
Using Secondary Inherited Characteristics During Reproductive Choice
to Replicate Allopatric Speciation
Gary B. Parker and Jay B. Nash
Department of Computer Science, Connecticut College, New London, U.S.A.
Keywords:
Allopatric, Sympatric, Speciation, Bio-Inspired, Genetic Algorithm, Artificial Life, Agent Modeling, Rule
Base, Evolutionary Computation.
Abstract:
The goal of this research is to create an environment where we can use evolutionary computation (EC) with
separate chromosomes in independent agents to replicate allopatric speciation, the process in which a species
diverges into two distinct, reproductively separate species based on geographic isolation. In previous work
done by Parker and Edwards, an environment and genetic algorithm were developed to simulate this type of
speciation. However, some aspects of the developed environment could be considered a priori knowledge. This
paper details a new system where agents do not have access to what we will refer to as “primary characteristics”
or characteristics that directly affect agent fitness and success. Characteristics that have no bearing on agent
fitness, referred to as "secondary characteristics", are solely used by the agents to determine reproductive
choice. This has a variety of benefits, most clearly that no a priori knowledge is used in the system. This
can result in two species that have identical primary, fitness affecting characteristics, but are reproductively
separated due to secondary, arbitrary characteristics. The reduction of knowledge available to the agents during
reproduction makes the system a better match for biological systems, but was expected to cause an increase
in cross species hybrids. However, it led to a higher degree of speciation than previous work on the topic.
As a result, this system improves upon the previous method used to simulate the natural process of allopatric
speciation via a genetic algorithm by reducing a priori knowledge and increasing efficacy.
1 INTRODUCTION
Speciation is the biological process of creating a new,
distinct species from an ancestral species. There are a
variety of definitions and rules for what makes a dis-
tinct species; for the purposes of this research, we
consider a species distinct if it will not produce off-
spring with individuals from a different species. Spe-
ciation is a key aspect of evolution (Coyne, 1992),
which requires that new species are developed from
ancestral species. In order for a new species to de-
velop, its population must be reproductively isolated
from the original species so that the two species can
evolve to have distinct characteristics. Although there
appears to be evidence for this found in the fossil
record, observations of it in nature are difficult to find
when species is defined as individuals who cannot re-
produce with other species. The Galapagos Finches
from separate islands are said to be separate species,
but they are still able to interbreed and produce vi-
able offspring. For our definition of species, which is
required for evolution, they should not be willing to
mate, even if allowed to intermingle within the same
environment.
The goal of this work is to demonstrate speciation
in a computational model, which is done by improv-
ing the results of a simulation developed by Parker
and Edwards (Parker and Edwards, 2019), and to do
so without using knowledge of an agent’s fitness dur-
ing the reproductive process. This results in gene
pools separated solely by the lack of interaction be-
tween the two species, instead of being separated by
factors that affect fitness. By doing this, we are more
closely replicating what might take place in nature.
In the research presented in this paper, we concen-
trate on the results of simulating allopatric speciation
(Mayr, 1963), which occurs when new species evolve
from a common ancestor due to geographic isolation.
Sympatric speciation, when a species diverges within
the same geographic area (Bolnick and Fitzpatrick,
2007), is planned for future research.
The ultimate objective of this research into repli-
cating speciation is to determine if it is possible to
simulate both allopatric and sympatric speciation us-
304
Parker, G. and Nash, J.
Using Secondary Inherited Characteristics Dur ing Reproductive Choice to Replicate Allopatric Speciation.
DOI: 10.5220/0013017900003837
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Joint Conference on Computational Intelligence (IJCCI 2024), pages 304-312
ISBN: 978-989-758-721-4; ISSN: 2184-3236
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.

ing a genetic algorithm (GA) with the individual
chromosomes, each within and controlling separate
agents. Standard GAs (Holland, 1975) consist of a
population of individuals that are intended to be a so-
lution to a given problem. The traits of these individ-
uals are encoded in chromosomes, which in our case
are constructed from a sequence of 0s and 1s so that
genetic operations such as crossover and mutation can
easily operate on them. In general, new individuals
are evolved by finding a fitness value for each cur-
rent individual, from which the highest scoring indi-
viduals are chosen as the basis for the next generation.
When two individuals reproduce, crossover is used to
combine their chromosomes into a new chromosome,
which is then used to create a new individual to be in-
troduced into the population. Our GA differs from a
standard GA, as instead of consisting of a population
of chromosomes, our GA consists of a population of
agents, each with its own set of chromosomes. In our
simulation, agents only reproduce when they are in
close proximity to each other; our simulation relies
on the fact that agents that are physically distant from
each other will reproduce less, if at all, regardless of
any other factors.
