
Neuron Labeling for Self-Organizing Maps Using a Novel
Example-Centric Algorithm with Weight-Centric Finalization
Willem S. van Heerden
a
Department of Computer Science, University of Pretoria, Pretoria, South Africa
Keywords:
Unsupervised Neural Networks, Self-Organizing Maps, Neuron Labeling, Data Science, Data Mining,
Exploratory Data Analysis.
Abstract:
A self-organizing map (SOM) is an unsupervised artificial neural network that models training data using
a map structure of neurons, which preserves the local topological structure of the training data space. An
important step in the use of SOMs for data science is the labeling of neurons, where supervised neuron labeling
is commonly used in practice. Two widely-used supervised neuron labeling methods for SOMs are example-
centric neuron labeling and weight-centric neuron labeling. Example-centric neuron labeling produces high-
quality labels, but tends to leave many neurons unlabeled, thus potentially hampering the interpretation or use
of the labeled SOM. Weight-centric neuron labeling guarantees a label for every neuron, but often produces
less accurate labels. This research proposes a novel hybrid supervised neuron labeling algorithm, which
initially performs example-centric neuron labeling, after which missing labels are filled in using a weight-
centric approach. The objective of this algorithm is to produce high-quality l abels while still guaranteeing
labels for every neuron. An empirical investigation compares the performance of the novel hybrid approach
to example-centric neuron labeling and weight-centric neuron labeling, and demonstrates the feasibility of the
proposed algorithm.
1 INTRODUCTION
Self-organizing maps (SOMs) are unsupervised learn-
ing neural networks (Kohonen, 2001), which have
been widely investigated in the literature (Kaski et al.,
1998; Oja et al., 2003; Poll ¨ a et al., 2009). SOMs ¨
have seen wid e use in da ta science, data mining, and
exploratory data ana lysis (van Heerden, 2017). Some
specific application areas include business analytics
(Bowen and Siegler, 2024), healthcare (Javed et al.,
2024), pandemic analysis (Galvan et al., 2021), as-
tronom y (Vantyghem et al., 2024), and environmental
research (Rosa et al., 2024). SOMs are discussed in
more detail within Section 2.
SOMs are made up of neurons, which to gether
model a training data set. Several SOM neu-
ron labeling techniques exist, each of which at-
taches typically text-based characterizations to a sub-
set of a SOM’s neurons. Neuron labeling often
plays an essential part in SOM-ba sed data analysis
(van Heerden and Engelbrech t, 2008). The quality of
SOM-based data science therefor e depends heavily
on the performance of the labeling algorithm that has
a
https://orcid.org/0000-0002-9736-7268
been chosen. Section 3 elaborates up on neuron label-
ing approaches.
Two of the most commonly used labeling meth-
ods are example-centric neuron labeling and weight-
centric neuron labe ling (Kohonen, 1989). Example-
centric neuron la beling is very accurate but leaves
some neur ons unlabeled, while weight-centric neuron
labeling lacks accuracy but guarantees a lab el for ev-
ery neuron (van Heerden, 2017).
Section 4 intro duces a novel SOM neuron labeling
algorithm , namely example-centric neuron labeling
with weight-centric finalization. This algorithm com-
bines the b est aspects of example-centric and weight-
centric neuron labeling, by guaranteeing high-quality
labels for every neuron of a SOM .
To demonstrate the advantages of the new algo-
rithm, an empirical analysis was conducted. Sec tion 5
presents the expe rimental results, which compare
the performan ce of the novel algorithm to example-
centric and weight-centric neuron labeling.
Finally, Section 6 concludes with a summary of
this work’s most important find ings. Additionally,
several avenues are suggested for future investigations
stemming from the research presented here.
508
van Heerden, W.
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization.
DOI: 10.5220/0013072000003837
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 16th International Joint Conference on Computational Intelligence (IJCCI 2024), pages 508-519
ISBN: 978-989-758-721-4; ISSN: 2184-3236
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
z
P
T
1
z
P
T
2
w
yx2
z
21
z
2I
z
11
z
12
.
.
.
w
yx1
D
T
w
yxI
.
.
.
. . .
z
1I
z
P
T
I
z
22
z
P
T
:
z
2
:
.
.
.
. . .
. . .
z
1
:
Neuron yx
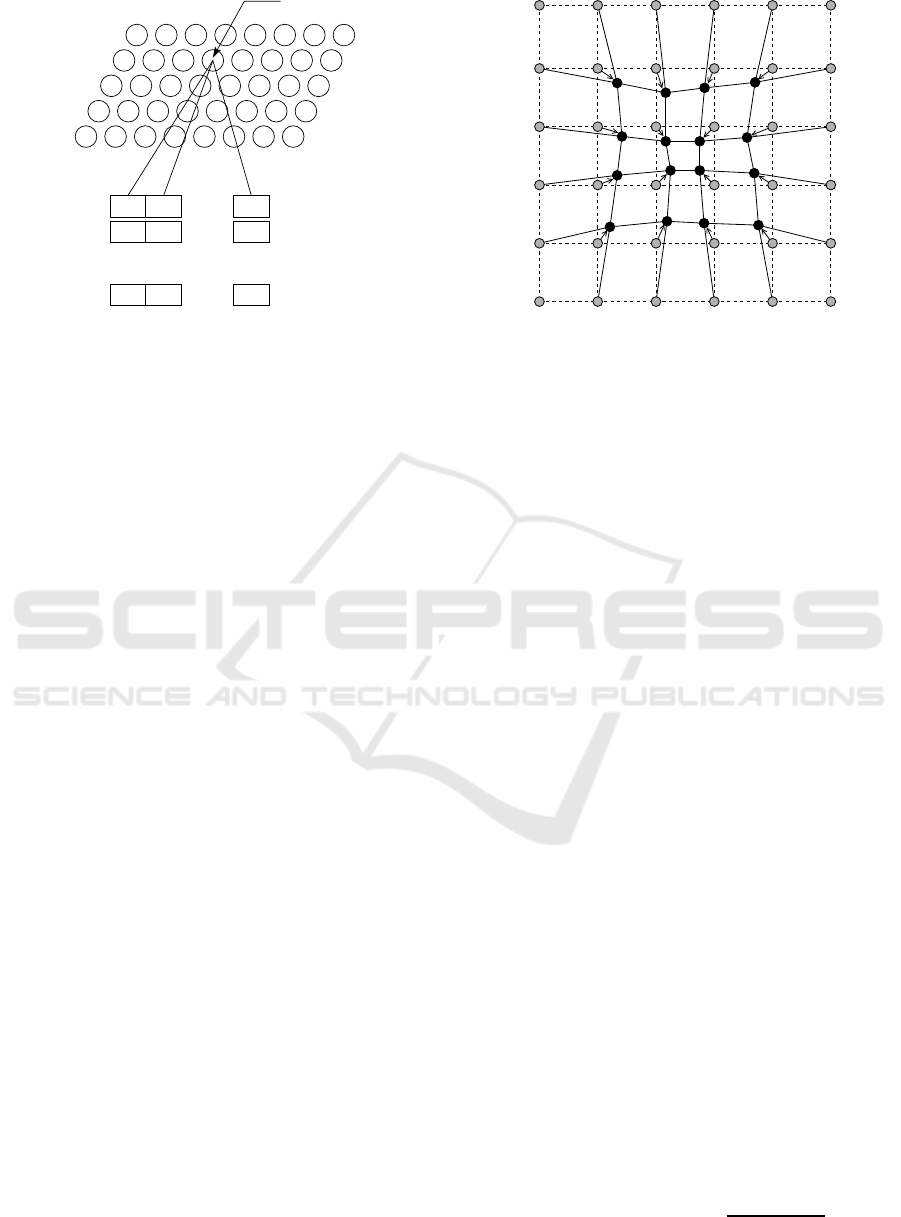
Figure 1: The architectural components making up a SOM.
2 SELF-ORGANIZING MAPS
Teuvo Kohonen intro duced the SOM , which is an un-
supervised n eural network based on th e associative
nature o f hu man cerebral cortices (Kohonen, 1982).
In contrast to supervised neural networks, the training
of a SOM does not require data classifications.
Figure 1 illustrates the architectural co mponents
of a SOM. Map training uses a training data set of
P
T
training examples, D
T
= {~z
1
,~z
2
,... ,~z
P
T
}. Each
training example is represented by an I-dimensional
vector of attribute values,~z
s
= (z
s1
,z
s2
,. .. ,z
sI
). Every
attribute value is denoted z
sv
and is a real value.
The central map structure of a SOM consists of a
grid of neurons with Y rows and X columns. Each
neuron, denoted n
yx
, is positioned at row y and col-
umn x of the g rid, and has a linked weight vector,
~w
yx
= (w
yx1
,w
yx2
,. .. ,w
yxI
). Every real valued weight,
w
yxv
, corresponds to z
sv
across every~z
s
in D
T
.
