
Optimizing Genetic Algorithms Using the Binomial Distribution
Vincent A. Cicirello
a
Computer Science, School of Business, Stockton University, 101 Vera King Farris Dr, Galloway, NJ, U.S.A.
Keywords:
Bernoulli Trials, Binomial Random Variable, Genetic Algorithm, Mutation, Uniform Crossover.
Abstract:
Evolutionary algorithms rely very heavily on randomized behavior. Execution speed, therefore, depends
strongly on how we implement randomness, such as our choice of pseudorandom number generator, or the
algorithms used to map pseudorandom values to specific intervals or distributions. In this paper, we observe
that the standard bit-flip mutation of a genetic algorithm (GA), uniform crossover, and the GA control loop
that determines which pairs of parents to cross are all in essence binomial experiments. We then show how
to optimize each of these by utilizing a binomial distribution and sampling algorithms to dramatically speed
the runtime of a GA relative to the common implementation. We implement our approach in the open-source
Java library Chips-n-Salsa. Our experiments validate that the approach is orders of magnitude faster than the
common GA implementation, yet produces solutions that are statistically equivalent in solution quality.
1 INTRODUCTION
The simulated evolutionary processes of genetic algo-
rithms (GA), and other evolutionary algorithms (EA),
are stochastic and rely heavily on randomized behav-
ior. Mutation in a GA involves randomly flipping bits.
Cross points in single-point, two-point, and k-point
crossover are chosen randomly. Uniform crossover
uses a process similar to mutation to randomly choose
which bits to exchange between parents. The decision
of whether to cross a pair of parents during a genera-
tion is random. Selection operators usually randomize
the selection of the population members that survive.
Random behavior pervades the EA. Thus, how
randomness is implemented significantly impacts per-
formance. Many have explored the effects of the
pseudorandom number generator (PRNG) on solution
quality (Krömer et al., 2018; Rajashekharan and Ve-
layutham, 2016; Krömer et al., 2013; Tirronen et al.,
2011; Reese, 2009; Wiese et al., 2005a; Wiese et al.,
2005b; Cantú-Paz, 2002), and others the effects of
PRNG on execution times (Cicirello, 2018; Nesmach-
now et al., 2015). Choice of PRNG isn’t the only
way to optimize an EA’s random behavior. Our open
source Java library Chips-n-Salsa (Cicirello, 2020)
optimizes randomness of its EAs and metaheuristics
beyond the PRNG, such as in the choice of algorithms
for Gaussian random numbers and random integers in
an interval. Section 2 further discusses related work.
a
https://orcid.org/0000-0003-1072-8559
This paper focuses on optimizing the randomness
of bit-flip mutation, uniform crossover, and the deter-
mination of which pairs of parents to cross. Formally,
all three of these are binomial experiments, with the
relevant GA control parameters serving as the success
probabilities for a sequence of Bernoulli trials. This
leads to our approach (Section 3) to each that replaces
explicit iteration over bits or the population with a bi-
nomial distributed random variable and an efficient
sampling algorithm.
Others, such as Ye et al, previously observed that
the number of bits mutated by the standard bit-flip
mutation follows a binomial distribution (Ye et al.,
2019). However, examining the source code of their
implementation (Ye, 2023) reveals that they gener-
ate binomial random variates by explicitly iterating
n times for a bit-vector of length n, generating a total
of n uniform variates in the process. Their approach
refactored where the explicit iteration occurs, but did
not eliminate it. Ye et al were not trying to optimize
the runtime of the traditional GA. Rather, their reason
for abstracting the mutated bit count was to enable ex-
ploring alternative mutation operators, such as their
normalized bit mutation (Ye et al., 2019) in which the
number of mutated bits follows a normal distribution
rather than a binomial.
In our approach, presented in detail in Section 3,
we eliminate the O(n) uniform random values gen-
erated during explicit iteration over bits. Instead,
we use an efficient algorithm for generating binomial
Cicirello, V.
Optimizing Genetic Algorithms Using the Binomial Distribution.
DOI: 10.5220/0013038300003837
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Joint Conference on Computational Intelligence (IJCCI 2024), pages 159-169
ISBN: 978-989-758-721-4; ISSN: 2184-3236
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
159

random variates (Kachitvichyanukul and Schmeiser,
1988) that requires only O(1) random numbers on av-
erage. Additionally, we use a more efficient sampling
algorithm for choosing which bits to mutate. Ours is
the first approach to use the binomial distribution in
this way to optimize the runtime of bit-flip mutation.
Furthermore, we also apply the technique to optimize
uniform crossover as well as the process of selecting
which pairs of parents to cross.
Our experiments (Section 4) demonstrate the mas-
sive gains in execution speed that result from this ap-
proach. We will see that the optimized bit-flip mu-
tation uses 70%-99% less time than the common im-
plementation. The optimized uniform crossover uses
61%-99% less time than the common implementa-
tion. A GA using the optimized decision of which
parents to cross, as well as the optimized operators,
uses 78%-97% less time than the common implemen-
tation. We implemented the approach in the open
source library Chips-n-Salsa (Cicirello, 2020) and re-
leased the code of the experiments as open source to
enable easily replicating the results. We wrap up with
a discussion in Section 5.
2 RELATED WORK
Several have shown that higher quality PRNGs that
pass more rigorous randomness tests do not lead to
better fitness, and that solution quality is insensitive
to PRNG (Rajashekharan and Velayutham, 2016; Tir-
ronen et al., 2011; Wiese et al., 2005a; Wiese et al.,
2005b; Cantú-Paz, 2002). For example, the ablation
study of Cantú-Paz shows that the PRNG used for se-
lection, crossover, and mutation does not affect fit-
ness, and that even using true random numbers does
not improve fitness (Cantú-Paz, 2002). Thus, choice
of PRNG has little, if any, impact on solution fitness.
Fewer have studied the impact of the PRNG on
execution times, but the research that exists shows po-
tential for massive efficiency gains. Nesmachnow et
al analyzed the effects of several implementation fac-
tors on the runtime of EAs implemented in C (Nes-
machnow et al., 2015). They showed 50%-60% im-
provement in execution times by using state-of-the-art
PRNGs such as R250 (Kirkpatrick and Stoll, 1981) or
Mersenne Twister (Matsumoto and Nishimura, 1998)
rather than the C library’s rand() function. Cicirello
showed that an adaptive permutation EA is 25%
faster (Cicirello, 2018) when using the PRNG Split-
Mix (Steele et al., 2014) rather than the classic lin-
ear congruential (Knuth, 1998), and by implementing
Gaussian mutation (Hinterding, 1995) with the zig-
gurat algorithm (Marsaglia and Tsang, 2000; Leong
Table 1: Notation shared across algorithm pseudocode.