This paper presents our results for modifying the
previous GA developed by Parker and Edwards to
remove various arbitrary factors, reduce the amount
of knowledge individuals have during reproduction,
reduce knowledge of potential mates’ primary char-
acteristics, and not arbitrarily penalize mating with
agents of different sizes (Parker and Edwards, 2019).
All of these changes are intended to make our system
better represent biological systems. Although we pre-
sumed these changes would result in less successful
speciation due to agents having less information for
decision making, we found that it instead removed the
existence of cross-species hybrids after speciation.
The motivation for this work is to both test biolog-
ical theories and to find aspects of speciation that can
be used to improve genetic algorithms. A simulation
that can accurately reflect speciation could provide in-
sight into real evolutionary processes. The current re-
search is solely concerned with replicating speciation.
However, it is possible that what is learned from this
could contribute to the performance of evolutionary
computation due to the species separating naturally
rather than being separated by a similarity function.
This is purely speculation and will be investigated in
a future work after further expansion of this research
into replicating speciation.
2 PREVIOUS WORKS
Speciation has been partially mimicked in other re-
search that mainly focuses on niching algorithms to
improve the ability of a GA to optimize a multimodal
function (Goldberg and Richardson, 1987). Gener-
ally, niching algorithms function by detecting a pop-
ulation of similar chromosomes and creating niche
populations manually. Although many ways have
been developed to maintain and create niches, we do
not consider them to replicate speciation as each niche
is determined by similar characteristics rather than by
the individuals themselves (Glibovets and Gulayeva,
2013). This artificially creates subpopulations, while
our research presents a possible way for individuals to
develop such subpopulations naturally via their own
mating preferences.
NeuroEvolution of Augmenting Topologies or
NEAT (Stanley and Miikkulainen, 2002) is similar to
our research in that it uses evolution to learn agent
controls and preferences; the learning is in real time,
and speciation is somewhat present. However, like
niching algorithms, NEAT uses a compatibility func-
tion to determine if two agents are part of the same
species instead of the agents themselves determining
what agents they interact with. That is not to say
this process does not have a basis in nature, how-
ever, since structurally different organisms are gen-
erally different species, regardless of the preference
of those organisms. NEAT uses historical data for
an agent to determine which gene corresponds to an-
other gene and which agents belong to which species.
Another feature of NEAT is fitness sharing, this pro-
cess allows agents of a similar species to share fitness
payoff, decreasing competition inside a species while
keeping high competition with other species. The sys-
tem described in this paper has a different goal; we
want species to develop naturally through reproduc-
tive isolation. Instead of restricting mating via a pro-
grammed function, the agents themselves will choose
not to mate with agents that have diverged into a dif-
ferent species. Additionally, the simulation does not
make agents share fitness payoff, although resource
scarcity tends to force one species to dominate its own
area. As fitness payoff is not shared between agents
of the same species, agents compete for food with all
other agents, regardless of species membership. This
encourages the formation of new species and the mu-
tation of existing species, as there is no advantage to
remain a part of an existing dominant species beyond
an increased chance of finding a compatible mate.
The work done on simulating allopatric specia-
tion focused on using purely biological processes to
optimize a GA. Unlike the Speciation Island Mode
Using Secondary Inherited Characteristics During Reproductive Choice to Replicate Allopatric Speciation
305

(Gustafson and Burke, 2006), a process which detects
new species and separates them to their own isolated
environment using a species barcode and common an-
cestry, this GA maintains the same grid and environ-
ment for all species, only using a temporary barrier to
simulate geographic and reproductive isolation. Ad-
ditionally, the species within this GA begin as a single
population that will naturally diverge into two. This
separates it from the idea of Cooperative Coevolu-
tionary Algorithms (Potter and Jong, 1994), as in that
system, the species are utilized to solve a problem to-
gether, while in the system described in this paper,
the agents are solely concerned with competition for
individual success and food resources.
Some research has been done specifically on sep-
arating species created by a GA via geographic isola-
tion, such as in a paper published by Wang Li et al.
(Li et al., 2012). This paper proposes a process in
which a standard GA is run to have a control species,
then the search domain is separated, and each sepa-
rated domain is evaluated to see if a specific species
is likely to evolve into a superior species. That pro-
cess has some close similarities with our speciation
simulation, although in that paper the agents are not
recombined. The process of agent recombination al-
lows us to see if the populations have naturally di-
verged far enough that the agents themselves consider
them different, rather than a predefined analysis func-
tion.
In addition to speciation, our simulation uses the
unique method of a Genetic Algorithm with Vary-
ing Population Size, or GAVaPS. This method im-
plements a way to have the population of a GA vary
by introducing aging into the agents (Arabas et al.,
1994). In GAVaPS, individuals die when their age ex-
ceeds a specific lifetime value. The paper discusses
different ways to determine a lifetime value for an in-
dividual, which remains constant as the agent ages.
