
Knowledge Graphs Can Play Together: Addressing Knowledge Graph
Alignment from Ontologies in the Biomedical Domain
Hanna Abi Akl
1,2,∗ a
, Dominique Mariko
3,∗
, Yann-Alan Pilatte
3,∗
, St
´
ephane Durfort
3,∗
,
Nesrine Yahiaoui
3,∗
and Anubhav Gupta
3,∗
1
Data ScienceTech Institute (DSTI), 4 Rue de la Coll
´
egiale, 75005 Paris, France
2
Universit
´
e C
ˆ
ote d’Azur, Inria, CNRS, I3S, France
3
Yseop, 4 Rue de Penthi
`
evre, 75008 Paris, France
hanna.abi-akl@dsti.institute, {dmariko, ypilatte, sdurfort, nyahiaoui, agupta}@yseop.com
Keywords:
Knowledge Graphs, Ontologies, Natural Language Processing, Information Extraction.
Abstract:
We introduce DomainKnowledge, a system that leverages a pipeline for triple extraction from natural text and
domain-specific ontologies leading to knowledge graph construction. We also address the challenge of align-
ing text-extracted and ontology-based knowledge graphs using the biomedical domain as use case. Finally, we
derive graph metrics to evaluate the effectiveness of our system compared to a human baseline.
1 INTRODUCTION
In the era of Large Language Models (LLMs), Knowl-
edge Graphs (KGs) have resurfaced to play an im-
portant role, whether as complements to LLM-based
technology to enhance predictions in Retrieval Aug-
mented Generation (RAG) models, or as standalone
systems that more faithfully capture factual informa-
tion (Pan et al., 2023b), (Peng et al., 2023), (Vogt
et al., 2022). The ongoing problem of hallucinations
in LLMs draws a line on their reliability and ques-
tions the interpretability and explainability of their
outputs. The inability to trust the responses of these
deep learning models leads to much hesitation in im-
plementing and deploying them in production, espe-
cially in sensitive domains such as healthcare (Pan
et al., 2023a).
KGs, on the other hand, have demonstrated their
staying power by circumventing the black-box mech-
anism of LLMs and offering open and traceable repre-
sentations of domain information (Pan et al., 2023a).
Their staying power is also strengthened by their in-
tegration with both deep learning solutions and more
classical frameworks like ontologies that provide for-
mal representations of knowledge (Pan et al., 2023b),
(Vogt et al., 2022). A major weakness they exhibit
however is their difficulty in integrating and align-
ing new knowledge. Unlike LLMs, which benefit
from fine-tuning to add new knowledge, ontologies,
a
https://orcid.org/0000-0001-9829-7401
∗
All authors contributed equally to this work.
and KGs by extension, require a lot of work in order
to enrich their representations in a single domain or
extend to a new one (Van Tong et al., 2021). This
weakness makes these technologies less transferable
on their own which is why they are often utilized as
components in larger systems that can benefit from
their advantages (Peng et al., 2023).
In this work, we present DomainKnowledge, a
system comprised of a workflow of information ex-
traction (IE) from unstructured text leading to the con-
struction of a consolidated domain KG. We showcase
strategies in our implementation to combine domain
knowledge from ontological sources and amount to a
generalized domain-specific KG mapping input text
entities to higher-order concepts. We also introduce
metrics inspired from graph theory to evaluate our
system. The rest of the work is structured as follows.
Section 2 presents related work in the literature. Sec-
tion 3 describes our methodology. In section 4 we
present our experimental setup. Section 5 discusses
our findings with an analysis of our results. Finally,
we conclude with future directions of work in section
6.
2 RELATED WORK
This section covers the literature pertaining to text-to-
graph extraction techniques as well as KG alignment
methods.
416
Abi Akl, H., Mariko, D., Pilatte, Y.-A., Durfor t, S., Yahiaoui, N. and Gupta, A.
Knowledge Graphs Can Play Together: Addressing Knowledge Graph Alignment from Ontologies in the Biomedical Domain.
DOI: 10.5220/0013015800003838
In Proceedings of the 16th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K 2024) - Volume 1: KDIR, pages 416-423
ISBN: 978-989-758-716-0; ISSN: 2184-3228
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

2.1 Knowledge Graph Construction
from Text
Research in IE shows different methods to construct
KGs from text. In their work, (Liu et al., 2022) sur-
veyed different methods for text information extrac-
tion from relation triples. They explored and com-
pared systems that capture relations in the form of
triples, spans and clusters using symbolic and deep
learning techniques at the syntactic and semantic lev-
els. (Kamp et al., 2023) compared rule-based open
IE engines to machine learning extraction systems
and found a trade-off between implementation and
precision. While rule-based systems exhibited bet-
ter overall performance in identifying and extracting
relations, they were much harder and more exhaus-
tive to implement than off-the-shelf machine learning
models.