The objective of SOM training is to adapt every
~w
yx
in the map stru c ture, so that they collectively
model D
T
. The model is a simp lified repr esentation
because D
T
is I-dimensional, wh ile the ma p is two-
dimensional. The map has two characteristics:
1. The map models the pr obability density function
of the data space represented by D
T
. Weight vec-
tors move towards denser parts of the data space
during train ing. Neurons th us model subsets o f
mutually similar da ta examples after training.
2. Data examples that are close together in the data
space are re presented by neurons that are posi-
tioned near each other in the map grid. The map
model there fore maintains the local topological
structure of th e data space represented by D
T
.
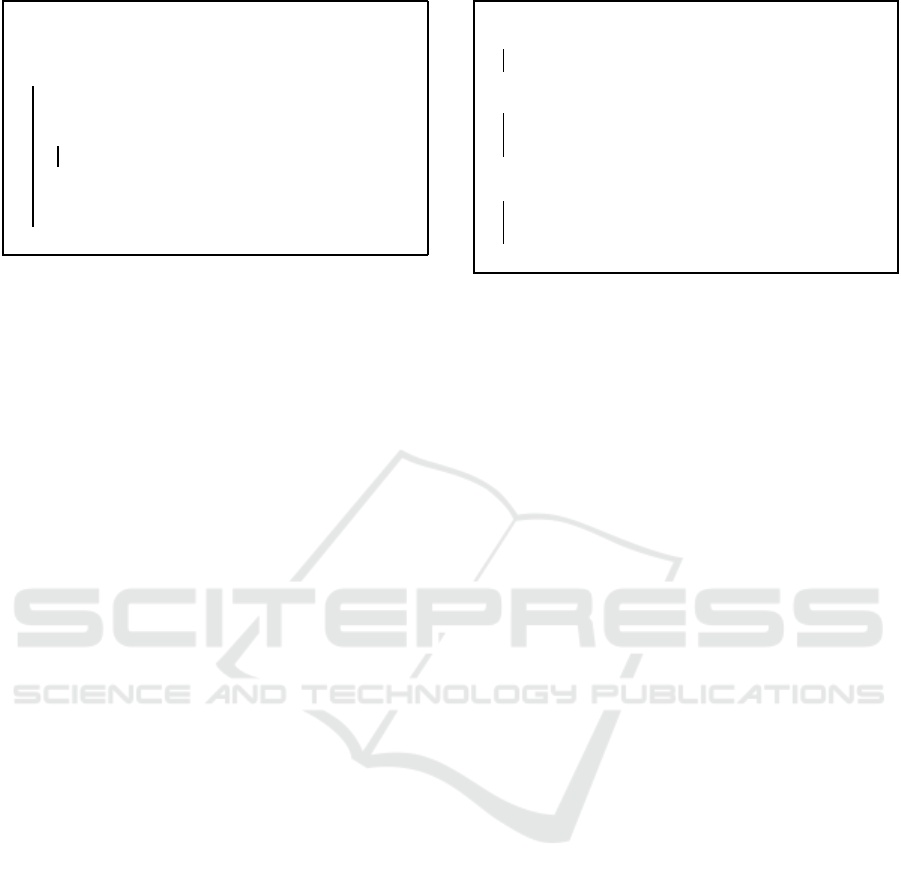
Figure 2 shows the resu lt of SOM training on syn-
thetic two-dimensional data. Gray circles represent
the original uniformly distributed positions of weight
x
x
x
x
x
x
x
Figure 2: SOM weight updates for two-dimensional data.
vectors in the map space, and dashed lines connect
the weig ht vectors of adjacen t neurons. Crosses show
the positions of training examples in the data space.
After training, the weight vectors shift to the loca-
tions of the black circles, where solid lines connect
the weight vectors of adjacent neurons. Characteris-
tic 1 is de monstrated by the drift of weight vectors to
the dense group of training exam ples. Training exam-
ples are also positioned close to the weight vectors of
adjacent neurons, illustrating characteristic 2.
Several SOM training algorithms exist (Engel-
brecht, 2007), which are all usable with the proposed
labeling approach. The experiments of Section 5 used
the original stochastic training procedure (Kohonen,
1982), shown in Algorithm 1, whic h is commonly
used and a good baseline for com parisons.
Stochastic training r epeatedly iterates over each
~z
s
∈ D
T
. The best matching unit (or BMU), denoted
n
ba
, with ~w
ba
closest to~z
s
, is defined as follows:
k~z
s
− ~w
ba
k
2
= min
∀yx
k~z
s
− ~w
yx
k
2
(1)
At the current training iteration, t, every ~w
yx
on the
map is updated relative to th e BMU, as follows:
~w
yx
(t + 1) = ~w
yx
(t) + ∆~w
yx
(t) (2)
The chan ge applied to a weight vecto r is composed of
an update for each w eight, as follows:
∆~w
yx
(t) =
∆w
yx1
(t),∆w
yx2
(t),... ,∆w
yxI
(t)
(3)
Finally, the change computed for a weight is defined
accordin g to the following e quation:
∆w
yxv
(t) = h
ba,yx
(t) ·
z
sv
− w
yxv
(t)
(4)
Here, h
ba,yx
(t) is the neighborhood function at itera-
tion t, which is u sually the smooth Ga ussian kernel:
h
ba,yx
(t) = η(t) · exp
−
kc
ba
− c
yx
k
2
2
2 ·
σ(t)
2
!
(5)
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization
509
initialize map grid
set current training iteration, t = 0
randomly shuffle D
T
repeat
choose unselected~z
s
∈ D
T
use Equation (1) to find BMU, n
ba
, for~z
s
forall ~w
yx
in the map do
update ~w
yx
using Equation (2)
end
update current training iteration, t = t + 1
if all~z
s
∈ D
T
selected, shuffle D
T
until a stopping condition is satisfied
Algorithm 1: Stochastic SOM training procedure.
In this equation, η(t) is the learning rate hyperparam-
eter and σ(t) is the kernel width hyp erparame te r, both
at iteration t, while kc
ba
− c
yx
k
2
is th e distance be-
tween the m a p coo rdinates of n
ba
and n
yx
.
For SOM trainin g to converge, the learning rate
and kernel width must both be monoton ic ally decreas-
ing functions of t. The repo rted experiments used an
exponential decay f unction for the learning rate:
η(t) = η(0) · e
−t/τ
1
(6)
Here, η(0) is the initial learning rate at the start of
training, and τ
1
is a constant governing the decay rate.
Similarly, the kernel width decays as follows:
σ(t) = σ(0) · e
−t/τ
2
(7)
The initial kernel width is σ(0 ), and τ
2
is a constant
affecting the rate of kern el width decay.
3 SOM NEURON LABELING
Neuron labeling attaches textual descriptions
to neurons, to represent neuron characteristics.
Neuron labels are often used during SOM-
based exploratory data analy sis, and are essen-
tial to automated rule extraction from SOMs
(van Heerden and Engelbrech t, 2016). Neuron label-
ing approaches are either supervised or unsupervised
(Azcarrag a et al., 200 8).
Supervised labeling uses classified examples from
a labeling data set (either the training set or separate
data). Algorithms in this category include example-
centric neuron labeling (Kohonen, 1989), example-
centric cluster labeling (Samarasinghe, 2007), and
weight-cen tric neuron labeling (Kohonen, 1989).
Unsupervised labeling is not based on cla ssified
labeling data. In the simplest instance, a human an-
alyst manually a ssigns ne uron labels (Corradini and
Gross, 1999). Algorithmic methods also exist, which
base labe ls either on the weight vectors of the map, or
train a SOM, m ap, with Y × X neurons
forall n
yx
in map do
define empty mapped example set, M
yx
end
forall labeling examples~z
s
do
use Equation (1) to find BMU, n
ba
, for~z
s
add~z
s
to M
ba
end
forall n
yx
in map do
find most common class, A
cls
, within M
yx
label n
yx
with A
cls
end
Algorithm 2: Example-centric neuron labeling.
the labelin g data in relation to the map neuro ns. These
unsuper vised algorithms include unique cluster label-
ing (Deboeck , 1999), unsupervised weight-based la-
beling (Serrano-Cinca, 1996; Lagus and Kaski, 1999;
van Heerden and Engelbrecht, 2012), and unsuper-
vised example -based labeling (Rauber and Merkl,
1999; Azcarraga et al., 2008).
Unsupervised labeling techniques are more flexi-
ble than their supervised c ounterparts. However, un-
supervised labelings are difficult to interpret objec-
tively, posing a problem for their empirical analysis.
As a consequence, most research focuses on super-
vised labeling algorithms, as this paper does.
Example-centric cluster labeling performs poorly,
particularly in the presence of heterogeneo us clusters
of weight vectors (van Heerden, 2017). This research
therefore focuses only on example-centric neuro n la-
beling and weight-centric neuron labeling, w hich are
elaborated upon in Sections 3.1 a nd 3.2.