Our research is similar to the idea of GAVaPS how-
ever differs in that the lifetime value is not predeter-
mined. Instead, age increases the amount of food an
agent must consume by increasing the amount of life
(energy) an agent loses each turn. Once the agent runs
out of energy, it dies. Thus, instead of determining an
agent’s lifetime value, the agent itself determines how
long it can live based on its fitness in its environment.
Other GA’s have been created to allow variable
population sizes, such as APGA, PRoFIGA, and the
PSO-GA hybrid algorithm (PGHA) (Shi et al., 2005).
The PGHA has some similarities to our research, pri-
marily in that “parents are neither dead after their
reproduction right away, nor living forever.” Agents
can, and often do, survive and outlive their children.
However, the longer they live, the higher the probabil-
ity they will die. While the PGHA uses actual prob-
ability to determine agent death, our speciation simu-
lation uses age as a factor that increases agent energy
consumption. In addition, in our work, selection is
solely based on the preferences of two agents in close
proximity.
3 ENVIRONMENT
A new model was created using the previous specia-
tion environment model developed by Parker and Ed-
wards as a baseline (Parker and Edwards, 2018). This
new model largely follows the aspects of the previ-
ously developed model, with functional changes con-
cerning agent chromosomes and reproductive func-
tions.
This model was designed to be as simple as possi-
ble to minimize the factors outside the agents’ direct
control while keeping enough complexity that specia-
tion could occur. The environment for this simulation
is a grid over which the agents can move, eat, and in-
teract with each other. This grid was designed to be
of variable size depending on testing requirements. In
the results reported in this paper, a 100 x 100 grid en-
vironment was used. Figure 1 shows what a 50 x 50
environment looks like with seeds before agents are
added.
Figure 1: Example 50x50 empty grid with random food
generation. A 100x100 grid was used in actual simulation
runs. Food is represented by a square icon, green icons rep-
resent large food, yellow icons represent medium food, and
red icons represent small food.
The grid is array based and uses discrete blocks
(spaces) which can be empty or be occupied by food
and/or an agent within the space. A seed is repre-
sented by a square of varying sizes depending on the
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
306

size of the seed, and agents are represented by spheres
of varying sizes and colors depending on the size and
RGB value of the agent. During each time cycle
(turn), the agents all select an action to perform that
turn, and some food is generated within the grid.
The food generated is of three possible sizes:
large, medium, and small. The density of food gener-
ation and the size of food generated can be specified
to determine the quantity and type of food added to
the grid per turn. Differing from the previous environ-
ment, the new environment is divided into four equal
sections: far left, center left, center right, and far right.
Food generation can be specified using probabilities
for each section and given a total amount of food to
generate. For example, the first two sections (far left
and center left) could have a 50/50 chance of gener-
ating 100 large or medium food, while the last two
sections (center right and far right) may have a 50/50
chance of generating 100 medium or small food. Hav-
ing the grid split up like this simulates differing envi-
ronments used to replicate geographical differences
like different climates or soil. Each time cycle, new
seeds are added to the grid based on the rules for each
section. The placement within a section is random;
however, each section generates a specific number of
seeds per turn. The food does not move, once placed
the seeds stay in that position until they are eaten by
an agent.
Figure 2: Example of the three different sizes of agents and
food. Agent color is determined by genes inside a chromo-
some while food color is static. Agents have a bold border
compared to the food and are circular instead of square.
4 AGENTS
Agents are able to move around the environment
freely (although they cannot move into a space oc-
cupied by another agent), eat seeds, and interact with
each other if two agents happen to occupy adjacent
spaces. The number of agents within the environment
at a given time is variable as it may increase or de-
crease depending on the variables of the environment:
food density and the type of food generated. Gener-
ally, the population size increases as the simulation
runs, due to agents becoming more fit, until it even-
tually reaches a steady state. The physical aspects
of each agent differ from previous research. While
agents still have a certain size and RGB value, they
also have four arbitrary characteristics represented by
a numerical value between 0 and 15. As new agents
are created, these physical characteristics, size, and
RGB value are inherited from the parents with a small
chance of mutation. An example of agent size and
RGB values can be seen in Figure 2.
Table 1: Agent Energy Gain From Seeds.
Agent Seed Size
Size Large Medium Small
Large 100 30 15
Medium 30 100 30
Small 15 30 100
The agents’ size varies between three values:
large, medium, and small. The size of the agent is the
only factor that determines the agent’s fitness for its
environment as it determines what size seed the agent
is most capable of eating (Table 1). For example, if
a large agent had to choose between eating a medium
or large seed, the best option would be the large seed.
The number of chromosomes in each agent has in-
creased from previous research done to replicate al-
lopatric speciation. The agents now have three chro-
mosomes in order to allow them to better differenti-
ate between themselves. The first chromosome dic-
tates agent behavior (discussed in the next section),
the second chromosome dictates the agents’ physical
characteristics as discussed previously, and the third
chromosome dictates an agent’s preferences for re-
production.