Natural language processing (NLP) methods like
sentence chunking, domain entity classification, re-
lation classification and sentence-to-graph techniques
to manipulate text directly through graph properties
have achieved promising results that exploit syntac-
tic and semantic text attributes through models built
on robust rule engines. These techniques, while ca-
pable of controlling the type of information to ex-
tract, have shown limitations when it comes to ex-
tending them to cover more exhaustive knowledge
(Chouham et al., 2023), (Dong et al., 2023), (Motger
and Franch, 2024), (Yu et al., 2022). Other research
geared toward machine and deep learning technol-
ogy combines these methods with classical NLP tech-
niques for better results. In their work, (Qian et al.,
2023) proposed an IE pipeline that combines pattern-
based, machine learning and LLM extractions that un-
dergo rule-based and machine learning scoring to de-
cide on keeping or discarding extracted information.
Transformer-based approaches have also been applied
to leverage embeddings information and transform
them to node properties in graphs constructed from
text (Friedman et al., 2022), (Melnyk et al., 2022).
These methods have showcased a better ability at cap-
turing text properties as node representations. Novel
hybrid systems making use of the availability of LLM
technology leveraged their prompting abilities to pro-
vide domain annotations for better information ex-
traction (Dunn et al., 2022), combine them with other
sources of knowledge like ontologies (Mihindukula-
sooriya et al., 2023), (Wadhwa et al., 2023) for better
coverage, and even use text generation techniques as
a comparative benchmark to identify viable relation
candidates for extraction (Hong et al., 2024).
2.2 Knowledge Graph Alignment
Several research avenues explore graph-based tech-
niques for KG alignment. (Zeng et al., 2021) sur-
vey distance-based and semantic matching scores for
effective entity alignment in KGs. In their work,
(Zhang et al., 2021) propose systems based on stacked
graph embeddings of different graph components like
neighboring entities and predicates to improve en-
tity alignment. Other methods focus on integrating
deep learning models to better express graph compo-
nent properties and answer the graph alignment prob-
lem. (Chaurasiya et al., 2022), (Dao et al., 2023) and
(Fanourakis et al., 2023) show that graph neural net-
works performed well in aligning different graph en-
tities when paired with distance-based graph features
and embeddings. In their work, (Yang et al., 2024)
show that LLMs could be leveraged to decompose the
alignment problem into multiple choice questions re-
ferring to sub-tasks to approximate the alignment of
entities with respect to neighboring nodes. (Trisedya
et al., 2023) propose a system composed of an at-
tribute aggregator and a node aggregator to combine
both node and relation properties and get better align-
ment predictions. (Zhang et al., 2023) showcase a
similar method aggregating property, relationship and
attribute triples to get a more complete representation
of entities and aid the entity alignment process.
Finally, neuro-symbolic systems aiming to com-
bine both classical rule-based techniques with sub-
symbolic architectures have also been proposed to
tackle the graph alignment problem. (Cotovio et al.,
2023) survey neural network architectures and rein-
forcement learning methods for better entity align-
ment predictions. In their work, (Xie et al., 2023)
convert different KGs into vector space embeddings
and combine them with graph neural networks to cre-
ate transitions and better delimit the best alignment
for a node entity. (Abi Akl, 2023) show the benefits of
using logic neural networks as reasoners with a rule-
set derived from upper ontologies in a hybrid system
to align entities from different KGs.
3 METHODOLOGY
The DomainKnowledge system proposes a data ac-
quisition and transformation pipeline that leverages
NLP and graph techniques to extract meaningful rela-
tionships from raw text and store them in graph struc-
tures to create a domain vocabulary. It consists of the
following components:
• An IE pipeline which handles the relationship ex-
traction. The IE component depends on the docu-
Knowledge Graphs Can Play Together: Addressing Knowledge Graph Alignment from Ontologies in the Biomedical Domain
417

ment or text extraction process that precedes it,
which should be capable of extracting raw text
and transforming it into a list of sentences, since
the IE pipeline identifies relationships at sentence
level.
• A knowledge storage system which references the
graph database storage and KG construction.