B(n, p) binomial distributed random integer
FlipBit(v, i) flips bit i of vector v
Length(v) simple accessor for length of v
Rand(a, b) random real in [a,b)
Rand(a) random integer in [0,a)
Sample(n, k) sample k distinct integers from
{0,1, ...,n −1}
⊕,∧,∨,¬ bitwise XOR, AND, OR, and NOT
et al., 2005) rather than the polar method (Knuth,
1998). There are also studies on the impact of PRNG
and related algorithmic components on the runtime
performance of randomized systems other than EAs,
such as Metropolis algorithms (Macias-Medri et al.,
2023). Others explored the effects of programming
language choice on GA runtime (Merelo-Guervós
et al., 2017; Merelo-Guervós et al., 2016).
The EAs of Chips-n-Salsa (Cicirello, 2020) op-
timize randomness in ways beyond the choice of
PRNG. For example, Chips-n-Salsa uses the algo-
rithm of Lemire for random integers in an inter-
val (Lemire, 2019) as implemented in the open source
library ρµ (Cicirello, 2022b), which is more than
twice as fast as Java’s built-in method due to a sig-
nificantly faster approach to rejection sampling. Such
bounded random integers are used by EAs in a vari-
ety of ways, such as the random cross sites for single-
point, two-point, and k-point crossover.
3 APPROACH
We now present the details of our approach. In Sec-
tion 3.1, we formalize the relationship between differ-
ent processes within a GA and binomial experiments.
In Section 3.2, we describe our approach to random
sampling. We derive our optimizations of random bit-
mask generation, mutation, and uniform crossover in
Sections 3.3, 3.4, and 3.5, respectively. We show how
to optimize the process of determining which pairs of
parents to cross, as well as putting all of the optimiza-
tions together in Section 3.6.
Table 1 summarizes the notation and functions
shared by the pseudocode of the algorithms through-
out this section. We assume that indexes into bit-
vectors begin at 0. The runtime of FlipBit(v, i),
Length(v), and Rand(a, b) is O(1). The runtime of
the bitwise operators is O(n) for vectors of length n.
Our bit-vector implementation utilizes an array of 32-
bit integers, enabling exploiting implicit parallelism,
such as for bitwise operations (e.g., a bitwise opera-
tion on two bit-vectors of length n requires
n
32
bitwise
operations on 32-bit integers).
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
160

3.1 Binomial Variates and the GA
A Bernoulli trial is an experiment with two possi-
ble outcomes, usually success and failure, and speci-
fied by success probability p. A binomial experiment
is a sequence of n independent Bernoulli trials with
identical p (Larson, 1982). The binomial distribution
B(n, p) is the discrete probability distribution of the
number of successes in an n-trial binomial experiment
with parameter p.
Our approach observes that GA mutation of a bit
vector of length n is a binomial experiment with n tri-
als, but the possible outcomes of each trial are “flip-
bit” and “keep-bit” instead of success and failure. The
number of bits flipped while mutating a bit vector of
length n must therefore follow binomial distribution
B(n, p
m
), where p
m
is the mutation rate. Uniform
crossover of a pair of bit vectors of length n is also
a binomial experiment with n trials, with the num-
ber of bits exchanged between parents following bino-
mial distribution B(n, p
u
), where p
u
is the probability
of exchanging a bit between the parents. Choosing
which pairs of parents to cross is likewise a binomial
experiment with n/2 trials for population size n, and
the number of crosses during a generation follows bi-
nomial distribution B(n/2, p
c
) for crossover rate p
c
.
We generate binomial random variates B(n, p)
with the BTPE algorithm (Kachitvichyanukul and
Schmeiser, 1988), whose runtime is O(1) (Cicirello,
2024b). This algorithm choice is critical to later anal-
ysis, as many alternatives do not have a constant run-
time (Kuhl, 2017; Knuth, 1998).
3.2 Sampling
To sample k distinct integers from the set
{0,1, ...,n − 1}, we implement Sample(n, k)
using a combination of reservoir sampling (Vitter,
1985), pool sampling (Goodman and Hedetniemi,
1977), and insertion sampling (Cicirello, 2022a),
choosing the most efficient based on k relative to n.
1 function Sample(n, k)
2 if k ≥
n
2
then
3 return ReservoirSample(n, k)
4 end
5 if k ≥
√
n then
6 return PoolSample(n, k)
7 end
8 return InsertionSample(n, k)
9 end
Algorithm 1: Random sampling.
Algorithms 1 and 2 provide pseudocode of the
details. See the original publications of the three sam-
1 function ReservoirSample(n, k)
2 s ← an array of length k
3 for i = 0 to k −1 do
4 s[i] ← i
5 end
6 for i = k to n −1 do
7 j ← Rand(i + 1)
8 if j < k then
9 s[ j] ← i
10 end
11 end
12 return s
13 end
14
15 function PoolSample(n, k)
16 s ← an array of length k
17 t ← an array of length n
18 for i = 0 to n −1 do
19 t[i] ← i
20 end
21 m ← n
22 for i = 0 to k −1 do
23 j ← Rand(m)
24 s[i] ←t[ j]
25 m ← m −1
26 t[ j] ← t[m]
27 end
28 return s
29 end
30
31 function InsertionSample(n, k)
32 s ← an array of length k
33 for i = 0 to k −1 do
34 v ← Rand(n −i)
35 j ← k −i
36 while j < k and v ≥s[ j] do
37 v ← v + 1
38 s[ j −1] ← s[ j]
39 j ← j + 1
40 end
41 s[ j −1] ← v
42 end
43 return s
44 end
Algorithm 2: Random sampling component algorithms.
pling algorithms for explanations for why each works.
Our composition (Algorithm 1) chooses from among
the three sampling algorithms (Algorithm 2) the one
that requires the least random number generation for
the given n and k. The runtime of reservoir sampling
and pool sampling is O(n), while the runtime of in-
sertion sampling is O(k
2
). Pool sampling and inser-
tion sampling each generate k random integers, while
Optimizing Genetic Algorithms Using the Binomial Distribution
161

reservoir sampling requires (n −k) random integers.
The runtime of the composite of these algorithms is
O(min(n,k
2
)), and requires min(k,n −k) random in-
tegers (Cicirello, 2022a). Minimizing random num-
ber generation is perhaps even more important than
runtime complexity, because although it is a constant
time operation, random number generation is costly.
3.3 Optimizing Random Bitmasks
Our algorithms for mutation and crossover rely on
random bitmasks of length n for probability p that
a bit is a 1. Algorithm 3 compares pseudocode
for the simple approach and the optimized binomial
approach. The runtime (worst, average, and best
cases) of the simple approach, SimpleBitmask(n,
p), is O(n). The runtime of the optimized version,
OptimizedBitmask(n, p), is likewise O(n), but the
only O(n) step initializes an all zero bit-vector in
line 12. Most of the cost savings is due to requiring
only O(n ·min(p,1 − p)) random numbers on aver-
age (the call to Sample() on line 14), compared to
SimpleBitmask(n, p), which always requires n ran-
dom numbers. When p is small, such as for mutation
where p is the mutation rate p
m
, the cost savings from
minimizing random number generation is especially
advantageous as we will see in the experiments.
