
Beyond Equality Matching: Custom Loss Functions for Semantics-Aware
ICD-10 Coding
Monah Bou Hatoum
1
, Jean Claude Charr
1
, Alia Ghaddar
2,3
, Christophe Guyeux
1
and David Laiymani
1
1
FEMTO-ST Institute, UMR 6174 CNRS, University of Franche-Comt
´
e, 90000 Belfort, France
2
Department of Computer Science, the International University of Beirut, Beirut P.O. Box 146404, Lebanon
3
Department of Computer Science, Lebanese International University, Beirut, Lebanon
{monah.bou
hatoum, jeanclaude.charr, christophe
˙
guyeux, davi
Keywords:
Deep Learning, Relevancy Comparison, Hierarchical Relationships, Semantic Similarity, Custom Loss
Function, Icd-10 Coding, Cosine Similarity, Medical Coding Automation, Machine Learning In Healthcare.
Abstract:
Background: Accurate ICD-10 coding is vital for healthcare operations, yet manual processes are ineffi-
cient and error-prone. Machine learning offers automation potential but struggles with complex relationships
between codes and clinical text. Objective: We propose a semantics-aware approach using custom loss func-
tions to improve accuracy and clinical relevance in multi-label ICD-10 coding by leveraging cosine similarity
to measure semantic relatedness between predicted and actual codes. Methods: Four custom loss functions
(True Label Cardinality Loss (TLCL), Predicted Label Cardinality Loss (PLCL), Balanced Harmonic Mean
Loss (BHML), and Weighted Harmonic Mean Loss (WHML)) were designed to capture hierarchical and se-
mantic relationships. These were validated on a dataset of 9.57 million clinical notes from 24 medical spe-
cialties, using binary cross-entropy (BCE) loss as a baseline. Results: Our approach achieved a test micro-F1
score of 88.54%, surpassing the 74.64% baseline, with faster convergence and improved performance across
specialties. Conclusion: Incorporating semantic similarity into the loss functions enhances ICD-10 code pre-
diction, addressing clinical nuances and advancing machine learning in medical coding.
1 INTRODUCTION
The International Classification of Diseases (ICD) is a
global standard for categorizing diseases, symptoms,
and medical procedures, critical for healthcare opera-
tions such as billing, quality control, and clinical re-
search (Otero Varela et al., 2021). Manual ICD-10
coding is inefficient, error-prone, and requires spe-
cialized knowledge (Mou et al., 2023; Zhou et al.,
2020), driving the adoption of machine learning to
automate this process (Esteva et al., 2019). However,
existing models struggle with the complexity and am-
biguity of medical data (Nayyar et al., 2021).
A significant limitation of current models is their
reliance on strict equality matching, penalizing pre-
dictions that deviate from exact matches (del Barrio
et al., 2020; Long, 2021; Mittelstadt et al., 2023).
This approach overlooks the clinical equivalence of
certain codes (e.g., Z01.8, Z01.9, Z48.8) and fails to
address hierarchical relationships in ICD-10, which
are vital for accurate representation. Conversely,
some codes (e.g., P74.31, P74.32) require strict speci-
ficity due to their distinct clinical implications (Ha-
toum et al., 2023). The ambiguity in clinical docu-
mentation further complicates this, as similar phras-
ing can correspond to different codes (Yu et al., 2023).
To overcome these challenges, we propose a
relevancy-based approach leveraging vector represen-
tations of ICD-10 codes and cosine similarity to mea-
sure semantic relatedness. This method assigns par-
tial credit for clinically valid predictions, enabling the
model to handle nuanced relationships between codes
effectively.
Our approach employs the Adam optimizer to ad-
dress sparse gradients and class imbalance in large-
scale datasets. We introduce four custom loss func-
tions: True Label Cardinality Loss (TLCL), Predicted
Label Cardinality Loss (PLCL), Balanced Harmonic
Mean Loss (BHML), and Weighted Harmonic Mean
Loss (WHML). These optimize both accuracy and
clinical relevance by capturing hierarchical and se-
mantic relationships while minimizing penalties for
166
Bou Hatoum, M., Charr, J. C., Ghaddar, A., Guyeux, C. and Laiymani, D.