The system workflow can be summarized in the fol-
lowing steps:
• Initiate a generic pipeline to identify and extract
relations from raw text
• Define a ruleset for meaningful relations
• Prune relations to conserve only meaningful ones
• Export relations into semantic graph structures
• Generate the domain vocabulary from graph rela-
tionships
3.1 System Overview
The user provides a number of documents from the
same domain (e.g., Pharmaceutical). The documents
are processed one by one as raw texts. The Domain-
Knowledge pipeline analyzes the texts as sentences
and extracts relationships as triples of the form (sub-
ject,relation,object). Relationships are identified with
the help of a domain ontology that emphasizes impor-
tant domain words to look out for, e.g., MedDRA for
Pharmaceutical. Once relationships are extracted, a
set of rules is applied to prune the bad ones. These
rules can vary from simple, e.g., eliminating relation-
ships with missing elements in the triples, to more
complex, e.g., evaluating the nature of the relation
like verbal versus non-verbal. The ruleset can also be
aided by the reference ontology to drop relationships
that contain no relevant terms in the subject and/or
object entities of the triple. The relationship matri-
ces are then concatenated into one matrix containing
all the relevant relationships from all the documents.
The matrix is then formatted into several files and ex-
ported in a way to preserve the following information:
• Each relationship is unique and is assigned a
unique identifier
• Each relationship triple has a clear subject, predi-
cate and object
• Each relationship clearly references the sentence
it is extracted from
• Repeated triples are kept
• Each relationship clearly references the document
it is extracted from
• Each document is unique and is assigned a unique
identifier
The exported information is then ingested into a graph
database that conserves the above-mentioned infor-
mation in a graph network. The graph network is
modeled as a subject/object node KG where nodes are
subject and object entities and edges are the relation
of the triple. Each node has properties associated with
it like its unique identifier, the sentence it is extracted
from, the name of the document it is extracted from,
the unique identifier of the document, the type of the
document (e.g., Clinical Study Report, Protocol) and
the domain of the document (e.g., Pharmaceutical).
Figure 1 shows the high-level architecture of our sys-
tem.
Figure 1: DomainKnowledge pipeline.
3.2 System Modules
3.2.1 Extractor
A plain text extractor keeping document layout based
on MuPDF
1
in Python.
3.2.2 Annotator
The Annotator’s output is based on Stanza’s
2
depen-
dency parser which provides a standardized way of
representing syntactic dependencies between words
in a sentence. Our system produces relations from
texts of documents using specific dependencies ap-
pearing in Stanza’s output. Two main relation types
were considered for extraction:
1. Verbal Relations: canonical verbal relations take
a verb as a cornerstone to build a triple (entity,
verb, entity) which can be transformed to (subject,
relation, object). The relations are described as
follows:
• root: the root of the sentence should usually be
the verb that is the main predicate. The root
usually has subject(s) and object(s), unless it is
intransitive or another verbal dependency inter-
feres.
• acl: behave like roots, but their subject already
has a dependency link to another verb (typi-
cally, as an object of the root, but not only).
1
https://shorturl.at/WXmwU
2
https://stanfordnlp.github.io/stanza/
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
418

• acl:relcl: adnominal relative clause introduced
by relative pronouns, which can either be their
subject or object, and reference another subject
or object in the context.
• advcl: adverbial clauses can have their own
subjects and objects, in which case they behave
like roots. If they modify nouns and have no
subject, they are linked to the verb they modify
and its subject.
2. Prepositional Relations: we use OpenIE
3
, an
open-source relation extraction tool, to build
prepositional relations from adpositions (e.g.,
’as’, ‘with’, ‘for’, etc.). Considering prepositions
as the pivot of the relation, subjects and objects
of verbal relations are split into smaller pieces
and can match better with ontology terms. We
use the dependency tag of the object entities of
these relations to identify them with the nmod tag.
The Annotator module is also in charge of con-
structing triples. Each sentence in the original text
is decomposed into entity-relation triples and stored
with metadata attributes such as document ID, sec-
tion ID (from the document layout), sentence ID, to-
kens positions and tokens POS tags. The triples are
sets of nodes and relations to be compared with the
value of the string data type available in the UMLS
metathesaurus
4
. Figure 2 shows the annotation logic.
Figure 2: Annotation flowchart.
3
https://shorturl.at/2VNh0
4
https://shorturl.at/5F0P8
3.2.3 Aggregator
The Aggregator relies on the National Library
of Medicine Unified Medical Language System
(UMLS® 2022AA) release. We consider the MR-
CONSO, MRSAB, MRSTY and MRREL data ta-
bles and reorganize their content into a graph data
model. We follow the UMLS data types as described
in the UMLS Metathesaurus Rich Release Format
5
and keep the data objects as nodes in the graph data
model. The data model consists of the following
nodes:
• AUI: atom
• CUI: concept
• LUI: term
• SUI: unique string
• TUI: semantic type
We preserve the relationships attributes as defined
in the original UMLS Metathesaurus
6
. We turn the
incoming relationships into direct links to find paths
between text NER nodes and UMLS nodes:
• CUI node has an atom node: CUI
HAS AUI
−−−−−→ AU I
• SUI node has an atom node: SUI
HAS AUI
−−−−−→ AU I
• SUI node has concept node: SUI
HAS CUI
−−−−−→ CUI
• CUI node has semantic type node: CUI
HAS STY
−−−−−→
TUI
The resulting data model is available in Figure 3.