1 function SimpleBitmask(n, p)
2 v ← an all 0 bit-vector of length n
3 for i = 0 to n −1 do
4 if Rand(0.0, 1.0) < p then
5 FlipBit(v, i)
6 end
7 end
8 return v
9 end
10
11 function OptimizedBitmask(n, p)
12 v ← an all 0 bit-vector of length n
13 k ←B(n, p)
14 indexes ← Sample(n, k)
15 for i ∈indexes do
16 FlipBit(v, i)
17 end
18 return v
19 end
Algorithm 3: Simple vs optimized bitmask creation.
Not shown in Algorithm 3 for presentation clar-
ity, our implementation of OptimizedBitmask(n, p)
treats p = 0.5 as a special case by generating 32 ran-
dom bits at a time with uniformly distributed random
32-bit integers. The OptimizedBitmask(n, p), as
presented in Algorithm 3, requires
n
2
random bounded
integers when p = 0.5, while this special case treat-
ment reduces this to
n
32
random integers.
3.4 Optimizing Mutation
Algorithm 4 compares the common approach to muta-
tion and our optimized approach. The p
m
is mutation
rate. Since the simple approach, SimpleMutation(v,
p
m
), iterates over all n bits, generating one random
number for each, its runtime is O(n). The runtime of
OptimizedMutation(v, p
m
) is likewise O(n) due to
the ⊕ of vectors of length n, and the initialization of
an all zero vector of length n. It is possible to elimi-
nate these O(n) steps with explicit iteration over only
the mutated bits instead of using a bitmask. However,
these steps are relatively inexpensive given the im-
plicit parallelism associated with utilizing 32-bit in-
tegers (e.g., bit operations on 32 bits at a time). The
real time savings comes from reducing random num-
ber generation, where OptimizedMutation(v, p
m
)
requires only O(n ·min(p
m
,1 − p
m
)) random num-
bers on average via the call to OptimizedBitmask(n,
p
m
) in line 12, while SimpleMutation(v, p
m
) re-
quires n. Since the mutation rate p
m
is generally
quite small, OptimizedMutation(v, p
m
) eliminates
nearly all random number generation.
1 function SimpleMutation(v, p
m
)
2 n ← Length(v) ;
3 for i = 0 to n −1 do
4 if Rand(0.0, 1.0) < p
m
then
5 FlipBit(v, i)
6 end
7 end
8 end
9 ;
10 function OptimizedMutation(v, p
m
)
11 n ← Length(v) ;
12 bitmask ← OptimizedBitmask(n, p
m
) ;
13 v ← v ⊕bitmask
14 end
Algorithm 4: Simple vs optimized mutation.
3.5 Optimizing Uniform Crossover
Algorithm 5 compares the simple uniform crossover,
determining the bits to exchange with iteration, ver-
sus our optimized version using our binomial trick.
The p
u
is uniform crossover’s parameter for the
per-bit probability of bit exchange. Both versions
are identical aside from how they generate a ran-
dom bitmask. The runtime of both is O(n) due
to the bitwise operations. The time savings for
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
162

OptimizedCrossover(v
1
, v
2
, p
u
) derives from op-
timizing bitmask creation in line 11, leading the opti-
mized uniform crossover to require O(n ·min(p
u
,1 −
p
u
)) random numbers, rather than the O(n) random
numbers required by the simple approach. For ex-
ample, if p
u
= 0.33 or p
u
= 0.67, then the optimiza-
tion requires only a third of the random numbers that
would be needed if explicit bit iteration was used.
1 function SimpleCrossover(v
1
, v
2
, p
u
)
2 n ← Length(v
1
)
3 bitmask ← SimpleBitmask(n, p
u
)
4 temp ← (v
1
∧bitmask) ∨(v
2
∧¬bitmask)
5 v
1
← (v
2
∧bitmask) ∨(v
1
∧¬bitmask)
6 v
2
← temp
7 end
8
9 function OptimizedCrossover(v
1
, v
2
, p
u
)
10 n ← Length(v
1
)
11 bitmask ← OptimizedBitmask(n, p
u
)
12 temp ← (v
1
∧bitmask) ∨(v
2
∧¬bitmask)
13 v
1
← (v
2
∧bitmask) ∨(v
1
∧¬bitmask)
14 v
2
← temp
15 end
Algorithm 5: Simple vs optimized uniform crossover
3.6 Optimizing a Generation
Algorithm 6 compares a simple implementation
of a generation with an optimized version. The
SimpleGeneration() explicitly iterates over the
n
2
pairs of possible parents (lines 4–9), gener-
ating a uniform random variate for each to de-
termine which parents produce offspring. The
OptimizedGeneration() generates a single bino-
mial random variate from B(⌊
n
2
⌋, p
c
), where p
c
is the
crossover rate, to determine the number of crossover
applications (line 18), and then iterates without need
for additional random numbers (lines 19–21).
Although the BTPE algorithm (Ka-
chitvichyanukul and Schmeiser, 1988) that we
use to generate binomial random variates utilizes re-
jection sampling (Flury, 1990), the average number of
rejection sampling iterations is constant, and thus the
average number of uniform variates needed by BTPE
to generate a binomial is also constant (Cicirello,
2024b). While SimpleGeneration() requires O(
n
2
)
uniform random variates, OptimizedGeneration()
requires only O(1) random numbers.
The OptimizedMutation() leads to additional
speed advantage. The pseudocode uses a generic
Crossover() operation rather than assuming any
specific operator. In the experiments, we consider
both uniform crossover, which we optimize, as well
as single-point and two-point crossover, which don’t
1 function SimpleGeneration(pop, p
c
, p
m
)
2 n ← Length(pop)
3 pop ← Selection(pop)
/* assume new population pop in
random order */
4 pairs ← ⌊
n
2
⌋
5 for i = 0 to pairs −1 do
6 if Rand(0.0, 1.0) < p
c
then
7 Crossover(pop
i
, pop
i+pairs
)
8 end
9 end
10 for i = 0 to n −1 do
11 SimpleMutation(pop
i
, p
m
)
12 end
13 end
14
15 function OptimizedGeneration(pop, p
c
,
p
m
)
16 n ← Length(pop)
17 pop ← Selection(pop)
/* assume new population pop in
random order */
18 pairs ← B(⌊
n
2
⌋, p
c
)
19 for i = 0 to pairs −1 do
20 Crossover(pop
i
, pop
i+pairs
)
21 end
22 for i = 0 to n −1 do
23 OptimizedMutation(pop
i
, p
m
)
24 end
25 end
Algorithm 6: Simple vs optimized generation.
have corresponding binomial optimizations. The
cross points of our single-point and two-point
crossover operators are selected using a more effi-
cient algorithm for bounded random integers (Lemire,
2019) than Java’s built-in method; and the pair of in-
dexes for two-point crossover are sampled using a
specially designed algorithm for small random sam-
ples (Cicirello, 2024a) that is significantly faster than
general purpose sampling algorithms such as those
utilized earlier in Section 3.2. However, the experi-
ments in this paper that use single-point and two-point
crossover use these same optimizations for both the
simple and optimized experimental conditions.