3.1 Example-Centric Neuron Labeling
Algorithm 2 represents example-centric neuron label-
ing. An initially empty set of mapped labeling exam-
ples is associated with each neuron on the map. Each
labeling example,~z
s
, is then mapped to its BMU us-
ing Equatio n (1), and ~z
s
is added to the labeling ex-
ample set of the BMU. Finally, each neu ron is labeled
using the most common classification appe aring in the
correspo nding labeling example set.
Example-centric neuron labeling leaves neurons
unlabeled when the associated labeling example sets
are empty. Suc h ne urons a re often referred to as inter-
polating units, and represent a large proportion of the
map in some instances (van He e rden, 2023).
Despite these u nlabeled neuro ns, example-centric
neuron la beling has been shown to outperform the
other supervised labeling methods when used as a
basis for simple classification tasks (van Heer den,
2017). It has also been observed that the presence of
NCTA 2024 - 16th International Conference on Neural Computation Theory and Applications
510

VIR SET
VER
VIR
VIR VER
SET SET SET SET SET
VER
VIR
VIR VIR
VIR
VIR
VER
VIR
VER
SET
VIR
VER
VER
SET SET
SET
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VER
VIR
VIR
VIR
VER
VIR
VER
VER
VIR
VER
VIR
VER
VER
VER
SET
VER
VER
VER
SET
SET
VER
VIR
VER
SET
VER
SET
VER
VER
SET
SET
VER
SET
SET
VER
VER
SET
SET
VER
VIR
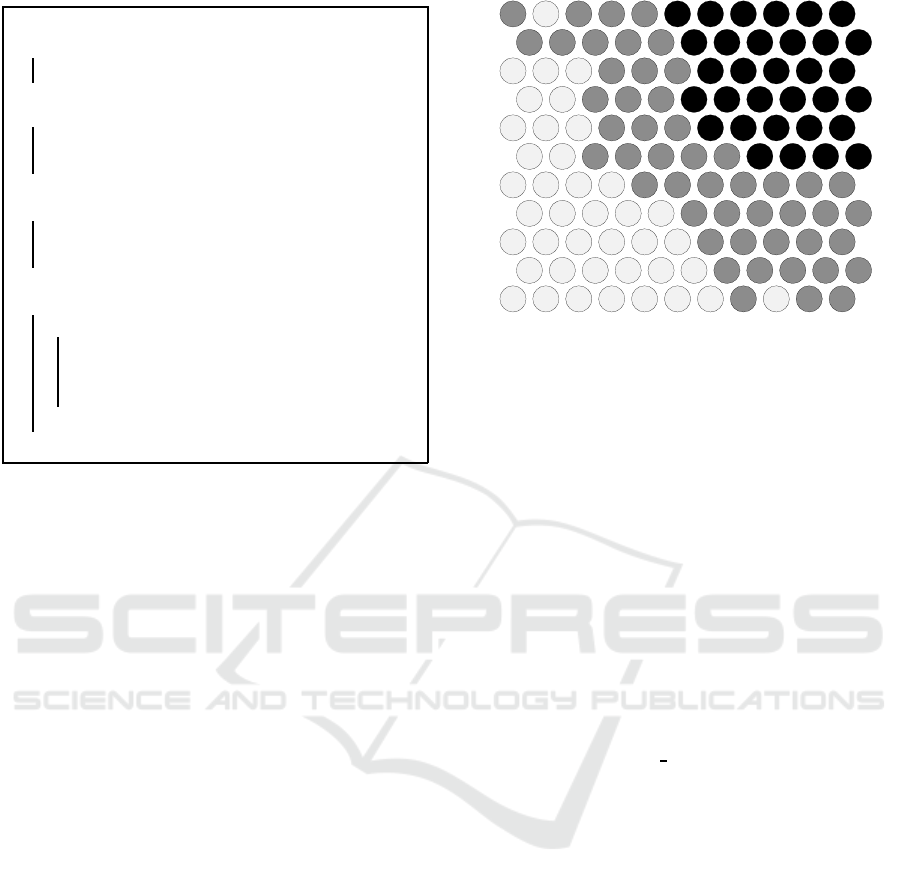
Figure 3: A SOM trained on the I ris data set and labeled
using example-centric neuron labeling.
unlabeled neurons is not strongly correlated to poor
label quality (van Heerden an d Engelbrecht, 2016).
While unlabeled neurons do not negatively
impact the quality of map characterization, th e
missing labels often hamper SOM-based ex-
plorator y data analysis performed by hum an experts
(van Heerden and Engelbrech t, 2016). This is
because the labeled map is broken up by u ncharacter-
ized areas, which obscur e a broader overview of the
model characteristics.
The outcomes of the labeling algorithms un-
der investigation are shown using an example SOM
that was trained on the well-known Ir is data set
(Fisher, 1936). Figure 3 shows the example SOM la-
beled using example-centric neuron labeling. T he la-
bels “SET”, “VER”, and “VIR” respectively represent
the Iris Setosa, Iris Versicolor, and Iris Virginica c la s-
sifications present in the data set. The abundance of
empty circle s in this visualization clearly illustrates
the large numbe r of unlabeled neurons left by the al-
gorithm, which account for 38.8% of the map.
3.2 Weight-Centric Neuron Labeling
Weight-ce ntric neuron labeling is outlined in Algo-
rithm 3. In contrast to example-centric neuron la-
beling, weight-centric neuro n labeling maps neurons
to da ta examples from a labeling set. Eac h neuron’s
weight vecto r is mapped to the closest labeling exam-
train a SOM, map, with Y × X neurons
forall n
yx
in map do
use Equation (8) to fin d BME,~z
e
, for ~w
yx
find class, A
cls
, associated with~z
e
label n
yx
with A
cls
end
Algorithm 3: Weight-centric neuron labeling.
VER VER SET SET
VER
VIR
VIR VER
VIR
VER SET SET SET SET SET
VIR VER
VIR
VIR VIR
VER
VIR
VER
VIR
VIR
VER
VIR
VER
VER
SET
VIR
VER
VER
SETVER SET
VER SET
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VER
VIR
VIR
VER
VIR
VIR
VER
VER
VIR
VER
VIR
VIR
VER
VER
VIR
VIR
VER
VER
VER
VIR
VER
VER
SET
VIR
VER
VER
SET
SET
VER
VER
VER
SET
VER
VER
VER
SET
SET
VER
VER
SET
SET
VER
VER
VER
SET
SET
VER
SET
SET
VER
VER
VER
SET
SET
VER
VER
SET
SET
VER
VER
VER
SET
SET
VER
VER
SET
SET
SET
Figure 4: A SOM trained on the I ris data set and labeled
using weight-centric neuron labeling.
ple,~z
e
, which is called the best m a tc hing example (or
BME), and is defined as follows:
k~w
yx
−~z
e
k
2
= min
∀s
k~w
yx
−~z
s
k
2
(8)
The neuron currently under consideration is then la-
beled with the classification of its BME.
Weight-ce ntric neuron labe ling is simpler than
example-centr ic neur on labeling, and does not require
the stor a ge and searchin g of mapped labeling exam-
ple sets. Additionally, the algor ithm gu a rantees a la-
bel for every neuron on a map, facilitating easier in-
terpretation for human data analysts.
Unfortu nately, neurons will receive poor weight-
centric labels if the BME match is not close. This re-
sults in a tendency for weight-centric neuron labeling
to produce less accurate labels than example-centric
neuron labeling (van Heerden, 2017).
Figure 4 depicts th e result of performing w e ight-
centric neuron labeling on the previously-mentioned
example SOM trained on the Iris data set. It is clear
that every neuron on th e map has received a label,
making the map more interpretable when compared
to the example-centric neuron labeling in Figure 3.
4 THE PROPOSED ALGORITHM
The proposed algorithm is referred to as example-
centric neuro n la beling with weight-centric finaliza-
tion, and hybr idizes example-centric neuron la beling
with weight-centric neuron labeling. The objective is
to generate labels that are accurate, while also guar-
anteeing labels for every ne uron o n a map.
Algorithm 4 represents the proposed technique,
which is a two-phase process. The first phase p er-
forms a norma l example- centric neuron labeling, in
which neurons that are BMUs at least once are la-
beled. This phase will typically produce high-quality
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization
511
train a SOM, map, with Y × X neurons
forall n
yx
in map do
define empty mapped example set, M
yx
end
forall labeling examples~z
s
do
use Equation (1) to fin d BMU, n
ba
, for~z
s
add~z
s
to M
ba
end
forall n
yx
in map do
find most common class, A
cls
, within M
yx
label n
yx
with A
cls
end
forall n
yx
in map do
if n
yx
has no associated label then
use Equation (8) to find BME,~z
e
, for ~w
yx
find class, A
cls
, associated with~z
e
label n
yx
with A
cls
end
end
Algorithm 4: Example-centric neuron labeling with weight-
centric finalization.
labels for o nly a subset of the map neurons. The
second phase then iterates over only the neurons that
have remained unlabeled, assigning a weight-centric
label to each. For e ach of these neurons, the classi-
fication of its BME becomes the neuron label. The
labels pro duced by the second phase will be of lower
quality, but will ensure a complete set of labels.