3.2.4 Merger
Outputs from the Annotator, i.e., entity-relation
triples, and the Aggregator, i.e., SUI objects, are
mapped with measures of semantic similarity using
the following algorithm:
• An exact matching measure using the Levenshtein
distance to compute a first similarity score.
• A semantic matching algorithm using cosine sim-
ilarity to compute a more refined evaluation of en-
tities that do not score highly on the exact match-
ing: each entity is mapped to a 512 dimensional
dense vector space, so the semantic matching al-
gorithm can draw similarities from the generated
vectors to find associations between two entities.
An additional Named Entity Recognition tagger,
i2b2
7
, is used to map long triples entities and SUI ob-
jects to augment the text-to-ontology mapping. Sub-
ject and object entities declared in extracted triples
5
https://shorturl.at/2HYBj
6
https://shorturl.at/iV97e
7
https://shorturl.at/aMN80
Knowledge Graphs Can Play Together: Addressing Knowledge Graph Alignment from Ontologies in the Biomedical Domain
419
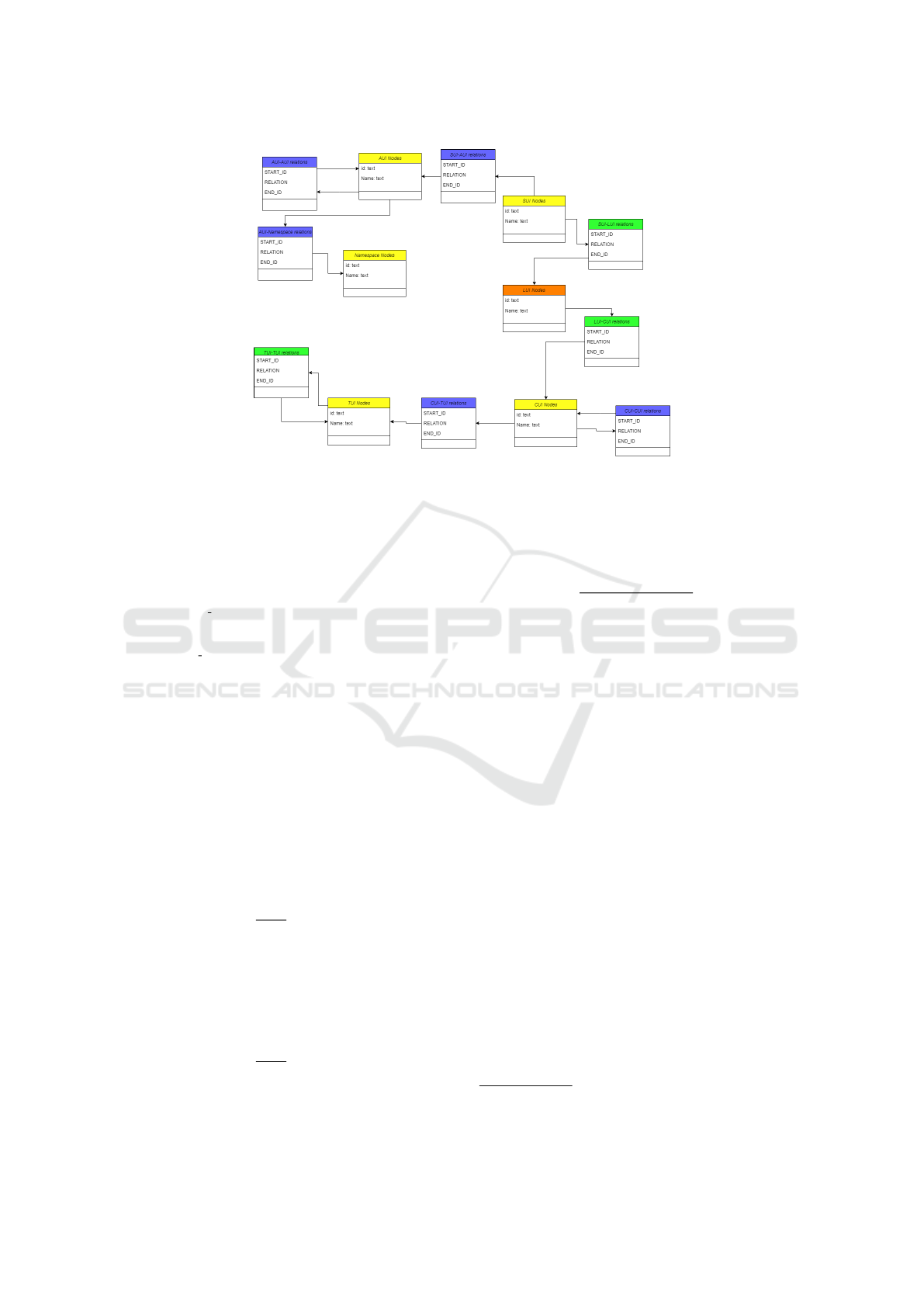
Figure 3: Graph data model.