4 EXPERIMENTS
We run the experiments on a Windows 10 PC with
an AMD A10-5700, 3.4 GHz processor and 8GB
memory, and we use OpenJDK 64-Bit Server VM
version 17.0.2. We implemented the optimized bit-
flip mutation, uniform crossover, and generation loop
Optimizing Genetic Algorithms Using the Binomial Distribution
163

Table 2: URLs for Chips-n-Salsa and experiments.
Chips-n-Salsa library
Source https://github.com/cicirello/Chips-n-Salsa
Website https://chips-n-salsa.cicirello.org/
Maven https://central.sonatype.com/artifact/org.
cicirello/chips-n-salsa
Experiments
Source https://github.com/cicirello/
optimize-ga-operators
Table 3: CPU time for 10
5
mutations for n = 1024.
CPU time (seconds) % less t-test
p
m
simple optimized time p-value
1/1024 0.856 0.00906 98.9% ∼ 0.0
1/512 0.857 0.0119 98.6% ∼ 0.0
1/256 0.860 0.0173 98.0% ∼ 0.0
1/128 0.866 0.0300 96.5% < 10
−291
1/64 0.877 0.0642 92.7% ∼ 0.0
1/32 0.899 0.140 84.5% < 10
−285
1/16 0.946 0.192 79.7% < 10
−321
1/8 1.05 0.255 75.7% < 10
−193
1/4 1.25 0.363 71.0% ∼ 0.0
within the open source library Chips-n-Salsa (Ci-
cirello, 2020), and use version 7.0.0 in the exper-
iments. All experiment source code is also open
source. Table 2 provides the relevant URLs.
The remainder of this section is organized as fol-
lows. Sections 4.1 and 4.2 present our experiments
with mutation and uniform crossover, respectively.
Then, in Section 4.3, we provide experiments com-
paring a fully optimized GA that optimizes the choice
of which pairs of parents to cross in addition to the
mutation and uniform crossover optimizations.
4.1 Mutation Experiments
In our mutation experiments, we consider bit-vector
length n ∈ {16,32,64, 128,256, 512,1024}, and mu-
tation rate p
m
∈ {
1
n
,
2
n
,. ..,
1
4
}. For each combination
(n, p
m
), we measure the CPU time to perform 100,000
mutations, averaged across 100 trials. We test the sig-
nificance of the differences between the simple and
optimized versions using Welch’s unequal variances
t-test (Welch, 1947; Derrick and White, 2016).
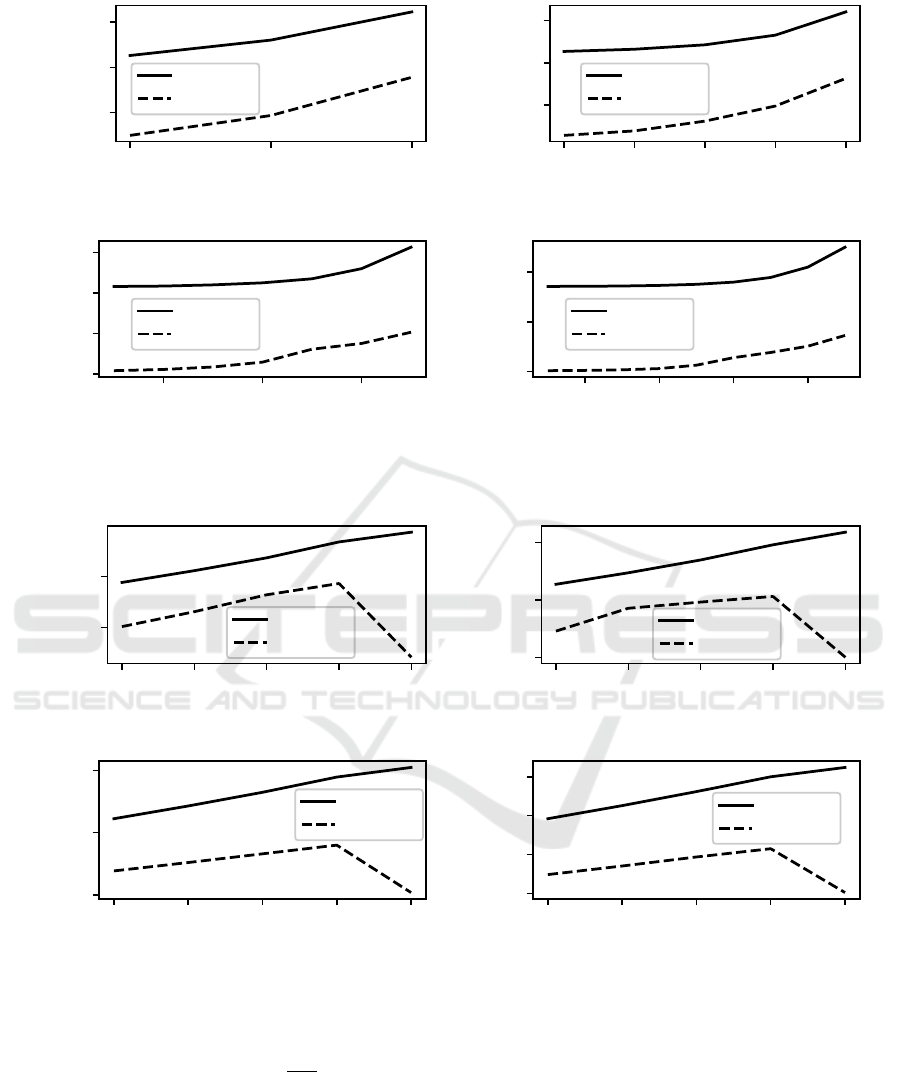
Figure 1 visualizes n ∈ {16, 64,256, 1024}. Ta-
ble 3 summarizes results for n = 1024. Data for all
other cases is found in the GitHub repository, and is
similar to the cases presented here.
The optimized mutation leads to massive perfor-
mance gains. For bit-vector length n = 1024, the opti-
mized mutation uses 71% less time for high mutation
rates. For typical low mutation rates, the optimized
Table 4: CPU time for 10
5
uniform crosses for n = 1024.