Beyond Equality Matching: Custom Loss Functions for Semantics-Aware ICD-10 Coding.
DOI: 10.5220/0013101000003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 166-174
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

clinically acceptable predictions.
Validated on a dataset of 9.57M clinical notes
spanning 24 specialties, our method demonstrated
significant improvements in micro-F1 scores, outper-
forming traditional binary cross-entropy loss. By en-
hancing automated ICD-10 coding, this approach has
the potential to improve healthcare efficiency, billing
accuracy, and clinical decision-making.
The rest of the paper is organized as follows: Sec-
tion 2 reviews related work, Section 3 details the pro-
posed loss functions, Section 4 presents the experi-
mental setup and results, Section 5 discusses findings,
and Section 6 concludes with future directions.
2 RELATED WORK
This section reviews recent advancements in natural
language processing (NLP) and their applications in
ICD-10 coding, focusing on large language models,
BERT-based architectures, custom loss functions, and
vector-based representations.
Recent developments in NLP have been driven by
large language models (LLMs) such as GPT-4 (Wu
et al., 2023), Claude 3 (Kurokawa et al., 2024),
and Gemini (Mihalache et al., 2024). These models
demonstrate remarkable capabilities in text process-
ing tasks (Kumari and Pushphavati, 2022). However,
their computational intensity and privacy concerns
have limited healthcare applications (Al-Bashabsheh
et al., 2023).
This has led to the adoption of more efficient mod-
els, particularly BERT (Bidirectional Encoder Repre-
sentations from Transformers) (Devlin et al., 2019),
which offers comparable effectiveness while requir-
ing fewer resources (Mohammadi and Chapon, 2020).
BERT-based models like ClinicalBERT (Alsentzer
et al., 2019) excel in capturing clinical context,
making them practical for automated coding solu-
tions (Grabner et al., 2022).
Parallel developments in custom loss func-
tions (Dinkel et al., 2019) have shown promise in
healthcare applications. These functions enhance
model performance by optimizing relationship dis-
covery rather than exact matching (Kulkarni et al.,
2024). Notable improvements have been demon-
strated in handling imbalanced datasets (Boldini et al.,
2022) and noisy medical records (Wang et al., 2019).
Recent work (Giyahchi et al., 2022) has shown
the effectiveness of custom loss functions in health-
care NLP tasks, while advances in vector-based rep-
resentations (Hatoum et al., 2024b) have improved
ICD-10 code prediction accuracy. Particularly, NNB-
SVR (Hatoum et al., 2024a) demonstrates a 12.73%
improvement through semantic vector representations
and cosine similarity evaluation.
Our work builds on these developments by com-
bining vector-based representations with custom loss
functions, addressing a gap in current research. This
approach moves beyond equality-based methods to
capture both semantic relationships between codes
and nuanced clinical information, potentially improv-
ing prediction accuracy and clinical relevance in ICD-
10 coding.
3 CUSTOM LOSS FUNCTIONS
FOR ICD-10 CODING
The complexity of ICD-10 coding necessitates a
more nuanced approach than traditional equality-
based methods. While conventional loss functions
penalize models for any mismatch between predicted
and true labels, these approaches overlook the hi-
erarchical and semantic relationships between ICD-
10 codes. To address this, we propose four cus-
tom loss functions that aim to capture these seman-
tic relationships while balancing the need for speci-
ficity and flexibility in predictions. These loss func-
tions are designed to integrate seamlessly with exist-
ing model architectures, ensuring they can be applied
to a wide variety of models without requiring struc-
tural changes. Our focus is on optimizing model per-
formance through these custom loss functions rather
than introducing new model architectures, ensuring
broad applicability across a wide range of existing and
future models in automated medical coding.