from sentences are declared as NER nodes in the con-
structed KG. The Merger outputs a KG construction
from a set of pre-configured semantic graphs, consis-
tent with the graph data model, adding the following
nodes and edges to the graph:
• Text node linked to another text node:
NER
T EXT LINK
−−−−−−−→ NER
• Text node matched to SUI node:
NER
HAS LEX ICAL
−−−−−−−−→ SU I
An example graph is presented in Figure 4.
3.3 Metrics
We define the following evaluation metrics:
• Coverage (CVRG). Let DT be the set of domain
tokens, i.e., any extracted entity from a given text
that is also linked, i.e., sharing a direct relation in
our KG, to an ontological concept from the do-
main. Let TT be the set of text tokens, i.e., any
extracted entity from the same text. The Cover-
age is defined as
|DT |
|T T |
× 100 (1)
• Mapping (MAPG). Let CT be the set of concept
tokens, i.e., any extracted entity from a given text
sharing the same syntactic (and semantic) name
as an ontological concept from the domain. The
Mapping is defined as
|CT |
|DT |
× 100 (2)
• Alignment (ALGT). Let r
NER−→TUI
be a direct
link from any extracted entity (NER) from a given
text to an ontological semantic type (TUI). Let
r
TUI
be a link from any source node to a TUI node,
i.e., r
TUI
= r
NER−→TUI
+ r
CUI−→TU I
. The Align-
ment is defined as
count(r
NER−→TUI
)
count(r
TUI
)
× 100 (3)
4 EXPERIMENTS
Experiments were performed on 52 Clinical Study
Reports (CSR) with the objective of finding direct re-
lationships between text entities, i.e., subject or ob-
ject of a triple (NER nodes), and ontological concepts
(CUI nodes) and semantic types (TUI nodes). We per-
form two experiments, each testing an algorithmic ap-
proach using the DomainKnowledge pipeline to ob-
tain an alignment from NER nodes to TUI nodes. All
experiments were hosted on an instance of Neo4j Au-
raDB
8
. The first experiment focuses on building sen-
tence clusters based on a sentence similarity score cal-
culated from the triples forming the sentences. The
intuition is that similar sentences will very likely be
paraphrases or rewording and will trace back to the
same higher-order ontological concepts. Grounding
these concepts makes the task of aligning NER and
TUI nodes easier. The experiment can be broken
down to the following steps:
1. The node2vec
9
embeddings is calculated for every
NER node.
8
https://tinyurl.com/yzxneyy5
9
https://tinyurl.com/55jc525f
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
420

Figure 4: Sample output graph.
2. A K-Nearest Neighbors (KNN)
10
clustering algo-
rithm is used to create pairwise clusters of NER
nodes using the node2vec embeddings as prop-
erty.
3. The resulting KNN similarity score knn score for
each pair of NER nodes is appended to the relation
in their triple if and only if they share a triple.
4. We define the sentence score for a sentence as
sentence score =
n
∑
i=1
knn score
i
(4)
where n is the number of relations of the
triples extracted from the sentence. All sen-
tences are compared and grouped based on
the sentence score. Sentences with equal sen-
tence scores underline similar sub-graphs from
NER to TUI nodes.
The final step is extracting the relevant NER and
TUI nodes from the different sentence group sub-
graphs. While this experiment shows promising re-
sults on a small batch of sentences, we lacked the re-
sources to handle the computational complexity of the
procedure on our ensemble of documents. We there-
fore did not report results for this method. The second
experiment targets ontology alignment directly using
the paths between NER, CUI and TUI nodes. The ex-
perimental setup is as follows:
1. The degree centrality DC
11
measure is calculated
for every NER, SUI, CUI and TUI node. Rela-
tions between SUI and AUI nodes are also con-
sidered for the SUI degree centrality calculation
10
https://tinyurl.com/yzx7dxv9
11
https://tinyurl.com/bddhh9e7
as they are considered additional information on
the representation of a concept. The calculations
are based on the following directed graph orienta-
tions:
• NER −→ SUI −→ AUI (a)
• NER −→ SUI −→ CUI −→ TUI (b)
The aim of this measure is to identify popular
nodes.