CPU time (seconds) % less t-test
p
u
simple optimized time p-value
0.1 0.962 0.245 74.6% ∼ 0.0
0.2 1.13 0.355 68.5% ∼ 0.0
0.3 1.31 0.471 64.0% < 10
−308
0.4 1.50 0.577 61.6% < 10
−312
0.5 1.62 0.0142 99.1% ∼ 0.0
mutation uses 96%–99% less time than the simple im-
plementation. All results are extremely statistically
significant with t-test p-values very near zero.
4.2 Uniform Crossover Experiments
We use the same bit-vector lengths n for the crossover
experiments as we did for the mutation experiments;
and we consider p
u
∈ {0.1, 0.2,0.3, 0.4,0.5} for the
per-bit probability of an exchange between parents.
We do not consider p
u
> 0.5 because any such p
u
has
an equivalent counterpart p
u
< 0.5 (e.g., exchanging
75% of the bits between the parents leads to the same
children as if we instead exchanged the other 25% of
the bits). For each combination (n, p
u
), we measure
the CPU time to perform 100,000 uniform crossovers,
averaged across 100 trials, and again test significance
with Welch’s unequal variances t-test.
Figure 2 visualizes n ∈ {16, 64,256, 1024}. Ta-
ble 4 summarizes results for n = 1024. The data for
all other cases is found in the GitHub repository, and
is similar to the cases presented here.
The optimized uniform crossover leads to similar
performance gains as seen with mutation. Specifi-
cally, for bit-vector length n = 1024, the optimized
uniform crossover uses approximately 60% to 75%
less time than the simple implementation, except for
the case of p
u
= 0.5 where the optimized version uses
99% less time. The performance of p
u
= 0.5 is espe-
cially strong due to our special case treatment when
generating random bitmasks (see earlier discussion in
Section 3). All results are extremely statistically sig-
nificant with t-test p-values very near zero.
4.3 GA Experiments
Unlike mutation and crossover, it is not feasible to
isolate the generation control logic of Algorithm 6
from the GA as it depends upon the genetic opera-
tors. Instead we experiment with a GA with simple
vs optimized operators. We use the OneMax prob-
lem (Ackley, 1985) for its simplicity since our focus
is on runtime performance of alternative implemen-
tations of operators, and use vector length n = 1024
bits. All experimental conditions use population size
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
164
2
−4
2
−3
2
−2
mutation rate p
m
(log scale)
0.010
0.015
0.020
CPU time (seconds)
simple
optimized
(a)
2
−6
2
−5
2
−4
2
−3
2
−2
mutation rate p
m
(log scale)
0.025
0.050
0.075
CPU time (seconds)
simple
optimized
(b)
2
−7
2
−5
2
−3
mutation rate p
m
(log scale)
0.0
0.1
0.2
0.3
CPU time (seconds)
simple
optimized
(c)
2
−9
2
−7
2
−5
2
−3
mutation rate p
m
(log scale)
0.0
0.5
1.0
CPU time (seconds)
simple
optimized
(d)
Figure 1: CPU time for 10
5
mutations vs mutation rate p
m
for lengths: (a) 16 bits, (b) 64 bits, (c) 256 bits, (d) 1024 bits.
0.1 0.2 0.3 0.4 0.5
uniform crossover bit-rate p
u
0.01
0.02
CPU time (seconds)
simple
optimized
(a)
0.1 0.2 0.3 0.4 0.5
uniform crossover bit-rate p
u
0.00
0.05
0.10
CPU time (seconds)
simple
optimized
(b)
0.1 0.2 0.3 0.4 0.5
uniform crossover bit-rate p
u
0.0
0.2
0.4
CPU time (seconds)
simple
optimized
(c)
0.1 0.2 0.3 0.4 0.5
uniform crossover bit-rate p
u
0.0
0.5
1.0
1.5
CPU time (seconds)
simple
optimized
(d)
Figure 2: CPU time for 10
5
uniform crosses vs uniform rate p
u
for lengths: (a) 16 bits, (b) 64 bits, (c) 256 bits, (d) 1024 bits.
100, stochastic universal sampling (Baker, 1987) for
selection, and mutation rate p
m
=
1
1024
, which leads
to an expected one mutated bit per population mem-
ber per generation. We consider crossover rates p
c
∈
{0.05,0.15,...,0.95}. Each GA run is 1000 genera-
tions, and we average results over 100 trials. We test
significance with Welch’s unequal variances t-test.
We consider four cases. Case (a) compares the
simple versions of the generation logic, mutation, and
uniform crossover (p
u
= 0.33) versus the optimized
versions of these. Case (b) is similar, but with uniform
crossover parameter p
u
= 0.49. We decided not to
use p
u
= 0.5 to avoid the extra strong performance
of our optimized bitmask generation for that special
case. Cases (c) and (d) are as the others, but with
single-point and two-point crossover, respectively.
Table 5 shows the results for case (a) uniform
crossover (p
u
= 0.33). The optimized approach
Optimizing Genetic Algorithms Using the Binomial Distribution
165
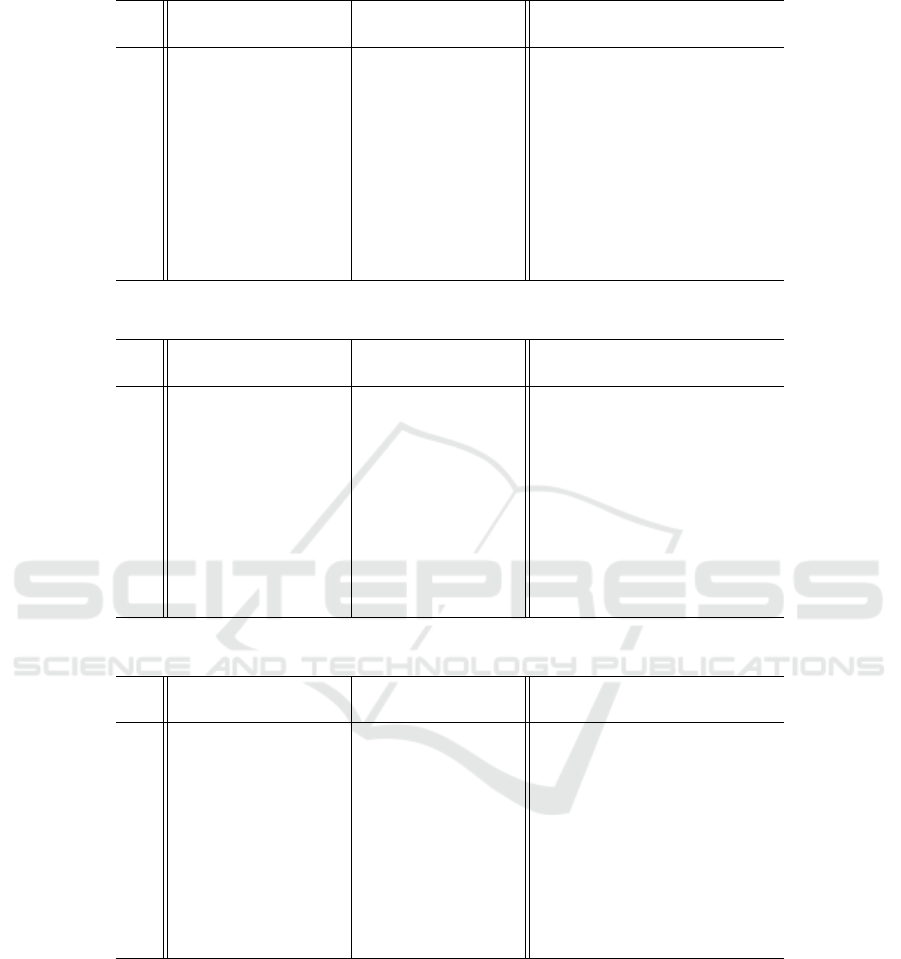
Table 5: CPU time and average solution of 1024-bit OneMax using 1000 generations with uniform crossover (p
u
= 0.33).