One of the possible actions of an agent is to mate
with another agent in an adjacent space. If both parent
agents agree to reproduce, a child agent is produced
and placed in the environment within three spaces of
the parent agents. Differing from the previous envi-
ronment, agent reproduction cannot fail due to dif-
ferences in agent size if two agents decide to repro-
duce. All criteria of both agents must be met for re-
production to occur. This chromosome dictates the
desired partner RGB, the desired values of the four
arbitrary characteristics, and the desired age (Figure
3). Notably different from the previous reproduction
system, size preference is absent from this decision-
making process. The lack of information about the
only factor impacting fitness (the primary characteris-
tic) forces the agents to use other aspects (secondary
characteristics) of a potential partner to determine if
reproduction will yield a successful offspring.
The first three bits of the chromosome dictate the
maximum desired color difference in a partner agent
Using Secondary Inherited Characteristics During Reproductive Choice to Replicate Allopatric Speciation
307
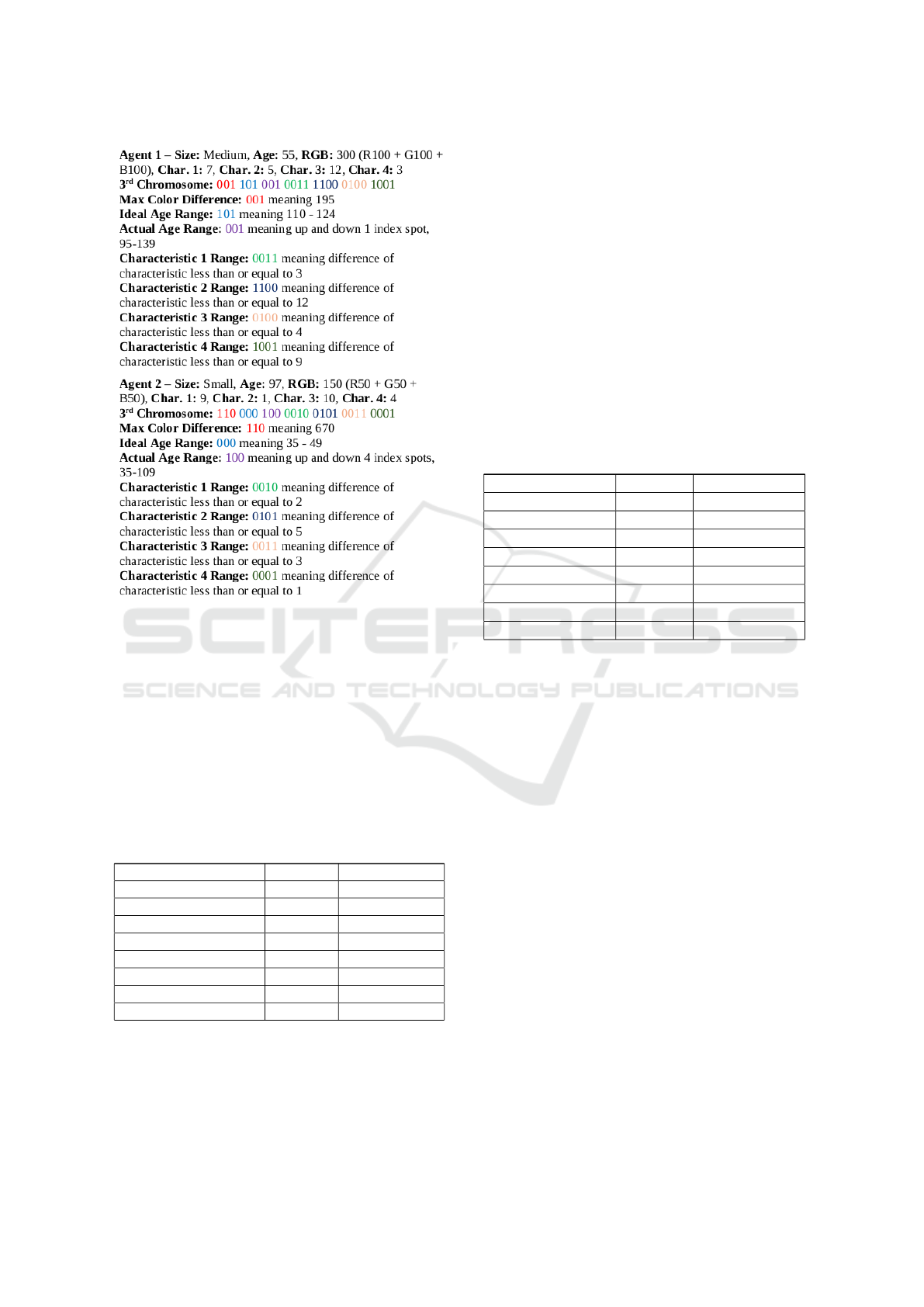
Figure 3: Example of two agents that would successfully re-
produce with each other given their chromosomes and spe-
cific information. Note that size, age, RGB, and arbitrary
characteristics are normally stored in a chromosome. How-
ever, they were taken out of binary format for the sake of
this example.