The labeling for the example Iris data set SOM,
which is p roduced by the pr oposed algorithm, is
shown in Figure 5. When comparing this labeling to
the standard weight-centric neuron labeling shown in
Figure 4, it is clear th at the labelings are very simi-
lar. The maps only differ in terms of two neuron la-
bels: the second from the left on the top row, and the
third from the right on the bottom row. By consulting
Figure 3 it is clear that these differences ar e due to
the example-centric neuron labeling performed dur-
ing the first labeling phase. It is hypothesized that
such differences constitute more accurate labels than
those produced by example-centric neur on labeling.
5 EXPERIMENTAL RESULTS
To explore the hypothesized advantage s of example-
centric neuro n la beling with weight-cen tric finaliza-
tion, a series of experiments were con ducted. These
experiments compared the perfo rmance of the novel
algorithm against ba sic example-centric and weight-
centric neuron labeling, when the algorithms were
used in SOM-based data c la ssification tasks.
VIR VER SET SET
VER
VIR
VIR VER
VIR
VER SET SET SET SET SET
VIR VER
VIR
VIR VIR
VER
VIR
VER
VIR
VIR
VER
VIR
VER
VER
SET
VIR
VER
VER
SETVER SET
VER SET
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VIR
VER
VIR
VIR
VER
VIR
VIR
VER
VER
VIR
VER
VIR
VIR
VER
VER
VIR
VIR
VER
VER
VER
VIR
VER
VER
SET
VIR
VER
VER
SET
SET
VER
VER
VER
SET
VER
VER
VER
SET
SET
VER
VER
SET
SET
VIR
VER
VER
SET
SET
VER
SET
SET
VER
VER
VER
SET
SET
VER
VER
SET
SET
VER
VER
VER
SET
SET
VER
VER
SET
SET
SET
Figure 5: A SOM trained on the I ris data set and labeled
using the proposed algorithm.
The standard stochastic tra ining algorithm pro-
duced SOMs for several b enchmar k data sets. Six
data sets were drawn f rom the UCI Machine Learning
Repository, namely the Iris data set (Fisher, 1936), the
ionosphere data set (Sigillito et al., 19 89), the th ree
monk’s problems data sets (Wnek, 1993), and the
glass identification data set (Ge rman, 1987). It should
be noted that the third monk’s problem set has 5%
classification noise added to the training data. The
data sets were preprocessed by using one-hot encod-
ing for nominal attributes (Bishop, 2006), and scaling
all attribute values to a [0.0, 1.0] range using min-max
normalization (Han et al., 2012).
The classification task was executed using the au-
thor’s open-source SOM an d labeling algorithm col-
lection (van Heerden, 2024), which is ba sed on Ko-
honen ’s original SOM
PAK r e ference implementa-
tion (Koho nen et al., 1996). Square maps were used
throughout, and all weight vectors we re initialized
using hyper cube initialization (Su et al., 19 99). The
SOM’s main stopping co ndition was a limit of 0.0001
on a 30-iteration moving average of the standard de-
viation compu te d fo r the map’s quantization error
(van Heerden, 2017). To ensure termination, training
was also stopped after 100 000 iterations.
Following map training, the appropriate neuron
labeling algorithm was applied. Fina lly, classifica-
tions wer e performed by mapping a data example to
its BMU. The label of the BMU was then applied as
the classification of the da ta example. Each classifi-
cation was compared to the actual classes of the data
example, and a misclassification occurred if a match
was not found. If the BMU was un la beled, the data
example re mained unclassified.
Hyperpa rameter tuning was pe rformed separately
for the three n e uron labeling algorithms a pplied to
each of the benchmark data sets. The tuning proce-
dure (van Heer den, 2017; Franken, 2009) used Sobol’
NCTA 2024 - 16th International Conference on Neural Computation Theory and Applications
512
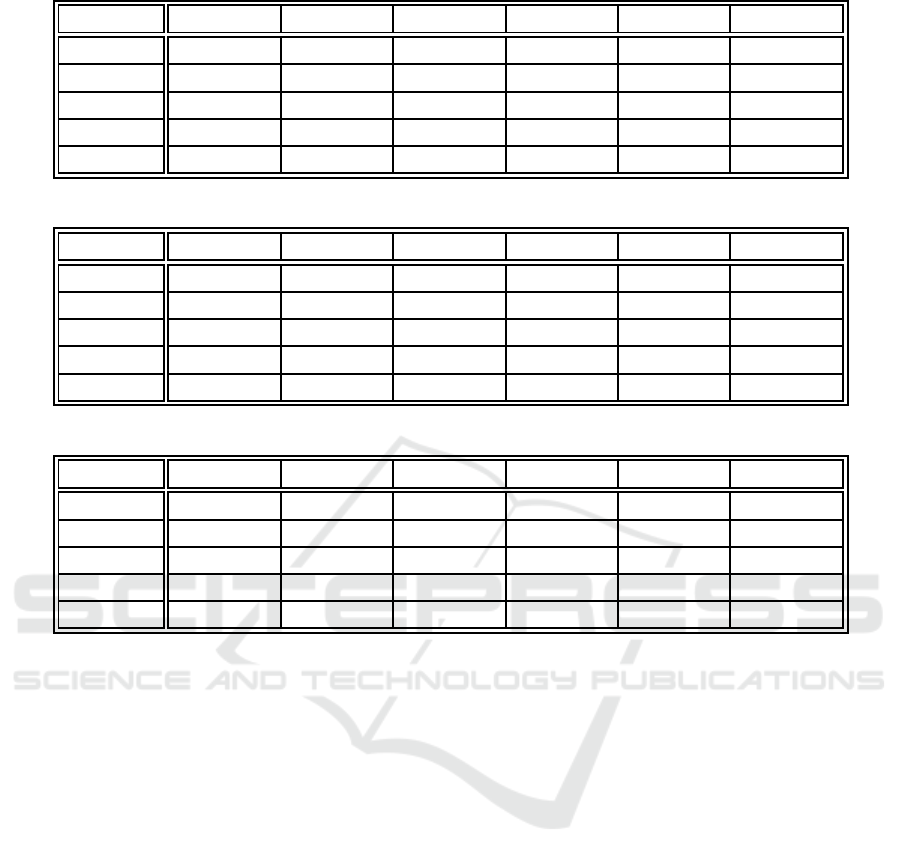
Table 1: Optimal hyperparameters per data set for example-centric neuron labeling.
Parameter Iris Ionosphere Monks 1 Monks 2 Monks 3 Glass
Y and X 5 7 14 15 11 6
η(0) 5.488 3.848 6.055 8.867 9.941 0.449
τ
1
1 432.617 1 209.961 849.609 1 365.234 577.148 430.664
σ(0) 2.119 1.818 10.445 1.348 3.029 1.948
τ
2
77.539 59.570 9.766 31.641 83.008 7.617
Table 2: Optimal hyperparameters per data set for weight-centric neuron labeling.
Parameter Iris Ionosphere Monks 1 Monks 2 Monks 3 Glass
Y and X 12 17 14 15 19 11
η(0) 4.609 0.918 6.563 8.867 9.629 0.508
τ
1
574.219 594.727 46.875 1 365.234 61.523 1 236.328
σ(0) 10.781 14.045 4.813 1.348 4.639 1.904
τ
2
96.094 87.305 34.375 31.641 67.383 5.859
Table 3: Optimal hyperparameters per data set for example-centric neuron labeling with weight-centric finalization.
Parameter Iris Ionosphere Monks 1 Monks 2 Monks 3 Glass
Y and X 11 15 15 16 14 13
η(0) 7.266 8.945 8.867 2.480 1.836 7.910
τ
1
363.281 908.203 1 365.234 1 391. 602 1 060.547 1 163.08 6
σ(0) 6.273 1.934 1.348 0.719 5.414 1.082
τ
2
16.406 58.984 31.641 2.930 55.078 75.195
sequences (Sobol’ , 1967) to generate configurations
that sample the parameter space uniformly. Ta-
bles 1 to 3 present the optimal hyperparameters for
example-centr ic neuron labeling, weig ht-centric neu-
ron labeling, and examp le-centric neuron labeling
with weight-centric finalization, respectively.