CPU time (seconds) % less t-test average solution t-test
p
c
simple optimized time p-value simple optimized p-value
0.05 0.869 0.0423 95.1% ∼ 0.0 687.33 686.57 0.541
0.15 0.942 0.0698 92.6% < 10
−279
709.20 709.88 0.611
0.25 1.01 0.0950 90.6% < 10
−259
715.94 714.62 0.328
0.35 1.07 0.121 88.7% ∼ 0.0 719.01 719.64 0.639
0.45 1.14 0.148 87.0% < 10
−314
720.77 721.57 0.512
0.55 1.21 0.174 85.6% ∼ 0.0 721.54 722.01 0.718
0.65 1.27 0.200 84.3% ∼ 0.0 722.97 724.94 0.158
0.75 1.34 0.224 83.3% ∼ 0.0 723.89 722.16 0.163
0.85 1.41 0.252 82.1% ∼ 0.0 725.60 726.18 0.655
0.95 1.47 0.277 81.2% ∼ 0.0 724.62 724.73 0.929
Table 6: CPU time and average solution of 1024-bit OneMax using 1000 generations with uniform crossover (p
u
= 0.49).
CPU time (seconds) % less t-test average solution t-test
p
c
simple optimized time p-value simple optimized p-value
0.05 0.875 0.0467 94.7% ∼ 0.0 690.05 689.17 0.454
0.15 0.956 0.0803 91.6% < 10
−312
711.34 712.19 0.572
0.25 1.03 0.114 88.9% ∼ 0.0 717.99 717.08 0.439
0.35 1.11 0.148 86.7% ∼ 0.0 720.71 722.44 0.162
0.45 1.19 0.182 84.8% ∼ 0.0 722.24 720.16 0.102
0.55 1.27 0.216 83.0% ∼ 0.0 723.10 723.38 0.826
0.65 1.35 0.250 81.5% ∼ 0.0 723.67 724.12 0.707
0.75 1.43 0.283 80.2% ∼ 0.0 725.33 724.64 0.582
0.85 1.51 0.318 79.0% ∼ 0.0 725.65 724.22 0.272
0.95 1.59 0.352 77.9% ∼ 0.0 724.85 724.44 0.761
Table 7: CPU time and average solution of 1024-bit OneMax using 1000 generations with single-point crossover.
CPU time (seconds) % less t-test average solution t-test
p
c
simple optimized time p-value simple optimized p-value
0.05 0.836 0.0283 96.6% ∼ 0.0 659.20 659.51 0.798
0.15 0.838 0.0286 96.6% ∼ 0.0 683.32 684.49 0.342
0.25 0.839 0.0306 96.4% < 10
−260
693.77 694.79 0.413
0.35 0.840 0.0308 96.3% ∼ 0.0 701.48 703.06 0.230
0.45 0.839 0.0325 96.1% < 10
−321
706.65 705.55 0.334
0.55 0.842 0.0323 96.2% ∼ 0.0 709.60 708.17 0.265
0.65 0.843 0.0342 95.9% ∼ 0.0 711.61 710.39 0.321
0.75 0.843 0.0355 95.8% < 10
−310
713.20 713.91 0.571
0.85 0.843 0.0366 95.7% < 10
−269
715.36 716.18 0.503
0.95 0.850 0.0373 95.6% < 10
−223
716.60 716.36 0.854
uses from approximately 81% less time for a high
crossover rate (p
c
= 0.95) to 95% less time for a
low crossover rate (p
c
= 0.05). The runtime differ-
ences are extremely statistically significant with t-test
p-values near 0.0.
Table 6 provides detailed results for case (b) uni-
form crossover (p
u
= 0.49). The results follow the
same trend as the previous case. The approach opti-
mized using the binomial distribution uses from ap-
proximately 78% less time for a high crossover rate
(p
c
= 0.95) to 95% less time for a low crossover rate
(p
c
= 0.05). The runtime differences are extremely
statistically significant with t-test p-values near 0.0.
Table 7 summarizes the results for case (c) single-
point crossover. Unlike uniform crossover, the speed
difference does not vary by crossover rate. Instead,
the optimized approach uses approximately 95% to
97% less time than the simple GA implementation
for all crossover rates. All runtime differences are ex-
tremely statistically significant with all t-test p-values
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
166
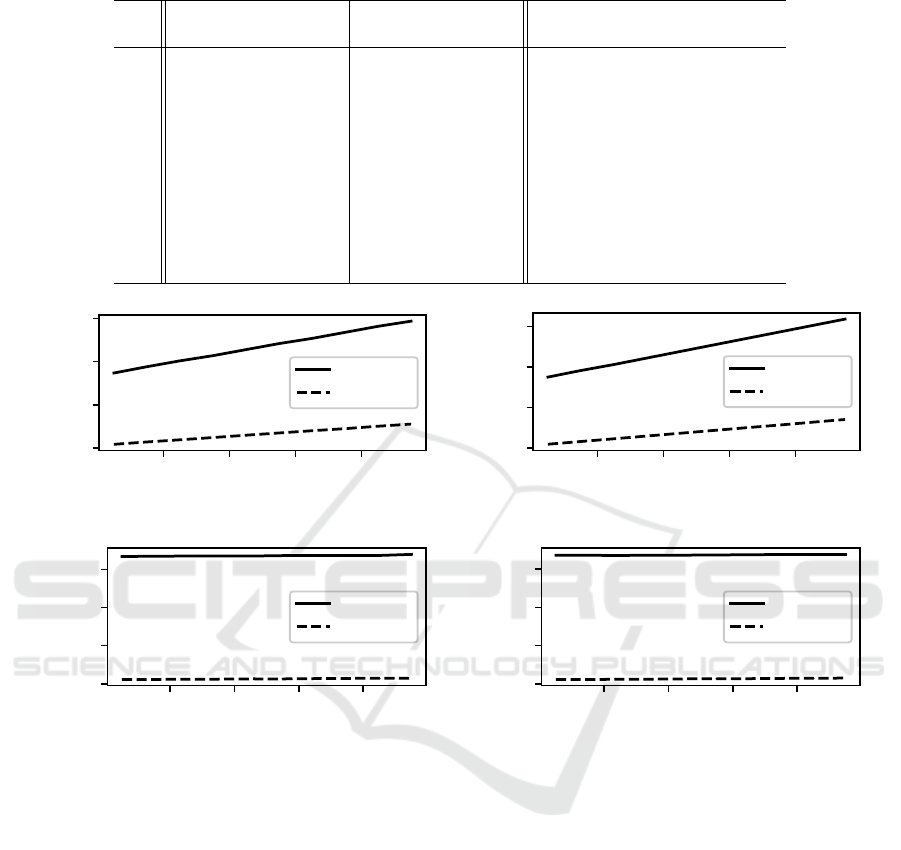
Table 8: CPU time and average solution of 1024-bit OneMax using 1000 generations with two-point crossover.