2. we define the weight w of a relation between 2
nodes A and B as the sum of their degree cen-
tralities DC
A
and DC
B
respectively. Formally,
w
AB
= w
BA
= w = DC
A
+ DC
B
.
3. For each NER node, we traverse the closed sub-
graphs respecting the path in (b) while opting for
the maximum total weight
W =
n
∑
i=1
w
i
(5)
where n is the total number of relations between a
NER node and a CUI node in a closed sub-graph.
4. We apply the same traversal algorithm to identify
the best direct relation between a CUI node and a
TUI node.
5. We finally use the results from the previous two
steps to find the best direct relation between a
NER node and a TUI node.
We evaluate our DomainKnowledge pipeline
against a human baseline consisting of clinical ana-
lysts from the biomedical domain who manually per-
form the alignment on the same dataset and report our
results.
Knowledge Graphs Can Play Together: Addressing Knowledge Graph Alignment from Ontologies in the Biomedical Domain
421
5 RESULTS
Of the 7407 extracted sentences over all documents,
a total of 172836 tokens were identified. From these
tokens, 131625 were relevant domain tokens covered
in triples, representing 76.16% of text coverage into
triples. To ensure these triples are viable, the relations
binding subject and object tokens had to be either ver-
bal or prepositional to rule out unusable triples. 16051
verbal or prepositional relations were extracted over
the text, which resulted in 13821 unique triples rep-
resenting approximately 10.50% of the total set of
extracted triples. This figure signifies that domain
concepts make up roughly 10% of a CSR, whereas
the remaining 90% are the different context windows
in which the domain vocabulary is used. A sum-
mary of the triple extraction process from our pipeline
is detailed in Table 1. From the extracted triples,
4002 NER objects are domain vocabulary that can
be mapped to UMLS concepts. An additional 3417
objects tagger by the NER tagger means a total of
53,67% of the extracted triple objects can be mapped
to UMLS concepts. The final KG yields 7151 indi-
rect links from NER to TUI nodes. Indirect links en-
compass any direct link from NER to CUI ot NER to
TUI directly. The calculations from our graph traver-
sal algorithm identify 1533 direct links from NER to
TUI, resulting in an alignment of 21,40%. Table 2
shows the details of the NER node alignment to the
domain ontology. Table 3 shows the performance of
our pipeline with respect to the human baseline. The
results show promise for our pipeline: it beats the hu-
man baseline on all metrics while retaining a good
domain coverage of the text. The mapping score indi-
cates the over half the extracted triples contains per-
tinent nodes that can be traced back to the domain
ontology, showcasing the effectiveness of our anno-
tation and extraction methods. The alignment score,
while relatively low, is encouraging when it comes
to finding higher-level concepts linked to the initial
document text. This opens the possibility to a wider
integration between domain ontologies and domain
texts, with potential possibilities to enhance the latter
with the former using the links between NER and TUI
to semi-automatically generate in-context text tem-
plates and enrich the document. It is worth noting
that the noticeable discrepancy in scores between the
metrics suggests issues that need to be addressed at
annotation and extraction level. Our pipeline still per-
forms poorly on adjectival relationships, identifying
acronyms (e.g., human arm versus ARM) and specific
wordings (e.g., 6 cycle versus cycle 6) which explains
the drops in scores between metrics.
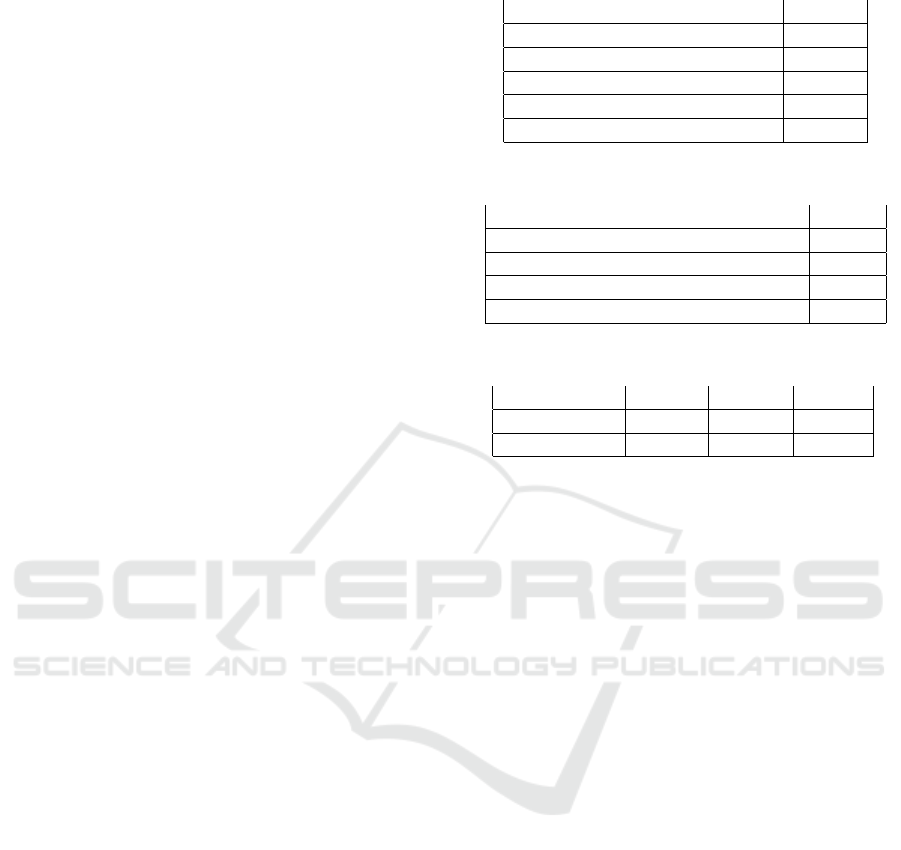
Table 1: Triple extraction summary.