For each algorithm and data set, a 30-fold cross-
validation was performed, and perfor mance measures
were recorded. To test whether performance mea-
sure differences wer e statistically significant, two-
tailed n on-parametric Wilcoxon signe d-rank hypoth-
esis tests (Wilcoxon, 1945) were performed with a
confidence level of 0.05. A Bonferroni correction
(Miller, 1981) was applied, to counteract the multi-
ple comparisons problem. The results tables tha t fol-
low report the mean and standa rd deviation for each
performance measure, as well as a hypothesis test p-
value. When a p-value indicates a significant p e rfor-
mance differenc e , the mean and standar d deviation of
the better-performing algor ithm are marked in bold.
Tables 4 and 5 respectively compare the overall
training set e rror perfor mance of the proposed ap-
proach against example-centric neuron labeling and
weight-cen tric neuron labeling. These errors could
be due to a combination of misclassified and unclas-
sified training set data examples. The tables clearly
illustrate that the proposed algorithm o utperforms
example-centr ic neuron labeling in all instances, and
weight-cen tric neuron labeling in all but one case
(for the third monk’s problem, which included train-
ing data noise, weight-centric neuron labelin g outper-
formed the proposed algorithm). The proposed ap-
proach generally did not outperform example-centr ic
neuron labeling by a very large margin, except in the
case of the ionosphere and glass identification data
sets. In contrast, weight-centric neuron labeling un-
derperformed by a substantial amount in the cases of
the ionosphere, glass identification, an d the first two
monk’s pro blems data sets.
To gain a deeper insight into whether any type of
classification error was more prevalent, Tables 6 and 7
summarize the training set errors due only to misclas-
sified data examples, when comparing the proposed
algorithm to exam ple-centric and weight-centric neu-
ron labelin g, respectively. The performance differ-
ences wer e exactly th e same as those observed for the
overall training set error, ind ic a ting that all classifica-
tion errors wer e due to incorrect c lassifications.
Tables 8 and 9 respectively present the differences
in performance when comparing the novel algorithm
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization
513
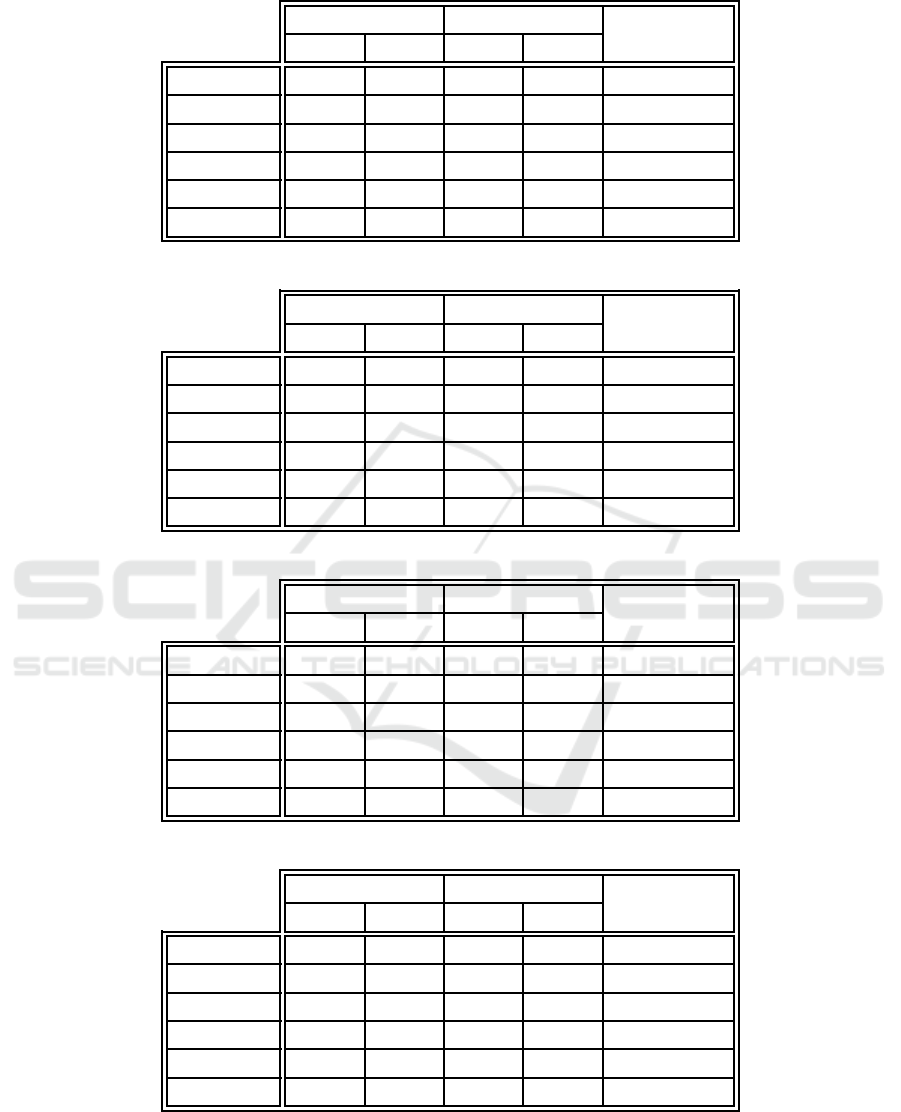
Table 4: Overall training error comparison between the proposed algorithm and example-centric neuron l abeling.
EC-WC EC
p-value
E
T
S
T
E
T
S
T
Iris 1.724 0.594 3 .655 0.705 3.725 × 10
−9
Ionosphere 3.647 0.845 10.137 1.221 1.863 × 10
−9
Monks 1 17.049 1.108 18.126 1.084 0.001
Monks 2 15.056 0.854 15.766 0.657 0.001
Monks 3 22.225 1.224 23.828 1.792 0.001
Glass 11.449 1.518 26.28 1.531 1.863 × 10
−9
Table 5: Overall training error comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
T
S
T
E
T
S
T
Iris 1.724 0.594 2 .575 0.700 2.265 × 10
−6
Ionosphere 3.647 0.845 15.216 1.62 6 1.863 × 10
−9
Monks 1 17.049 1.108 27.974 2.416 1.863 × 10
−9
Monks 2 15.056 0.854 23.947 2.693 1.863 × 10
−9
Monks 3 22.225 1.224 19.769 1.873 5.307 × 10
−6
Glass 11.449 1.518 27.778 2.720 1.863 × 10
−9
Table 6: Training misclassified error comparison between the proposed algorithm and example-centric neuron labeling.
EC-WC EC
p-value
E
TM
S
TM
E
TM
S
TM
Iris 1.724 0.594 3 .655 0.705 3.725 × 10
−9
Ionosphere 3.647 0.845 10.137 1.22 1 1.863 × 10
−9
Monks 1 17.049 1.108 18.126 1.084 0.001
Monks 2 15.056 0.854 15.766 0.657 0.001
Monks 3 22.225 1.224 23.828 1.792 0.001
Glass 11.449 1.518 26.28 1.531 1.863 × 10
−9
Table 7: Training mi sclassified error comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
TM
S
TM
E
TM
S
TM
Iris 1.724 0.594 2 .575 0.700 2.265 × 10
−6
Ionosphere 3.647 0.845 15.216 1.62 6 1.863 × 10
−9
Monks 1 17.049 1.108 27.974 2.416 1.863 × 10
−9
Monks 2 15.056 0.854 23.947 2.693 1.863 × 10
−9
Monks 3 22.225 1.224 19.769 1.873 5.307 × 10
−6
Glass 11.449 1.518 27.778 2.720 1.863 × 10
−9
NCTA 2024 - 16th International Conference on Neural Computation Theory and Applications
514
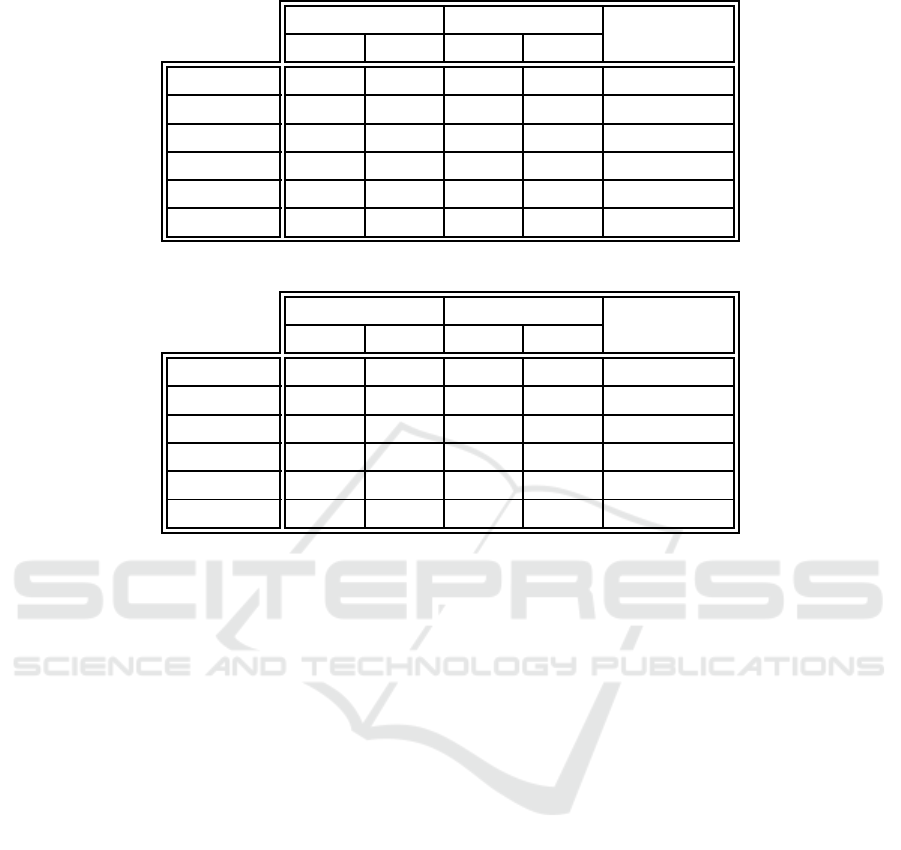
Table 8: Training unclassified error comparison between the proposed algorithm and example-centric neuron labeling.