CPU time (seconds) % less t-test average solution t-test
p
c
simple optimized time p-value simple optimized p-value
0.05 0.840 0.0291 96.5% < 10
−316
666.82 666.08 0.526
0.15 0.840 0.0292 96.5% ∼ 0.0 692.26 692.32 0.959
0.25 0.839 0.0306 96.4% < 10
−296
702.23 703.29 0.394
0.35 0.841 0.0313 96.3% ∼ 0.0 708.16 709.99 0.124
0.45 0.841 0.0333 96.0% < 10
−301
712.22 712.39 0.897
0.55 0.842 0.0338 96.0% ∼ 0.0 714.05 713.80 0.845
0.65 0.844 0.0347 95.9% < 10
−311
718.86 715.63 0.00829
0.75 0.845 0.0364 95.7% ∼ 0.0 718.60 717.78 0.504
0.85 0.845 0.0377 95.5% < 10
−318
719.54 718.86 0.611
0.95 0.846 0.0383 95.5% ∼ 0.0 720.30 721.55 0.320
0.2 0.4 0.6 0.8
crossover rate p
c
0.0
0.5
1.0
1.5
CPU time (seconds)
simple
optimized
(a)
0.2 0.4 0.6 0.8
crossover rate p
c
0.0
0.5
1.0
1.5
CPU time (seconds)
simple
optimized
(b)
0.2 0.4 0.6 0.8
crossover rate p
c
0.00
0.25
0.50
0.75
CPU time (seconds)
simple
optimized
(c)
0.2 0.4 0.6 0.8
crossover rate p
c
0.00
0.25
0.50
0.75
CPU time (seconds)
simple
optimized
(d)
Figure 3: CPU time for 1024-bit OneMax using 1000 generations with population size 100 vs crossover rate p
c
for crossover
operators: (a) uniform (p
u
= 0.33), (b) uniform (p
u
= 0.49), (c) single-point, (d) two-point.
very near 0.0.
Table 8 summarizes the results for case (d) two-
point crossover. The trend is the same as in the case
of single-point crossover. The binomially optimized
approach uses approximately 95% to 97% less time
than the basic implementation, and does not vary by
crossover rate. All runtime differences are extremely
statistically significant with all t-test p-values very
near 0.0.
Tables 5, 6, 7, and 8 also show average OneMax
solutions (i.e., number of 1-bits) to demonstrate that
the optimized approach does not statistically alter GA
problem-solving behavior. For any given crossover
operator and crossover rate p
c
, there is no statistical
significance in solution quality between the optimized
and simple approach (i.e., high p-values).
Figure 3 visualizes the results for all four cases.
When uniform crossover is used, runtime increases
with crossover rate. The graphs appear to show con-
stant runtime when either single-point (Figure 3c) or
two-point crossover (Figure 3d) is used. However,
runtime is increasing in these cases, just very slowly
as seen in the detailed results from Tables 7 and 8.
The performance differential between the simple and
optimized approaches doesn’t vary by crossover rate
when single-point (Figure 3c) or two-point crossover
(Figure 3d) are used, with the optimized approach
consistently using 95% less time; while the perfor-
mance differential between the optimized and simple
approaches does vary with crossover rate when uni-
form crossover is used (Figures 3a and 3b).
Optimizing Genetic Algorithms Using the Binomial Distribution
167

5 CONCLUSION
In this paper, we demonstrated how we can signifi-
cantly speed up the runtime of some GA operators,
including the common bit-flip mutation and uniform
crossover, by observing that such operators define bi-
nomial experiments (i.e., sequence of Bernoulli tri-
als). This enables replacing explicit iteration that gen-
erates a random floating-point value for each bit to
determine whether to flip (for mutation) or exchange
(for crossover), with the generation of a single bino-
mial random variate to determine the number of bits
k, and an efficient sampling algorithm to choose the k
bits to mutate or cross. As a consequence, costly ran-
dom number generation is significantly reduced. A
similar approach is also seen for the generation logic
that determines the number of parents to cross.
The technique is not limited to these opera-
tors, and is applicable for any operator that is con-
trolled by some probability p of including an ele-
ment in the mutation or cross. For example, sev-
eral evolutionary operators for permutations (Ci-
cirello, 2023) operate in this way, including uni-
form order based crossover (Syswerda, 1991), order
crossover 2 (Syswerda, 1991; Starkweather et al.,
1991), uniform partially matched crossover (Cicirello
and Smith, 2000), uniform scramble mutation (Ci-
cirello, 2023), and uniform precedence preservative
crossover (Bierwirth et al., 1996). We adapt this ap-
proach in our implementations of all of these evolu-
tionary permutation operators in the open source li-
brary Chips-n-Salsa (Cicirello, 2020).
REFERENCES
Ackley, D. H. (1985). A connectionist algorithm for genetic
search. In ICGA, pages 121–135.
Baker, J. (1987). Reducing bias and inefficiency in the se-
lection algorithm. In ICGA, pages 14–21.
Bierwirth, C., Mattfeld, D. C., and Kopfer, H. (1996). On
permutation representations for scheduling problems.
In PPSN, pages 310–318.
Cantú-Paz, E. (2002). On random numbers and the per-
formance of genetic algorithms. In GECCO, pages
311–318.
Cicirello, V. A. (2018). Impact of random number genera-
tion on parallel genetic algorithms. In Proceedings of
the Thirty-First International Florida Artificial Intelli-
gence Research Society Conference, pages 2–7. AAAI
Press.
Cicirello, V. A. (2020). Chips-n-Salsa: A java li-
brary of customizable, hybridizable, iterative, parallel,
stochastic, and self-adaptive local search algorithms.
Journal of Open Source Software, 5(52):2448.
Cicirello, V. A. (2022a). Cycle mutation: Evolving per-
mutations via cycle induction. Applied Sciences,
12(11):5506.