Object Count
Sentences 7407
Tokens in sentences 172836
Tokens covered in triples 131625
Verbal or prepositional relations 16051
Unique triple objects 13821
Table 2: Alignment Summary.
Object Count
Unique NER objects linked to UMLS 4002
I2b2 NER objects linked to UMLS 3417
NER to CUI/TUI indirect links 7151
NER to TUI direct links 1533
Table 3: Comparative results of our methodology.
Method CVRG MAPG ALGT
Baseline 68.00 40.00 10.00
Our Pipeline 76.16 53.67 21.40
6 CONCLUSION
We introduce a system for domain information ab-
straction from text and ontology alignment for a more
effective KG creation. Our method has the advan-
tage of providing good text-to-triple coverage while
maintaining strict semantic consistency for overlap-
ping tokens, which allows better mapping and align-
ment to higher-order domain ontologies. Our exper-
iments show the need to expand the annotation and
extraction processes of our system in order to han-
dle edge cases in unstructured text and capture triples
more faithfully. In future work, we will target enhanc-
ing the triple extraction process from text by making
the annotator more flexible with handling edge cases
like acronyms or sentence rewordings. We will in-
tegrate features like coreference resolution to capture
more fine-grained triples and improve KG construc-
tion. we will also aim to evaluate our system against
other architectures like LLMs and widen the scope of
our experimentation to include other types of biomed-
ical documents (e.g., Protocols) as well as extend it to
other domains like finance.
REFERENCES
Abi Akl, H. (2023). The path to autonomous learners. In
Science and Information Conference, pages 808–830.
Springer.
Chaurasiya, D., Surisetty, A., Kumar, N., Singh, A., Dey,
KDIR 2024 - 16th International Conference on Knowledge Discovery and Information Retrieval
422

V., Malhotra, A., Dhama, G., and Arora, A. (2022).
Entity alignment for knowledge graphs: progress,
challenges, and empirical studies. arXiv preprint
arXiv:2205.08777.
Chouham, E. M., Espejel, J. L., Alassan, M. S. Y., Dahhane,
W., and Ettifouri, E. H. (2023). Entity identifier: A
natural text parsing-based framework for entity rela-
tion extraction. arXiv preprint arXiv:2307.04892.
Cotovio, P. G., Jimenez-Ruiz, E., and Pesquita, C. (2023).
What can knowledge graph alignment gain with
neuro-symbolic learning approaches? arXiv preprint
arXiv:2310.07417.
Dao, N.-M., Hoang, T. V., and Zhang, Z. (2023).
A benchmarking study of matching algorithms for
knowledge graph entity alignment. arXiv preprint
arXiv:2308.03961.
Dong, K., Sun, A., Kim, J.-J., and Li, X. (2023). Open
information extraction via chunks. arXiv preprint
arXiv:2305.03299.
Dunn, A., Dagdelen, J., Walker, N., Lee, S., Rosen, A. S.,
Ceder, G., Persson, K., and Jain, A. (2022). Struc-
tured information extraction from complex scientific
text with fine-tuned large language models. arXiv
preprint arXiv:2212.05238.
Fanourakis, N., Efthymiou, V., Kotzinos, D., and
Christophides, V. (2023). Knowledge graph em-
bedding methods for entity alignment: experimen-
tal review. Data Mining and Knowledge Discovery,
37(5):2070–2137.
Friedman, S., Magnusson, I., Sarathy, V., and Schmer-
Galunder, S. (2022). From unstructured text to causal
knowledge graphs: A transformer-based approach.
arXiv preprint arXiv:2202.11768.