EC-WC EC
p-value
E
TU
S
TU
E
TU
S
TU
Iris 0.000 0.000 0 .000 0.000 N/A
Ionosphere 0.000 0.000 0.000 0.000 N /A
Monks 1 0.000 0.000 0 .000 0.000 N/A
Monks 2 0.000 0.000 0 .000 0.000 N/A
Monks 3 0.000 0.000 0 .000 0.000 N/A
Glass 0.000 0.000 0.000 0.000 N/A
Table 9: Training unclassified error comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
TU
S
TU
E
TU
S
TU
Iris 0.000 0.000 0 .000 0.000 N/A
Ionosphere 0.000 0.000 0.000 0.000 N /A
Monks 1 0.000 0.000 0 .000 0.000 N/A
Monks 2 0.000 0.000 0 .000 0.000 N/A
Monks 3 0.000 0.000 0 .000 0.000 N/A
Glass 0.000 0.000 0.000 0.000 N/A
to the standard example-centric and weigh t-centric
methods, in ter ms of the training set error due only to
unclassified data examp les. Here, it is observable that
no training examples were left unclassified by any of
the algorithms. All the labeling a pproaches therefore
fully modeled the u nderlying training data.
In terms of algorithm analysis, training set per-
formance is not a realistic representation of the real-
world behavior that can be expected from an algo-
rithm. As a result, the next three sets of comparisons
focus o n test set e rror, computed from data examples
that were not presented during SOM training.
The differences in overall test set classification
performance, when contrasting the new algorithm
and the basic example-centric and weight-centric la-
beling techniques, are illustrated in Tables 10 and
11. No statistically significant differences in test er-
ror performance were observed when comparing the
proposed method to example-cen tric neuron label-
ing. This implies that example-centric neuron la-
beling with weight-centric finalization classifies ex-
amples as well as example-centric labeling alone.
This is expe c te d, because the first phase of the hy-
brid technique is based on example-c entric labeling.
The weig ht-centric finalization thus does not inter-
fere with classification accuracy. Weight-centric la-
beling performed significantly worse than the novel
algorithm by large margins in half of the data sets
(the ionosphere and first two mon k’s proble ms). In
the other three data sets, no significant performance
difference was obser ved. The example-centric basis
of the hybrid method is responsible for this behavior.
Tables 12 and 13 juxtapose the test set error due
to only misclassifications. While very small perfor-
mance differences were observable when comparing
the novel approach to example-centric labeling, none
were statistically significant. The compa rison against
weight-cen tric labeling prod uced very similar results
to those observed for th e overall test error.
The test set e rror due to unclassified examples is
compare d in Tables 14 and 15. No statistically sig-
nificant differences were observed when comparing
the new algorithm against either example-centric or
weight-cen tric neuron labeling. This supports the ob-
servation that classification errors were due mostly to
misclassifications, w hich was made when considering
the training set error performance.
Finally, Tables 16 and 17 compare the percentage
of neuron s th a t were le ft unlabeled by th e algorithms.
The example-centric method produced significantly
more unlabeled neu rons than the proposed algorithm.
The observed d ifferences were generally large, with
example-centr ic neuron labeling leaving ne arly half
the neurons for the first two monk’s prob le ms u nchar-
acterized. The combined approach and weight-ce ntric
labeling of course both lef t no neurons unlabeled.
When viewed holistically, example-centric label-
ing with weight-centric finalization classified data ex-
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization
515
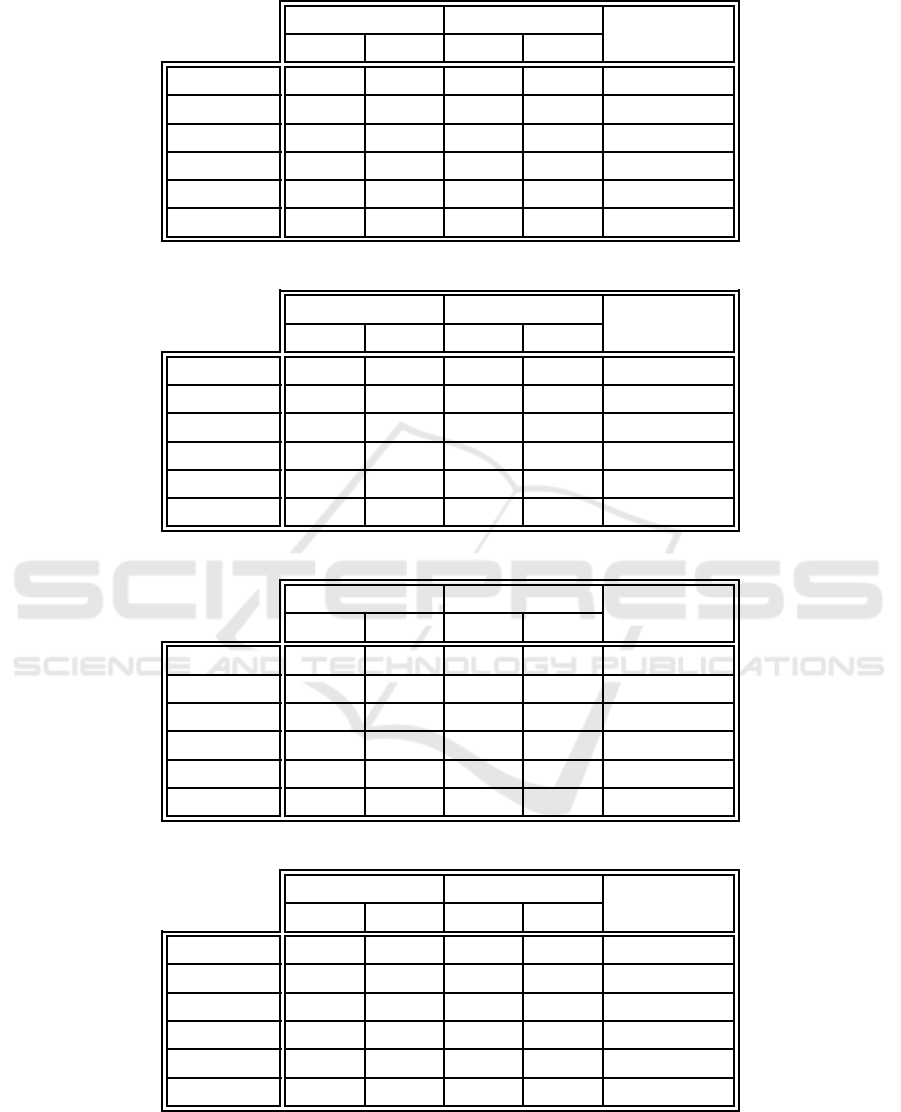
Table 10: Overall test error comparison between the proposed algorithm and example-centric neuron labeling.
EC-WC EC
p-value
E
G
S
G
E
G
S
G
Iris 2.667 6.915 4 .000 8.137 0.688
Ionosphere 8.788 9.086 11.515 10.9 23 0.124
Monks 1 19.286 12.178 20.00 0 12.358 0.532
Monks 2 19.762 10.559 20.23 8 10.116 0.866
Monks 3 26.190 9.807 26.429 12.178 0.959
Glass 25.714 14.236 31.905 18 .639 0. 187
Table 11: Overall test error comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
G
S
G
E
G
S
G
Iris 2.667 6.915 3 .333 7.581 1.000
Ionosphere 8.788 9.086 15.152 12.0 16 0.002
Monks 1 19.286 12.178 31.66 7 11.817 2.365 × 10
−4
Monks 2 19.762 10.559 29.76 2 9.395 3.052 × 10
−5
Monks 3 26.190 9.807 27.619 12.118 0.842
Glass 25.714 14.236 33.333 17 .729 0. 024
Table 12: Test misclassified error comparison between the proposed algorithm and example-centric neuron labeling.