(Table 2). This color difference is the sum of the dif-
ferences between each of the three RGB values. The
maximum possible color difference is 765 (3 * 255)
and the minimum difference is 0. This range is found
using the formula MAXDIFF = 100 + (95 * BITVAL).
Table 2: Bit Combination for the Maximum Color Differ-
ence Between Two Agents.
Max Color Difference Bit Value Decimal Value
100 0 0 0 0
195 0 0 1 1
290 0 1 0 2
385 0 1 1 3
480 1 0 0 4
575 1 0 1 5
670 1 1 0 6
765 1 1 1 7
The next six bits of the reproductive preference
chromosome decide the desired age of a mate. An
individual agent’s age is determined by how many
time cycles that agent has been alive. The age of
reproductive fertility was defined to be between the
ages of 35-154. The desired age range of a possi-
ble mate was encoded into a six-bit string by using
the first three bits to define the starting range of 15,
such as 65-79, and then using the next three bits to
expand that range by some number of index values,
in this case one index expansion would result in an
actual range of 50-94 (Table 3). The formula for the
lower bound of the initial age range, using the first
three bits, is: LOWERAGE = 35 + (15 * BITVAL)
and UPPERAGE= 49 + (15 * BITVAL). In order to
find the age range index increase specified by the
second three bits, the formula is: ACTUALLOWER
= MAX(35,LOWERAGE-15*BITVAL) and ACTU-
ALUPPER = MIN(154,UPPERAGE+15*BITVAL).
Table 3: Bit Combination for the Initial Age Range of a
Partner Agent.
Initial Age Range Bit Value Decimal Value
35-49 0 0 0 0
50-64 0 0 1 1
65-79 0 1 0 2
80-94 0 1 1 3
95-109 1 0 0 4
110-124 1 0 1 5
125-139 1 1 0 6
140-154 1 1 1 7
The remaining 16 bits of the chromosome define
the preferred values of four arbitrary, inherited char-
acteristics each agent has. Reflecting that each char-
acteristic has a total possible range of 0-15, these
16 bits are divided into four preferences of four bits
each. Each preference has a corresponding charac-
teristic and is a value between 0-15. This value is
the maximum difference two agents can have for that
characteristic for them to be willing to mate. For each
characteristic, we take the absolute difference for that
characteristic between an agent and a possible partner,
if that difference is less than or equal to the agents’
preference for that characteristic, then the agents are
willing to mate.
5 AGENT CONTROLLER
The actions of each agent are controlled by a rule-
based system, which is made up of a set of different
rules (antecedent/consequent) that control the agent’s
actions. If the antecedent of a rule is true, then the
rule fires and the action in the consequent is taken.
Agents only have one possible action per time cycle,
which means they must somehow decide on the best
possible action they can take per turn. Our research
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
308

Table 4: An Example Of The Action Chromosome That Decides The Priorities For The Rule Based Agent Controller.
Rule Free Reproduce Large Seed Medium Seed Small Seed Large Seed Medium Seed Small Seed
Base Space No Eat No Eat No Eat Eat Eat Eat
Chromosome 1 1 0 1 0 1 1 1 1 1 1 0 1 0 1 0 0 0 0 0 1 0 1 0 0 1 1 1 1 0 0 0
Decimal Value 13 7 14 10 0 10 7 8
The Complete Action Chromosome Is: 11010111111010100000101001111000
uses priority ordering, learned by the overall GA, to
decide which rules have the highest priority. If there
is a tie in priority, then a random tied rule is chosen.
The agents are capable of performing various ac-
tions: move to a free space, reproduce with an adja-
cent agent, move to a large seed space and eat it, move
to a medium seed space and eat it, move to a small
seed space and eat it, move to a large seed space and
do not eat it, move to a medium seed space and do not
eat it, and to move to a small seed space and do not eat
it. During each time cycle, the immediate surround-
ings of an agent (one space up, down, left, and right)
are analyzed to see which rules are possible to fire.
Once the possible actions are determined, the agent
fires the highest priority action possible to perform,
with a tie broken by random choice.
An agent starts with 100 energy. All actions carry
a base cost of the square root of the agents’ age, with
reproduction carrying an additional cost of 80 energy.
The additional cost of reproduction is only levied
upon production of a child agent, since if both agents
decide to reproduce; there is no chance of failure.