3.1 Definitions
Consider a set of n samples X = {x
i
}
n
i=1
with true-
label sets Y = {y
i
}
n
i=1
and predicted-label sets
ˆ
Y =
{ ˆy
i
}
n
i=1
, where sets y
i
and ˆy
i
represent respectively the
true and predicted sets of labels for sample x
i
. Let Λ =
{λ
j
}
m
j=1
be the set of all unique ICD-10 codes, where
m is the total number of unique codes. Each ICD-
10 code λ
j
∈ Λ is mapped to a d-dimensional vector
representation v
j
= f (λ
j
) through a function f : Λ →
R
d
. We denote |y
i
| as the cardinality of the true-labels
set y
i
, and | ˆy
i
| as the cardinality of the predicted-labels
set ˆy
i
. Therefore, the sets y
i
and ˆy
i
can be expressed
as:
y
i
= {y
i j
}
|y
i
|
j=1
ˆy
i
= { ˆy
i j
}
| ˆy
i
|
j=1
where j is the j-th label in the true label set y
i
and the
j-th label in predicted-label set ˆy
i
for sample x
i
.
Beyond Equality Matching: Custom Loss Functions for Semantics-Aware ICD-10 Coding
167

3.2 Formulation
Each true label y
i j
∈ y
i
and predicted label ˆy
i j
∈ ˆy
i
are mapped to their vector representations v
i j
= f (y
i j
)
and ˆv
i j
= f ( ˆy
i j
) respectively. The cosine similarity
between these vector representations is calculated as:
cos(v
i j
, ˆv
i j
) =
v
⊤
i j
ˆv
i j
∥v
i j
∥
2
∥ ˆv
i j
∥
2
where ∥v
i j
∥
2
and ∥ ˆv
i j
∥
2
are the L
2
norms of v
i j
and
ˆv
i j
respectively. Let τ be a tunable threshold hyper-
parameter in the range [0,1] that controls the strict-
ness of the matching criteria, with lower values al-
lowing more dissimilar vectors to match and higher
values requiring stronger similarity to be considered
as a match.
The binary indicator δ
i j
, which determines
whether the predicted and true label vectors are con-
sidered relevant, is defined as:
δ
i j
=
(
1, if cos(v
i j
, ˆv
i j
) ≥ τ
0, otherwise
It is worth mentioning that we chose cosine simi-
larity to measure semantic relatedness between ICD-
10 code vectors due to its proven effectiveness in text
classification and information retrieval tasks, partic-
ularly in medical domains (Al-Anzi and AbuZeina,
2020). Cosine similarity offers several key advan-
tages: invariance to document length, computational
efficiency for sparse data (common in medical cod-
ing), and an intuitive interpretable scale. More-
over, it captures semantic relationships effectively by
comparing vector directions rather than magnitudes,
enabling detection of nuanced connections between
ICD-10 codes (Silva et al., 2024). These properties
make cosine similarity especially well-suited for en-
hancing our ICD-10 code prediction model, allowing
us to capture both semantic and hierarchical relation-
ships between codes efficiently.
Having defined the core elements, we now in-
troduce four custom loss functions that utilize these
similarities to optimize ICD-10 coding predictions.
These loss functions provide a framework that bal-
ances the need to capture all relevant codes while min-
imizing irrelevant predictions.
3.3 Custom Loss Functions
3.3.1 True Label Cardinality Loss (TLCL)
TLCL encourages the model to predict all true labels
by assigning equal weight to each one. This loss func-
tion is particularly useful when recall is prioritized, as
it ensures the model captures as many relevant codes
as possible. However, this emphasis on recall means it
may not strongly penalize irrelevant predictions. The
TLCL is computed as:
T LCL = −
1
n
n
∑
i=1
1
|y
i
|
|y
i
|
∑
j=1
(1 − δ
i j
)
where n is the number of samples, |y
i
| is the num-
ber of true labels for sample i, and δ
i j
is the binary
indicator of whether the true and predicted label vec-
tors match. This formulation ensures that each true
label contributes equally to the loss, regardless of the
total number of true labels for a given sample.
3.3.2 Predicted Label Cardinality Loss (PLCL)
PLCL focuses on precision by giving equal weight to
each predicted label, regardless of the number of true
labels. This approach helps avoid irrelevant predic-
tions, making it ideal for scenarios where avoiding
false positives is critical. However, it may not suf-
ficiently reward predicting the full set of true labels.
