overall performance. To handle some of these issues
for our future research we intend to focus on
expanding the dataset to include more diverse cases,
further improving the model architecture, and
integrating additional preprocessing techniques to
enhance segmentation accuracy. Moreover, extensive
clinical trials are necessary to validate the efficacy of
the proposed methodology in real-world clinical
environments.
5 CONCLUSIONS
In this study, we emphasize the critical role of deep
learning-based CAD systems in the detection of lung
cancer using CT datasets, highlighting the importance
of early detection in improving patient survival rates.
We employed the LungQuant automated system for
segmenting the lung region and demonstrated the
generalization of this algorithm with different
datasets, achieving an average of 90% DSC with
Luna-16, in comparison to the 96% reported in the
original study. We then applied CLAHE
preprocessing to reduce noise and enhance tissue
details in the lung parenchyma. These pre-processed
images were input into an Attention Res-UNet for the
nodule segmentation task, resulting in DSC scores of
85%, 83%, and 81% for the training, validation, and
test sets, respectively. The model achieved average
sensitivity and specificity metrics of 0.86 and 0.92,
with an average FPR of 2.25 per scan. Our findings
indicate that attention mechanisms and residual
blocks significantly enhance segmentation
performance, even in complex scenarios. This work
underscores the transformative potential of deep
learning and explainable AI in lung cancer diagnosis,
advocating for their integration into clinical practice
to improve patient outcomes. For future work, we aim
to further refine the model to reduce the false positive
rate per scan, thereby enhancing its clinical utility and
reliability.
ACKNOWLEDGEMENTS
Research partly supported by: Artificial Intelligence
in Medicine (next AIM, https://www.pi.infn.it/aim)
project, INFN-CSN5; PNRR - M4C2 -
Partenariato Esteso ”FAIR - Future Artificial
Intelligence Research” - Spoke 8, funded by the
European Commission under the NextGeneration
EU programme; the European Union
NextGenerationEU through the Italian Ministry
of University and Research under PNRR M4C2-I1.3
Project PE 00000019 ”HEAL ITALIA” to Maria
Evelina Fantacci and Arman Zafaranchi CUP
I53C22001440006; European Union -
NextGenerationEU through the Italian Ministry of
University and Research under PNRR - M4C2-I1.5 -
Project ECS00000017 “Tuscany Health Ecosystem
(THE)” - CUP I53C21000350006.
The views and opinions expressed are those of the
authors only and do not necessarily reflect those of
the European Union or the European Commission
Neither the European Union nor the European
Commission can be held responsible for them.
REFERENCES
Barca, P., Palmas, F., Fantacci, M. E., & Caramella, D.
(2018). Evaluation of the adaptive statistical iterative
reconstruction algorithm in chest CT (Computed
Tomography) a preliminary study toward its
employment in low dose applications, also in
conjunction with CAD (Computer Aided Detection).
HEALTHINF 2018 - 11th International Conference on
Health Informatics, Proceedings; Part of 11th
International Joint Conference on Biomedical
Engineering Systems and Technologies, BIOSTEC
2018, 5, 688–694. https://doi.org/10.5220/
0006750706880694
Durstenfeld, M. S., Sun, K., Tahir, P., Peluso, M. J., Deeks,
S. G., Aras, M. A., Grandis, D. J., Long, C. S., Beatty,
A., & Hsue, P. Y. (2022). Use of Cardiopulmonary
Exercise Testing to Evaluate Long COVID-19
Symptoms in Adults: A Systematic Review and Meta-
analysis. In JAMA Network Open (Vol. 5, Issue 10, pp.
E2236057–E2236057). American Medical
Association. https://doi.org/10.1001/jamanetworkopen.
2022.36057
Explainability Applied to a Deep-Learning Based Algorithm for Lung Nodule Segmentation
137

Fang, X., Kruger, U., Homayounieh, F., Chao, H., Zhang,
J., Digumarthy, S. R., Arru, C. D., Kalra, M. K., & Yan,
P. (2021). Association of AI quantified COVID-19
chest CT and patient outcome. International Journal of
Computer Assisted Radiology and Surgery, 16(3), 435–
445. https://doi.org/10.1007/s11548-020-02299-5
Gozes, O., Frid-Adar, M., Greenspan, H., Browning, P. D.,
Zhang, H., Ji, W., ... & Siegel, E. (2020). Rapid ai
development cycle for the coronavirus (covid-19)
pandemic: Initial results for automated detection &
patient monitoring using deep learning ct image
analysis. arXiv preprint arXiv:2003.05037.Greenspan,
H., San José Estépar, R., Niessen, W. J., Siegel, E., &
Nielsen, M. (2020). Position paper on COVID-19
imaging and AI: From the clinical needs and
technological challenges to initial AI solutions at the lab
and national level towards a new era for AI in
healthcare. Medical Image Analysis, 66.
https://doi.org/10.1016/j.media.2020.101800
Houda, I., Dickhoff, C., Uyl-de Groot, C. A., Reguart, N.,
Provencio, M., Levy, A., Dziadziuszko, R., Pompili, C.,
Di Maio, M., Thomas, M., Brunelli, A., Popat, S.,
Senan, S., & Bahce, I. (2024). New systemic treatment
paradigms in resectable non-small cell lung cancer and
variations in patient access across Europe. The Lancet
Regional Health - Europe, 38, 100840.
https://doi.org/10.1016/j.lanepe.2024.100840
Karimkhani, H., Attariabad, A., & Vahed, H. (2022). High
sensitive plasmonic sensor with simple design of the
ring and the disk resonators. Optical and Quantum
Electronics, 54(6). https://doi.org/10.1007/s11082-
022-03736-2
Kyriakopoulou, M. (2020). Histogram Equalization on
Medical Images: CLAHE implementation on CT
images.