Conversely, if either agent decides not to reproduce,
there is no chance of reproduction. As agents age, the
cost of each action (a cost that must be paid per time
cycle as agents must take an action) increases, mean-
ing that the agent must eat more food to survive as
it ages. If an agent eats a seed, it will get a certain
amount of energy, as shown in Table 1. For example,
if we had a large agent with 200 energy, 100 age, and
it moved to and ate a large seed, the resulting energy
would be 200 + 30 – sqrt(100) = 220.
An example of an action chromosome and its re-
sultant priorities can be seen in Table 4. This table de-
picts a single agents’ chromosome broken down into
each action gene. The chromosome is split into eight
four-bit sections, and the action that has the highest
priority and is possible for the agent to perform is cho-
sen.
According to this example, the agent would most
want to move to a large seed space and not eat the food
(pseudocode for the algorithm can be seen in Algo-
rithm 1). However, if one of the adjacent spaces does
not contain a large seed, the agent cannot perform this
action. If this is the case, the next highest priority rule
will be checked to see if it can be fired. In this exam-
ple, the agent would then check if there is an open free
space to move to. This process of finding the highest
rule to fire continues until a rule is found that can be
fired; there is at least one rule that can always be fired.
6 AGENT CHARACTERISTICS
It is important to note the difference in how we treat
certain characteristics of each agent. Characteristics
that affect agent energy gain, and therefore fitness, are
what we call “primary characteristics.” The only char-
acteristic that meets this definition is the size of each
agent. This primary characteristic is not available to
other agents when deciding reproductive preference.
Therefore, the agents must decide reproduction based
on other characteristics. These other characteristics
are called “secondary characteristics” and have no im-
pact on agent energy gain or fitness. These character-
istics include the RGB value of an agent, the age of an
agent, and four arbitrary characteristics that are repre-
sented by 4-bit strings with a decimal value between
0-15. These secondary characteristics are available
to the agents during reproduction and are what the
agents use to decide if they want to mate with any
adjacent agent.
All these characteristics are inherited during re-
production with a small chance of mutation. Because
these values are inherited, over time and as more com-
mon ancestors appear, agents begin to hold similar
characteristics to other agents in their proximity. Us-
ing this, agents can effectively determine if an agent
is a good mate or if the agent is a different species and
not a good mate.
7 GENETIC ALGORITHM
The properties of the rule-based system, the reproduc-
tive preferences, and the agent characteristics are all
specified by a GA. This allows the agents to pass their
preferences and characteristics to their children. The
GA operates on three chromosomes (one for actions,
one for preferences, and one for characteristics), each
made up of 1s and 0s. Selection in our system is based
on physical proximity in our environment (adjacent
spaces) of the two individuals and if they both agree to
reproduce depending on the other’s RGB value, age,
and arbitrary characteristics. Specifically, size is not
Using Secondary Inherited Characteristics During Reproductive Choice to Replicate Allopatric Speciation
309

Algorithm 1: Pseudocode for the rule-based system
that decides agent movement and actions. The infor-
mation about each agent is stored in an agent class
and parsed each time cycle to decide agent actions
with updated information.
for each agent in the environment do
Get the adjacent spaces (maximum 4);
for each action do
Determine weight for the action;
if action is possible then
Add action to priority queue with
weight;
if action added is the same weight as
other action then
Randomly choose action to be
placed higher in queue;
end
end
end
View action at top of priority queue;
if action is reproduce then
Attempt to reproduce;
if mate did not agree to reproduce or no
mate is found then
Remove reproduce from priority
queue;
else
Create new agent;
Place new agent in random space
within 3 spaces of the parent agent;
end
else
Select and fire rule at top of priority
queue;
if rule can move agent to more than one
space then
Randomly decide space to move
agent to;
else
Remove action from priority queue
and go back to view action at top of
priority queue step;
end
end
Reduce energy of agent in regard to age;
if reproduced successfully then
Reduce energy of agent by 80;
end
Add energy to agent if food consumed;
Add one age to agent;
Update agent information for new position
and time;
end
a factor in reproductive choice, even though size is
the only determining factor of an agent’s ability to
acquire energy and thus survive. The idea of selec-
tion based on agent fitness does not fully apply here,
as any agent can attempt to reproduce with any other
agent. Fitter agents do have a higher chance of re-
production because they are more likely to consume
more food, have more energy for reproduction, and
will survive longer.
Notably, this simulation does not have a fitness
function nor are the agents evaluated for fitness. In-
stead, as the agents evolve, they will determine their
own preferences for what makes a good potential
mate. These preferences are evolved over time and
are passed down via standard crossover to offspring in
a similar way to that of the action chromosome. This
process is what we rely on for the agents to eventually
speciate, as over time the agents may develop specific
preferences that are only met by their own species.
Chromosomes inherited from the parents are pro-
duced through single point crossover with a 100%
chance of crossover, and mutation, which is bitwise
with a very low probability (1/300) of mutating a ran-
dom bit in the chromosome.