The PLCL is calculated as:
PLCL = −
1
n
n
∑
i=1
1
| ˆy
i
|
| ˆy
i
|
∑
j=1
(1 − δi j)
where | ˆy
i
| is the number of predicted labels for
sample i. This loss function penalizes each incorrect
prediction equally, encouraging the model to make
more conservative predictions to minimize false posi-
tives.
3.3.3 Balanced Harmonic Mean Loss (BHML)
BHML combines TLCL and PLCL using the harmonic
mean, creating a balance between precision and re-
call. This ensures that the model emphasizes both
predicting all true labels and avoiding irrelevant pre-
dictions. BHML is defined as:
BHML =
2
1
T LCL
+
1
PLCL
This formula is based on the harmonic mean of two
elements, which is generally defined for n elements
as H =
n
∑
n
i=1
1
x
n
(Ferger, 1931). The harmonic mean
gives more weight to the smaller value, ensuring that
the model does not overly prioritize either recall or
precision at the expense of the other.
3.3.4 Weighted Harmonic Mean Loss (WHML)
WHML introduces a weighting parameter α to fine-
tune the balance between precision and recall. This
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
168
flexibility allows for a more tailored optimization
strategy depending on the specific characteristics of
the dataset or the clinical application used. WHML is
computed as:
W HML =
1
α
T LCL
+
1−α
PLCL
were α ∈ [0, 1] controls the balance between TLCL
and PLCL. The behavior of WHML varies based on
the value of α:
• When α = 0, WHML is equivalent to PLCL, fo-
cusing entirely on precision.
• When 0 < α < 0.5, the model prioritizes precision
(PLCL) over recall, but still considers both.
• When α = 0.5, WHML is equivalent to BHML,
providing a balanced approach between precision
and recall.
• When 0.5 < α < 1, the model prioritizes recall
(TLCL) over precision, but still considers both.
• When α = 1, WHML is equivalent to TLCL, fo-
cusing entirely on recall.
WHML serves as a generalized version of the
other loss functions, encompassing TLCL, PLCL, and
BHML as special cases. By adjusting α, we can adapt
the loss function to the specific medical coding re-
quirements, providing a unified framework that can
be tailored to various ICD-10 coding scenarios.
3.4 Loss Function Selection and Impact
The choice of loss function significantly impacts the
model’s behavior during training. TLCL improves re-
call by encouraging the prediction of all relevant la-
bels. PLCL enhances precision by reducing false pos-
itives. BHML and WHML offer balanced approaches,
with WHML providing additional flexibility through
its weighting parameter α. The flexibility of these
custom loss functions allows practitioners to tailor the
model’s optimization strategy based on the specific
clinical context, whether prioritizing capturing all rel-
evant diagnoses or minimizing incorrect predictions.
Ultimately, these custom loss functions enable the
development of models that are more aligned with
real-world ICD-10 coding needs, improving both the
efficiency and accuracy of medical coding systems.
By incorporating semantic similarity into the loss
function, we ensure that clinically relevant but imper-
fect matches are appropriately handled, advancing the
state of automated ICD-10 coding.
4 EXPERIMENTS AND RESULTS
This section evaluates the performance of our pro-
posed custom loss functions for multi-label ICD-10
code prediction, demonstrating the value of leverag-
ing vector code similarities and label cardinalities to
improve clinical relevance.