Lizzi, F., Agosti, A., Brero, F., Cabini, R. F., Fantacci, M.
E., Figini, S., Lascialfari, A., Laruina, F., Oliva, P.,
Piffer, S., Postuma, I., Rinaldi, L., Talamonti, C., &
Retico, A. (2022). Quantification of pulmonary
involvement in COVID-19 pneumonia by means of a
cascade of two U-nets: training and assessment on
multiple datasets using different annotation criteria.
International Journal of Computer Assisted Radiology
and Surgery, 17(2), 229–237.
https://doi.org/10.1007/s11548-021-02501-2
Lizzi, F., Postuma, I., Brero, F., Cabini, R. F., Fantacci, M.
E., Lascialfari, A., Oliva, P., Rinaldi, L., & Retico, A.
(2023). Quantification of pulmonary involvement in
COVID-19 pneumonia: an upgrade of the LungQuant
software for lung CT segmentation. European Physical
Journal Plus, 138(4). https://doi.org/10.1140/epjp/
s13360-023-03896-4
Mohamed, E., García Martínez, D. J., Hosseini, M. S.,
Yoong, S. Q., Fletcher, D., Hart, S., & Guinn, B. A.
(2024). Identification of biomarkers for the early
detection of non-small cell lung cancer: a systematic
review and meta-analysis. Carcinogenesis, 45(1–2), 1–
22. https://doi.org/10.1093/carcin/bgad091
Murphy, K., van Ginneken, B., Schilham, A. M. R., de
Hoop, B. J., Gietema, H. A., & Prokop, M. (2009). A
large-scale evaluation of automatic pulmonary nodule
detection in chest CT using local image features and k-
nearest-neighbour classification. Medical Image
Analysis, 13(5), 757–770. https://doi.org/10.1016/j.
media.2009.07.001
Peters, A. A., Decasper, A., Munz, J., Klaus, J., Loebelenz,
L. I., Hoffner, M. K. M., Hourscht, C., Heverhagen, J.
T., Christe, A., & Ebner, L. (2021). Performance of an
AI based CAD system in solid lung nodule detection on
chest phantom radiographs compared to radiology
residents and fellow radiologists. Journal of Thoracic
Disease, 13(5), 2728–2737. https://doi.org/10.
21037/jtd-20-3522
Scapicchio, C., Ballante, E., Brero, F., Cabini, R. F.,
Chincarini, A., Fantacci, M. E., ... & Retico, A. (2023).
Integration of a Deep Learning-Based Module for the
Quantification of Imaging Features into the Filling-in
Process of the Radiological Structured Report.
In HEALTHINF (pp. 663-670).
Scapicchio, C., Chincarini, A., Ballante, E., Berta, L., Bicci,
E., Bortolotto, C., Brero, F., Cabini, R. F., Cristofalo,
G., Fanni, S. C., Fantacci, M. E., Figini, S., Galia, M.,
Gemma, P., Grassedonio, E., Lascialfari, A., Lenardi,
C., Lionetti, A., Lizzi, F., … Retico, A. (2023). A
multicenter evaluation of a deep learning software
(LungQuant) for lung parenchyma characterization in
COVID-19 pneumonia. European Radiology
Experimental, 7(1). https://doi.org/10.1186/s41747-
023-00334-z
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R.,
Parikh, D., & Batra, D. (2016). Grad-cam: Why did you
say that? visual explanations from deep networks via
gradient-based localization. Grad-CAM: Visual
Explanations from Deep Networks via Gradient-Based
Localization, 17, 331–336.
http://arxiv.org/abs/1610.02391
Silva, M., Picozzi, G., Sverzellati, N., Anglesio, S.,
Bartolucci, M., Cavigli, E., Deliperi, A., Falchini, M.,
Falaschi, F., Ghio, D., Gollini, P., Larici, A. R.,
Marchianò, A. V, Palmucci, S., Preda, L., Romei, C.,
Tessa, C., Rampinelli, C., & Mascalchi, M. (2022).
Low-dose CT for lung cancer screening: position paper
from the Italian college of thoracic radiology.
Radiologia Medica, 127(5), 543–559.
https://doi.org/10.1007/s11547-022-01471-y
Sudre, C.H., Li, W., Vercauteren, T., Ourselin, S., Jorge
Cardoso, M. (2017). (2017). Generalised Dice Overlap
as a Deep Learning Loss Function for Highly
Unbalanced Segmentations. Springer, vol 10553(Deep
Learning in Medical Image Analysis and Multimodal
Learning for Clinical Decision Support).
EXPLAINS 2024 - 1st International Conference on Explainable AI for Neural and Symbolic Methods
138