Hong, Z., Chard, K., and Foster, I. (2024). Combin-
ing language and graph models for semi-structured
information extraction on the web. arXiv preprint
arXiv:2402.14129.
Kamp, S., Fayazi, M., Benameur-El, Z., Yu, S., and Dreslin-
ski, R. (2023). Open information extraction: A review
of baseline techniques, approaches, and applications.
arXiv preprint arXiv:2310.11644.
Liu, P., Gao, W., Dong, W., Huang, S., and Zhang, Y.
(2022). Open information extraction from 2007 to
2022–a survey. arXiv preprint arXiv:2208.08690.
Melnyk, I., Dognin, P., and Das, P. (2022). Knowl-
edge graph generation from text. arXiv preprint
arXiv:2211.10511.
Mihindukulasooriya, N., Tiwari, S., Enguix, C. F., and Lata,
K. (2023). Text2kgbench: A benchmark for ontology-
driven knowledge graph generation from text. In
International Semantic Web Conference, pages 247–
265. Springer.
Motger, Q. and Franch, X. (2024). Nlp-based re-
lation extraction methods in re. arXiv preprint
arXiv:2401.12075.
Pan, J. Z., Razniewski, S., Kalo, J.-C., Singhania, S., Chen,
J., Dietze, S., Jabeen, H., Omeliyanenko, J., Zhang,
W., Lissandrini, M., et al. (2023a). Large language
models and knowledge graphs: Opportunities and
challenges, 2023. arXiv preprint arXiv:2308.06374.
Pan, S., Luo, L., Wang, Y., Chen, C., Wang, J., and Wu,
X. (2023b). Unifying large language models and
knowledge graphs: A roadmap, 2023. arXiv preprint
arXiv:2306.08302.
Peng, C., Xia, F., Naseriparsa, M., and Osborne, F. (2023).
Knowledge graphs: Opportunities and challenges. Ar-
tificial Intelligence Review.
Qian, K., Belyi, A., Wu, F., Khorshidi, S., Nikfarjam, A.,
Khot, R., Sang, Y., Luna, K., Chu, X., Choi, E.,
et al. (2023). Open domain knowledge extraction for
knowledge graphs. arXiv preprint arXiv:2312.09424.
Trisedya, B. D., Salim, F. D., Chan, J., Spina, D., Scholer,
F., and Sanderson, M. (2023). i-align: an interpretable
knowledge graph alignment model. Data Mining and
Knowledge Discovery, 37(6):2494–2516.
Van Tong, V., Huynh, T. T., Nguyen, T. T., Yin, H.,
Nguyen, Q. V. H., and Huynh, Q. T. (2021). In-
complete knowledge graph alignment. arXiv preprint
arXiv:2112.09266.
Vogt, L., Kuhn, T., and Hoehndorf, R. (2022). Semantic
units: Organizing knowledge graphs into semantically
meaningful units of representation [internet].
Wadhwa, S., Amir, S., and Wallace, B. C. (2023). Revisiting
relation extraction in the era of large language models.
arXiv preprint arXiv:2305.05003.
Xie, F., Zeng, X., Zhou, B., and Tan, Y. (2023). Improv-
ing knowledge graph entity alignment with graph aug-
mentation. In Pacific-Asia Conference on Knowledge
Discovery and Data Mining, pages 3–14. Springer.
Yang, L., Chen, H., Wang, X., Yang, J., Wang, F.-Y., and
Liu, H. (2024). Two heads are better than one: Inte-
grating knowledge from knowledge graphs and large
language models for entity alignment. arXiv preprint
arXiv:2401.16960.
Yu, B., Zhang, Z., Li, J., Yu, H., Liu, T., Sun, J., Li, Y., and
Wang, B. (2022). Towards generalized open informa-
tion extraction. arXiv preprint arXiv:2211.15987.
Zeng, K., Li, C., Hou, L., Li, J., and Feng, L. (2021). A
comprehensive survey of entity alignment for knowl-
edge graphs. AI Open, 2:1–13.
Zhang, R., Su, Y., Trisedya, B. D., Zhao, X., Yang, M.,
Cheng, H., and Qi, J. (2023). Autoalign: fully au-
tomatic and effective knowledge graph alignment en-
abled by large language models. IEEE Transactions
on Knowledge and Data Engineering.
Zhang, R., Trisedy, B. D., Li, M., Jiang, Y., and Qi, J.
(2021). A benchmark and comprehensive survey on
knowledge graph entity alignment via representation
learning. arXiv preprint arXiv:2103.15059.
Knowledge Graphs Can Play Together: Addressing Knowledge Graph Alignment from Ontologies in the Biomedical Domain
423