The GA used in our research is similar to a steady-
state GA, except that we do not use standard chromo-
some replacement, old individuals die when out of en-
ergy, and new individuals are added to the population
with no regard to the overall size of the population.
The size of the population does have a “soft cap” as
only so much food is generated per time cycle, but
that is the only limiting factor on the size of the pop-
ulation.
8 RESULTS
As in previous research, initial tests were done to
check the model (Parker and Edwards, 2018). These
tests showed that agent populations could evolve to
optimize for the available food sources and adjust
when both locations and types of food changed.
Tests were then created to replicate an environ-
ment that would allow allopatric speciation. A pop-
ulation of randomly generated agents was generated
on the grid while medium sized food was generated in
the center of the grid to encourage the agents to form a
single species. As expected, all large and small agents
died out during the formation of this initial popula-
tion, while a population of medium agents was estab-
lished in the center of the grid. Once this population
was fully established (around 5,000 time cycles), a
physical barrier was placed in the center of the grid,
dividing the existing population in half.
At the same time, food generation was changed.
On the left side of the wall, some medium food was
produced for a limited period while the majority of
food produced was changed to small food. On the
right side, a similar process was followed, with the
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
310

Table 5: Results of Various Runs With Different Parameters.
Run Total Time Total Reproduction Total Total Attempts Total Number Of Run
Cycles Attempts Reproductions At Hybridization Hybrids Created Information
Full 1 40,000 24,214,134 586,723 789,205 0 Standard Run
Full 2 40,000 26,562,194 644,462 1,386,087 0 Only Large Food Produced
RGB 1 40,000 15,191,461 553,952 67,650 1,358 Reproduce on Age/Color
only change being that the majority of food would
now be large. After a period of adjustment (roughly
another 10,000 time cycles) all medium food genera-
tion was stopped.
As the agents evolved to adapt to their new en-
vironment, a population of small agents were estab-
lished on the left side of the wall while a population of
large agents were established on the right side of the
wall. These two populations were allowed to evolve
independently for a period of time (15,000 time cy-
cles), then the barrier was removed, and the two pop-
ulations were allowed to interact freely for a period
of time (10,000 time cycles) as food generation was
changed so that both large and small food was gener-
ated in the center of the grid.
When the barrier was removed, agents were
tagged to see if they would reproduce with a popu-
lation that was not their own (a different species). If
an agent of the left-side population reproduced with
an agent of the right-side population, their offspring
was called a first-degree hybrid.
If this first-degree hybrid (or any other degree of
hybrid) mated with any other agent in the population,
their offspring was called an n-th degree hybrid. We
reason that first-degree hybrids could exist with dis-
tinct species (such as a horse and donkey making a
mule). However, for a successful test, the first-degree
hybrid should never reproduce, meaning no n-th de-
gree hybrids.
We ran a series of runs starting from the random
agent generation stage and from there recorded the
number of hybrids produced. While many runs pro-
duced hybrids, in approximately 20% of runs the two
populations separated far enough that no first-degree
hybrids were produced (and therefore no n-th degree
hybrids) [Table 5]. This successfully showed that our
method of using arbitrary characteristics could repli-
cate allopatric speciation without agents knowing the
fitness value of other agents (as they could not see
each other’s size value).
A second test was designed to see the impact agent
size had in the separation of these populations. This
test was identical to the first, with the change that both
sides of the barrier would generate large food. This
was done so that the agents on both sides of the bar-
rier would evolve to be large agent populations. In-
stead of a small agent population and a large agent
population, there would be two large agent popula-
tions. During this test, the agents were able to fully
speciate in some cases and produce no first-degree hy-
brids when the two species were reintroduced [Table
5]. This test concluded that agent size had no impact
on agent reproductive choice and that agents of the
same size were able to successfully speciate.
A third test was designed to find the impact of
adding the four arbitrary characteristics to the char-
acteristic chromosome. This test was identical to the
first test; however, the agents were slightly modi-
fied. The agent’s reproductive choice function was
changed to no longer consider the four character-
istics on the end of the characteristic chromosome
and solely determine reproductive choice based on
agent color and age. Based on the result that the
agents could not successfully speciate in a reasonable
amount of time without considering the four arbitrary
characteristics, we believe that the number of fea-
tures considered during reproductive choice directly
affects the difficulty of speciation. A notable aspect
of this test was that because the agents were less spe-
cific with their choice of mate, they had lower overall
energy and therefore attempted to reproduce less of-
ten. This led to a much lower number of hybridiza-
tion attempts than the previous tests while still hav-
ing roughly the same total overall reproductions. Al-
though we believe that speciation is theoretically still
possible within the parameters of this test, the less-
ened number of features considered for reproduction
seems to have increased the difficulty for the agents to
speciate, as the agents were never able to successfully
speciate in any of the trials that were conducted.