EC-WC EC
p-value
E
GM
S
GM
E
GM
S
GM
Iris 2.667 6.915 3 .333 7.581 1.000
Ionosphere 8.788 9.086 11.212 10.3 19 0.191
Monks 1 19.286 12.178 18.81 0 12.226 0.965
Monks 2 19.762 10.559 19.52 4 9.177 0.971
Monks 3 26.190 9.807 25.952 12.082 0.734
Glass 25.714 14.236 30.476 19 .031 0. 314
Table 13: Test misclassified error comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
GM
S
GM
E
GM
S
GM
Iris 2.667 6.915 3 .333 7.581 1.000
Ionosphere 8.788 9.086 15.152 12.0 16 0.002
Monks 1 19.286 12.178 31.66 7 11.817 2.365 × 10
−4
Monks 2 19.762 10.559 29.76 2 9.395 3.052 × 10
−5
Monks 3 26.190 9.807 27.619 12.118 0.842
Glass 25.714 14.236 33.333 17 .729 0. 024
NCTA 2024 - 16th International Conference on Neural Computation Theory and Applications
516
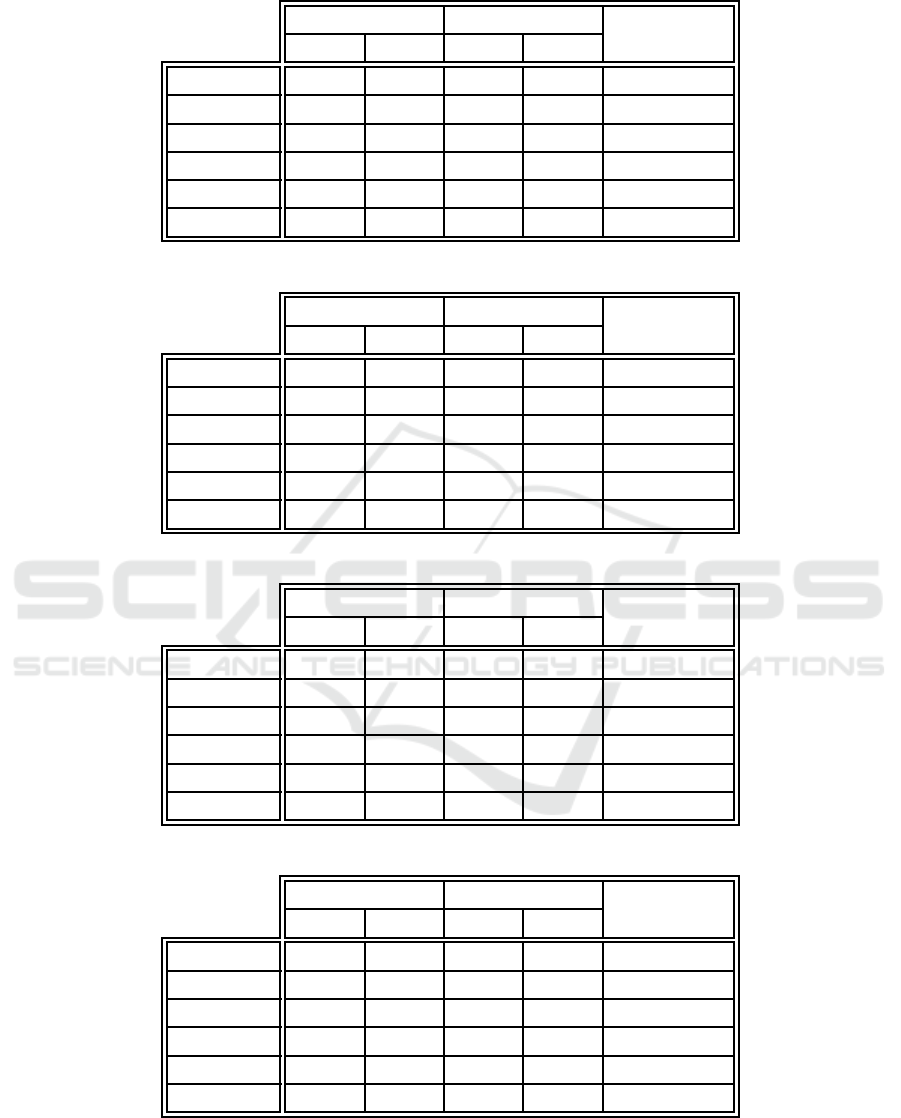
Table 14: Test unclassified error comparison between the proposed algorithm and example-centric neuron labeling.
EC-WC EC
p-value
E
GU
S
GU
E
GU
S
GU
Iris 0.000 0.000 0 .667 3.651 1.000
Ionosphere 0.000 0.000 0.303 1.660 1.000
Monks 1 0.000 0.000 1 .190 3.294 0.125
Monks 2 0.000 0.000 0 .714 2.179 0.250
Monks 3 0.000 0.000 0 .476 1.812 0.500
Glass 0.000 0.000 1.429 4.359 0.250
Table 15: Test unclassified error comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
GU
S
GU
E
GU
S
GU
Iris 0.000 0.000 0 .000 0.000 N/A
Ionosphere 0.000 0.000 0.000 0.000 N /A
Monks 1 0.000 0.000 0 .000 0.000 N/A
Monks 2 0.000 0.000 0 .000 0.000 N/A
Monks 3 0.000 0.000 0 .000 0.000 N/A
Glass 0.000 0.000 0.000 0.000 N/A
Table 16: Unlabeled neuron percentage comparison between the proposed algorithm and example-centric neuron labeling.
EC-WC EC
p-value
E
U
S
U
E
U
S
U
Iris 0.000 0.000 19.467 3. 104 1.863 × 10
−9
Ionosphere 0.000 0.000 3.810 1.912 3.7 25 × 1 0
−9
Monks 1 0.000 0.000 43.469 3. 065 1.863 × 10
−9
Monks 2 0.000 0.000 49.807 1. 885 1.863 × 10
−9
Monks 3 0.000 0.000 32.700 4. 967 1.863 × 10
−9
Glass 0.000 0.000 8.241 3.961 7.45 1 × 10
−9
Table 17: Unlabeled neuron percentage comparison between the proposed algorithm and weight-centric neuron labeling.
EC-WC WC
p-value
E
U
S
U
E
U
S
U
Iris 0.000 0.000 0 .000 0.000 N/A
Ionosphere 0.000 0.000 0.000 0.000 N /A
Monks 1 0.000 0.000 0 .000 0.000 N/A
Monks 2 0.000 0.000 0 .000 0.000 N/A
Monks 3 0.000 0.000 0 .000 0.000 N/A
Glass 0.000 0.000 0.000 0.000 N/A
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization
517

amples no worse than basic example-centric label-
ing, a nd had better classification performance than
normal weight-centric labeling. At the same time,
example-centr ic labeling with weight-centric finaliza-
tion clearly maintained a high er perc entage of labeled
neurons than example-centric neuron labeling.
The proposed algorithm underperformed in terms
of training misclassification error on the single data
set with injec te d noise (the third monk’s problem),
when compare d to weight-ce ntric neuron labeling.
This type of performance shortfall is, however, not
clear when considering test e rror. A more thorough
investigation into the performance of the algorithms
in the presence of noise is deferred to future studies.
6 CONCLUSIONS
This paper introduces a novel supervised neuron la-
beling algorithm that succe ssfully comb ines the ad-
vantages of both example-centric and weight-centric
neuron labeling. If only label q uality is imp ortant, the
proposed technique is largely equivalent to example-
centric labeling , with the latter bein g preferable due
to a less complex algorithm. However, if humans are
to analyze a SOM’s labels, the proposed hybrid ap-
proach is preferable becau se it maintains a fully la-
beled map wh ile offering high-q uality labels.
Future work will focus on a lternative ways in
which high-quality complete SOM labelings can be
achieved. Three approaches are under consideration:
• An approa ch (Li and Eastman, 2006) that has
been briefly proposed, but not experime ntally ex-
plored, characterizes each unlabeled neuron us-
ing the mean distance between the weight vector
of the unlabeled neuron and the weight vectors
of neuron groups tha t are labeled with the sam e
class. The unlabeled neuron r eceives the label of
the class with the smallest mean distance.
• The propagation of labels from characterized neu-
rons to adjacent uncharacterized neurons will be
investigated. A possible basis for this is neuron
proxim ity graphs (Herrmann and Ultsch, 2007).
• Sem i-supervised SOMs learn the distribution of
classification attributes ac ross the map, without
biasing training (Kiviluoto and Bergius, 1997).
Labels will be based on the learned cla ssification
attributes for each neuron.