Cicirello, V. A. (2022b). ρµ: A java library of randomiza-
tion enhancements and other math utilities. Journal of
Open Source Software, 7(76):4663.
Cicirello, V. A. (2023). A survey and analysis of evo-
lutionary operators for permutations. In 15th Inter-
national Joint Conference on Computational Intelli-
gence, pages 288–299.
Cicirello, V. A. (2024a). Algorithms for generating small
random samples. Software: Practice and Experience,
pages 1–9.
Cicirello, V. A. (2024b). On the average runtime of an open
source binomial random variate generation algorithm.
arXiv preprint arXiv:2403.11018 [cs.DS].
Cicirello, V. A. and Smith, S. F. (2000). Modeling ga per-
formance for control parameter optimization. In Pro-
ceedings of the Genetic and Evolutionary Computa-
tion Conference (GECCO-2000), pages 235–242.
Derrick, B. and White, P. (2016). Why welch’s test is type
I error robust. Quantitative Methods for Psychology,
12(1):30–38.
Flury, B. D. (1990). Acceptance–rejection sampling made
easy. SIAM Review, 32(3):474–476.
Goodman, S. E. and Hedetniemi, S. T. (1977). Introduction
to the Design and Analysis of Algorithms, chapter 6.3
Probabilistic Algorithms, pages 298–316. McGraw-
Hill, New York, NY, USA.
Hinterding, R. (1995). Gaussian mutation and self-adaption
for numeric genetic algorithms. In IEEE CEC, pages
384–389.
Kachitvichyanukul, V. and Schmeiser, B. W. (1988). Bino-
mial random variate generation. CACM, 31(2):216–
222.
Kirkpatrick, S. and Stoll, E. P. (1981). A very fast shift-
register sequence random number generator. Journal
of Computational Physics, 40(2):517–526.
Knuth, D. E. (1998). The Art of Computer Program-
ming, Volume 2, Seminumerical Algorithms. Addison-
Wesley, 3rd edition.
Krömer, P., Platoš, J., and Snášel, V. (2018). Evaluation
of pseudorandom number generators based on residue
arithmetic in differential evolution. In Intelligent Net-
working and Collaborative Systems, pages 336–348.
Krömer, P., Snášel, V., and Zelinka, I. (2013). On the use
of chaos in nature-inspired optimization methods. In
IEEE SMC, pages 1684–1689.
Kuhl, M. E. (2017). History of random variate generation.
In Winter Simulation Conference, pages 231–242.
Larson, H. J. (1982). Introduction to Probability Theory
and Statistical Inference. Wiley, 3rd edition.
Lemire, D. (2019). Fast random integer generation in an in-
terval. ACM Transactions on Modeling and Computer
Simulation, 29(1):3.
Leong, P. H. W., Zhang, G., Lee, D.-U., Luk, W., and Vil-
lasenor, J. (2005). A comment on the implementation
of the ziggurat method. Journal of Statistical Soft-
ware, 12(7):1–4.
ECTA 2024 - 16th International Conference on Evolutionary Computation Theory and Applications
168

Macias-Medri, A., Viswanathan, G., Fiore, C., Koehler, M.,
and da Luz, M. (2023). Speedup of the metropolis pro-
tocol via algorithmic optimization. Journal of Compu-
tational Science, 66:101910.
Marsaglia, G. and Tsang, W. W. (2000). The ziggurat
method for generating random variables. Journal of
Statistical Software, 5(8):1–7.
Matsumoto, M. and Nishimura, T. (1998). Mersenne
twister: A 623-dimensionally equidistributed uniform
pseudo-random number generator. ACM Transactions
on Modeling and Computer Simulation, 8(1):3–30.
Merelo-Guervós, J.-J., Blancas-Álvarez, I., Castillo, P. A.,
Romero, G., García-Sánchez, P., Rivas, V. M., García-
Valdez, M., Hernández-Águila, A., and Román, M.
(2017). Ranking programming languages for evolu-
tionary algorithm operations. In Applications of Evo-
lutionary Computation, pages 689–704.
Merelo-Guervós, J.-J., Blancas-Álvarez, I., Castillo, P. A.,
Romero, G., Rivas, V. M., García-Valdez, M.,
Hernández-Águila, A., and Romáin, M. (2016). A
comparison of implementations of basic evolutionary
algorithm operations in different languages. In IEEE
CEC, pages 1602–1609.
Nesmachnow, S., Luna, F., and Alba, E. (2015). An em-
pirical time analysis of evolutionary algorithms as
c programs. Software: Practice and Experience,
45(1):111–142.
Rajashekharan, L. and Velayutham, C. S. (2016). Is dif-
ferential evolution sensitive to pseudo random number
generator quality?–an investigation. In Intelligent Sys-
tems Technologies and Applications, pages 305–313.
Reese, A. (2009). Random number generators in genetic al-
gorithms for unconstrained and constrained optimiza-
tion. Nonlinear Analysis: Theory, Methods & Appli-
cations, 71(12):e679–e692.
Starkweather, T., McDaniel, S., Mathias, K., Whitley, D.,
and Whitley, C. (1991). A comparison of genetic se-
quencing operators. In ICGA, pages 69–76.
Steele, G. L., Lea, D., and Flood, C. H. (2014). Fast split-
table pseudorandom number generators. In OOPSLA,
pages 453–472.
Syswerda, G. (1991). Schedule optimization using genetic
algorithms. In Handbook of Genetic Algorithms. Van
Nostrand Reinhold.
Tirronen, V., Äyrämö, S., and Weber, M. (2011). Study on
the effects of pseudorandom generation quality on the
performance of differential evolution. In Adaptive and
Natural Computing Algorithms, pages 361–370.
Vitter, J. S. (1985). Random sampling with a reservoir.
ACM Trans on Mathematical Software, 11(1):37–57.
Welch, B. L. (1947). The generalization of student’s prob-
lem when several different population varlances are
involved. Biometrika, 34(1-2):28–35.
Wiese, K., Hendriks, A., Deschenes, A., and Youssef, B.
(2005a). Significance of randomness in p-rnapredict:
a parallel evolutionary algorithm for rna folding. In
IEEE CEC, pages 467–474.
Wiese, K. C., Hendriks, A., Deschênes, A., and Youssef,
B. B. (2005b). The impact of pseudorandom number
quality on p-rnapredict, a parallel genetic algorithm
for rna secondary structure prediction. In GECCO,
pages 479–480.
Ye, F. (2023). IOHalgorithm. https://github.com/
IOHprofiler/IOHalgorithm (accessed Sept 30, 2024).
Ye, F., Doerr, C., and Bäck, T. (2019). Interpolating local
and global search by controlling the variance of stan-
dard bit mutation. In IEEE CEC, pages 2292–2299.
Optimizing Genetic Algorithms Using the Binomial Distribution
169