In the results of our testing, the agents were able
to evolve into separate species when a sufficient num-
ber of features were considered during reproduction,
regardless of how those features affect fitness. The
number of features seems to have directly affected the
difficulty the agents had in speciating and determining
if a potential mate was a member of a different species
or not.
9 CONCLUSION
The replication of allopatric speciation has been
achieved previously by Parker and Edwards, how-
ever, in that work it was recognized that agent selec-
Using Secondary Inherited Characteristics During Reproductive Choice to Replicate Allopatric Speciation
311

tion would need to be expanded in order to achieve a
more complete replication of speciation (Parker and
Edwards, 2019). In this paper, we have been able to
conclude that the goal of replicating allopatric spe-
ciation using solely agent selectivity and influential
characteristics has been achieved. Our system was,
in fact, more complete than previous work as when
the populations were reintroduced, in some cases no
first-degree hybrids were produced whatsoever.
This increased completeness in speciation was
possibly because in the previous work, agents would
solely consider the size of a prospective mate due to
size being the primary fitness defining characteristic.
As this characteristic could randomly mutate, it would
be possible that this mutation could lead to an agent
that has the same size as a different species but was
not actually a member of that species. The simulation
described in this paper removes this possibility as the
chance of a random mutation affecting many different
characteristics in such a way that the agent would be
mistaken as a different species would be highly un-
likely.
Additionally, this new system of simulating al-
lopatric speciation has the added benefit of being ca-
pable of producing separate species from agents of the
same size. This allows agents to evolve the features
that best fit their environment, without compromising
the features used to decide mate selection. This also
decouples the number of possible species from the
number of different environments. In this system, a
large number of reproductively separate species could
develop from identical starting conditions.
In future work, we would like to explore the pos-
sibility of speciation without a hard barrier, such as
in the concept of ring species, where a chain of in-
tergrading species encircles a barrier and the terminal
species coexist without interbreeding. This alteration
would allow simulation of an alternative way for real
world speciation to occur. We also plan to attempt
sympatric speciation to see if there are conditions that
could make it possible.
REFERENCES
Arabas, J., Michalewicz, Z., and Mulawka, J. (1994).
Gavaps - a genetic algorithm with varying population
size. In Proceedings of the First IEEE Conference on
Evolutionary Computation. IEEE World Congress on
Computational Intelligence, pages 73–78. IEEE.
Bolnick, D. I. and Fitzpatrick, B. M. (2007). Sympatric
speciation: Models and empirical evidence. An-
nual Review of Ecology, Evolution, and Systematics,
38(1):459–487.
Coyne, J. A. (1992). Genetics and speciation. Nature,
355(6360):511–515.
Glibovets, N. and Gulayeva, N. (2013). A review of niching
genetic algorithms for multimodal function optimiza-
tion. Cybernetics and Systems Analysis, 49(6):815–
820.
Goldberg, D. E. and Richardson, J. (1987). Genetic algo-
rithms with sharing for multimodal function optimiza-
tion. In Proceedings of the Second International Con-
ference on Genetic Algorithms on Genetic Algorithms
and Their Application, page 41–49, USA. L. Erlbaum
Associates Inc.
Gustafson, S. and Burke, E. K. (2006). The speciating is-
land model: An alternative parallel evolutionary algo-
rithm. Journal of Parallel and Distributed Computing,
66(8):1025–1036.
Holland, J. H. (1975). Adaptation in natural and artificial
systems: An introductory analysis with applications to
biology, control, and artificial intelligence. U Michi-
gan Press.
Li, W., Li, B., and Zhang, Q. (2012). Genetic algorithm
with geographic speciation. In 2012 8th International
Conference on Natural Computation, pages 665–668,
Chongqing, China.
Mayr, E. (1963). Animal Species and Evolution. Harvard
University Press.
Parker, G. and Edwards, T. (2018). A simple environment
for research in speciation. In Proceedings of the AAAI
2018 Fall Symposium on Artificial Intelligence and
Natural Systems.
Parker, G. B. and Edwards, T. B. (2019). Using a genetic
algorithm to replicate allopatric speciation. In 2019
IEEE Congress on Evolutionary Computation (CEC).
IEEE.
Potter, M. A. and Jong, K. A. D. (1994). A cooperative
coevolutionary approach to function optimization. In
Davidor, Y., Schwefel, H.-P., and Männer, R., editors,
Parallel Problem Solving from Nature — PPSN III,
volume 866 of Lecture Notes in Computer Science,
pages 249–257. Springer, Berlin, Heidelberg.
Shi, X., Liang, Y., Lee, H., and Wang, L. (2005). An im-
proved ga and a novel pso-ga-based hybrid algorithm.
Information Processing Letters, 93(5):255–261.
Stanley, K. O. and Miikkulainen, R. (2002). Evolving Neu-
ral Networks through Augmenting Topologies. Evo-
lutionary Computation, 10(2):99–127.
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
312