Finally, the general perform ance characteristics of the
supervised labeling algorithms will be investigated in
greater d e pth. T he scalability of supervised labeling
algorithm s to very large datasets and maps will be of
interest. It will also be informative to analyze the per-
formance characteristics of the algorithms in the pres-
ence of classification and attribute noise.
REFERENCES
Azcarraga, A., Hsieh, M.-H., Pan, S.-L., and Setiono, R.
(2008). Improved SOM labeling methodology for
data mining applications. In Soft Computing for
Knowledge Discovery and Data Mining, pages 45–75.
Springer. doi:10.1007/978-0-387-69935-6
3.
Bishop, C. M. (2006). Pattern Recognition and Machine
Learning. Springer.
Bowen, F. and Siegler, J. (2024). Self-organizing maps:
A novel approach to identify and map business clus-
ters. Journal of Management Analytics, 11(2):228–
246. doi:10.1080/23270012.2024.2306628.
Corradini, A. and Gross, H .-M. (1999). A hy-
brid stochastic-connectionist architecture for gesture
recognition. In Proceedings of ICIIS, pages 336–341.
doi:10.1109/ICIIS.1999.810286.
Deboeck, G. (1999). Public domain vs. commercial tools
for creating neural self-organizing maps. PC AI,
13(1):27–33.
Engelbrecht, A. P. (2007). Computational Intelli-
gence: An Introduction. Wiley, 2nd edition.
doi:10.1002/9780470512517.
Fisher, R. A. (1936). Iris data set. UCI Machine Learning
Repository. doi:10.24432/C56C76.
Franken, N. ( 2009). Visual exploration of algorithm param-
eter space. In Proceedings of CEC, pages 389–398.
doi:10.1109/CEC.2009.4982973.
Galvan, D., Effting, L., Cremasco, H., and Conte-Junior,
C. A. (2021). The spread of the COVID-19 out-
break in Brazil: An overview by Kohonen Self-
Organizing Map networks. Medicina, 57(3):1–19.
doi:10.3390/medicina57030235.
German, B. (1987). Glass identification data set. UCI Ma-
chine Learning Repository. doi:10.24432/C5WW2P.
Han, J., Kamber, M., and Pei, J. (2012). Data Mining: Con-
cepts and Techniques. Morgan Kaufmann, 3rd edition.
doi:10.1016/C2009-0-61819-5.
Herrmann, L. and Ultsch, A. (2007). Label prop-
agation for semi-supervised learning in Self-
Organizing Maps. In Proceedings of WSOM.
doi:10.2390/biecoll-wsom2007-113.
Javed, A., Rizzo, D. M., Lee, B. S., and Gramling, R.
(2024). S O MTimeS: S elf organizing maps for time
series clustering and its application to serious illness
conversations. Data Mining and Knowledge Discov-
ery, 38:813–839. doi:10.1007/s10618-023-00979-9.
Kaski, S., Kangas, J., and Kohonen, T. (1998). B ibliography
of Self-Organizing Map (SOM) papers: 1981–1997.
Neural Computing Surveys, 1:102–350.
Kiviluoto, K. and Bergius, P. (1997). Analyzing financial
statements with the Self-Organizing Map. In Proceed-
ings of WSOM, pages 362–367.
NCTA 2024 - 16th International Conference on Neural Computation Theory and Applications
518

Kohonen, T. (1982). Self-organizing formation of topolog-
ically correct feature maps. Biological Cybernetics,
43(1):59–69. doi:10.1007/BF00337288.
Kohonen, T. (1989). Self-Organization and As-
sociative Memory. Springer, 3rd edition.
doi:10.1007/978-3-642-88163-3.
Kohonen, T. (2001). Self-Organizing Maps. Springer-
Verlag, 3rd edition. doi:10.1007/978-3-642-56927-2.
Kohonen, T., Hynninen, J., Kangas, J., and Laaksonen, J.
(1996). SOM
PAK: The Self-Organizing Map pro-
gram package. Technical Report A31, Helsinki Uni-
versity of Technology.
Lagus, K. and Kaski, S. (1999). Keyword selection
method for characterizing text document maps. In
Proceedings of ICANN, volume 1, pages 371–376.
doi:10.1049/cp:19991137.
Li, Z. and Eastman, J. R. (2006). The nature and clas-
sification of unlabelled neurons in t he use of Ko-
honen’s Self-Organizing Map for supervised clas-
sification. Transactions in GIS, 10(4):599–613.
doi:10.1111/j.1467-9671.2006.01014.x.
Miller, Jr, R. G. (1981). Simultaneous Statisti-
cal Inference. Springer-Verlag, 2nd edition.
doi:10.1007/978-1-4613-8122-8.
Oja, M., Kaski, S., and Kohonen, T. (2003). Bibliography
of Self-Organizing Map (SOM) papers: 1998–2001
addendum. Neural Computing Surveys, 3:1–156.
P¨oll¨a, M., Honkela, T., and Kohonen, T. (2009). Bibliog-
raphy of Self-Organizing Map (SOM) papers: 2002-
2005 addendum. Technical Report T KK -ICS-R23,
Helsinki University of Technology.
Rauber, A. and Merkl, D. (1999). The SOMLib digital li-
brary system. In Proceedings of ECDL, pages 323–
342. doi:10.1007/3-540-48155-9
21.
Rosa, A. H., Stubbings, W. A., Akinrinade, O. E., Gon-
tijo, E. S. J., and Harrad, S. (2024). Neural net-
work for evaluation of the impact of the UK COVID-
19 national lockdown on atmospheric concentrations
of PAHs and PBDEs. Environmental Pollution,
341:122794. doi:10.1016/j.envpol.2023.122794.
Samarasinghe, S. (2007). Neural Networks for Applied
Sciences and Engineering: From Fundamentals to
Complex Pattern Recognition. Auerbach Publica-
tions, B oca Raton, Florida, United States of America.
doi:10.1201/9780849333750.
Serrano-Cinca, C. (1996). Self organizing neural networks
for financial diagnosis. Decision Support Systems,
17(3):227–238. doi:10.1016/0167-9236(95)00033-X.
Sigillito, V., Wing, S., H utt on, L ., and Baker, K. (1989).
Ionosphere data set. UCI Machine Learning Reposi-
tory. doi:10.24432/C5W01B.
Sobol’, I. M. (1967). On the distribution of points
in a cube and the approximate evaluation of
integrals. USSR Computational Mathemat-
ics and Mathematical Physics, 7(4):86–112.
doi:10.1016/0041-5553(67)90144-9.
Su, M.-C., Liu, T.-K., and Chang, H.-T. (1999). An efficient
initialization scheme for the self-organizing feature
map algorithm. In Proceedings of IJCNN, volume 3,
pages 1906–1910. doi:10.1109/IJCNN.1999.832672.
van H eerden, W. S. (2017). Self-organizing feature maps for
exploratory data analysis and data mining: A practical
perspective. Master’s thesis, University of Pretoria.
van Heerden, W. S. (2023). Automatic distance-based inter-
polating unit detection and pruning in self-organizing
maps. In Proceedings of SSCI, pages 1298–1303.
doi:10.1109/SSC I52147.2023.10372025.
van Heerden, W. S. (2024). SOMLib.
https://github.com/wvheerden/SOMLib.
van Heerden, W. S. and Engelbrecht, A. P. (2008). A
comparison of map neuron labeling approaches
for unsupervised self-organizing feature maps.
In Proceedings of IJCNN, pages 2139–2146.
doi:10.1109/IJCNN.2008.4634092.
van Heerden, W. S. and Engelbrecht, A. P. (2012). Unsuper-
vised weight-based cluster labeling for self-organizing
maps. In Proceedings of WSOM, pages 45–54.
doi:10.1007/978-3-642-35230-0
5.
van Heerden, W. S. and Engelbrecht, A. P. (2016). An
investigation into the effect of unlabeled neurons
on Self-Organizing Maps. In Proceedings of SSCI.
doi:10.1109/SSC I.2016.7849938.
Vantyghem, A. N., Galvin, T. J., Sebastian, B., O’Dea,
C., Gordon, Y. A., Boyce, M., Rudnick, L., Pol-
sterer, K., Andernach, H., Dionyssiou, M., Venkatara-
man, P., Norris, R., Baum, S., Wang, X. R., and
Huynh, M. (2024). Rotation and flipping invari-
ant self-organizing maps wi th astronomical images:
A cookbook and application to the VLA Sky Sur-
vey QuickLook images. Astronomy and Computing,
47:100824. doi:10.1016/j.ascom.2024.100824.
Wilcoxon, F. (1945). Individual comparisons by rank-
ing methods. Biometrics Bulletin, 1(6):80–83.
doi:10.2307/3001968.
Wnek, J. (1993). Monk’s problems data sets. UCI Machine
Learning Repository. doi:10.24432/C5R30R.
Neuron Labeling for Self-Organizing Maps Using a Novel Example-Centric Algorithm with Weight-Centric Finalization
519