4.1 Dataset
The dataset comprises 9.57M clinical notes collected
over three years from a private hospital. As shown
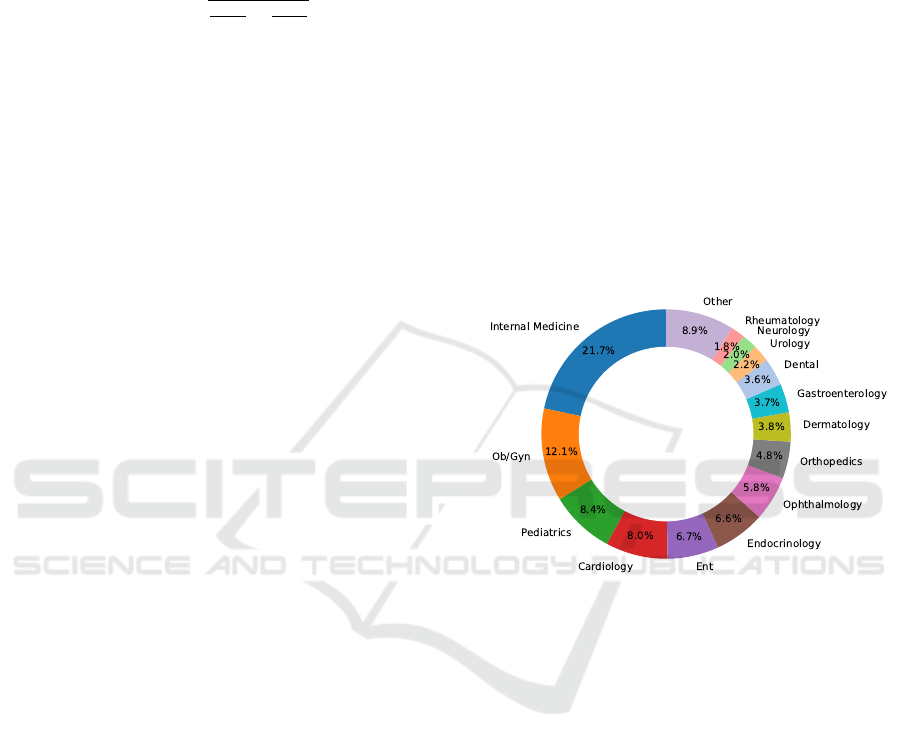
in Figure 1, it is imbalanced, with Internal Medicine
(21.71%) and OB/GYN (12.06%) being the most rep-
resented specialties, while others like Neurology are
less prevalent. This imbalance poses challenges for
predictive models to perform well across all special-
ties.
Figure 1: Distribution of the dataset across medical spe-
cialties, highlighting significant representation of Internal
Medicine and OB/GYN.
To ensure data quality, clinical notes were prepro-
cessed using a tool that unified medical terms, ex-
panded abbreviations, normalized dates, and trans-
formed investigational values into categorical data.
These steps improved data consistency and reliability
for ICD-10 prediction models (Hatoum et al., 2023).
Variability in physician writing styles, includ-
ing terminology and phrasing, was mitigated through
standardization. The dataset, containing 3,100 unique
ICD-10 codes, was in English. Strict privacy mea-
sures ensured data confidentiality, with all processing
performed within the hospital’s secure infrastructure,
adhering to privacy regulations.
4.2 Setup
Clinical notes were tokenized using the BertTok-
enizer, and the pretrained ClinicalBERT model was
used as the embedding layer(Alsentzer et al., 2019),
chosen for its effectiveness in capturing domain-
Beyond Equality Matching: Custom Loss Functions for Semantics-Aware ICD-10 Coding
169
specific language patterns to enhance ICD-10 code
predictions.
ICD-10 labels were converted into a binary matrix
(9.57M x 3,100) using scikit-learn’s MultiLabelBina-
rizer. Data was split into 5 folds for cross-validation.
The model, implemented with Keras and TensorFlow,
used ClinicalBERT as the embedding layer followed
by a dense output layer with sigmoid activations. Key
hyperparameters are summarized in Table 1.
Table 1: Key hyperparameters used in the classification ex-
periments.
Parameter Value
Embedding Layer ClinicalBERT
Optimizer Adam
Learning Rate 0.001
Batch Size 32
Early Stopping Patience 5
Number of Folds (Cross-Validation) 5
Number of Epochs (max) 50
Cosine Similarity Threshold 0.76
The Adam optimizer was selected for its effi-
ciency in handling sparse gradients and large-scale
datasets, which is critical for high-dimensional ICD-
10 tasks with class imbalance. The model was first
trained using binary cross-entropy (BCE) loss (Zhang
and Sabuncu, 2018) as a baseline. We then evaluated
the proposed custom loss functions (TLCL, PLCL,
BHML) and WHML with α ∈ 0.25, 0.75, where α =
0.75 achieved the highest F1-micro score. Optimal α
values may vary depending on dataset characteristics.
4.3 Results
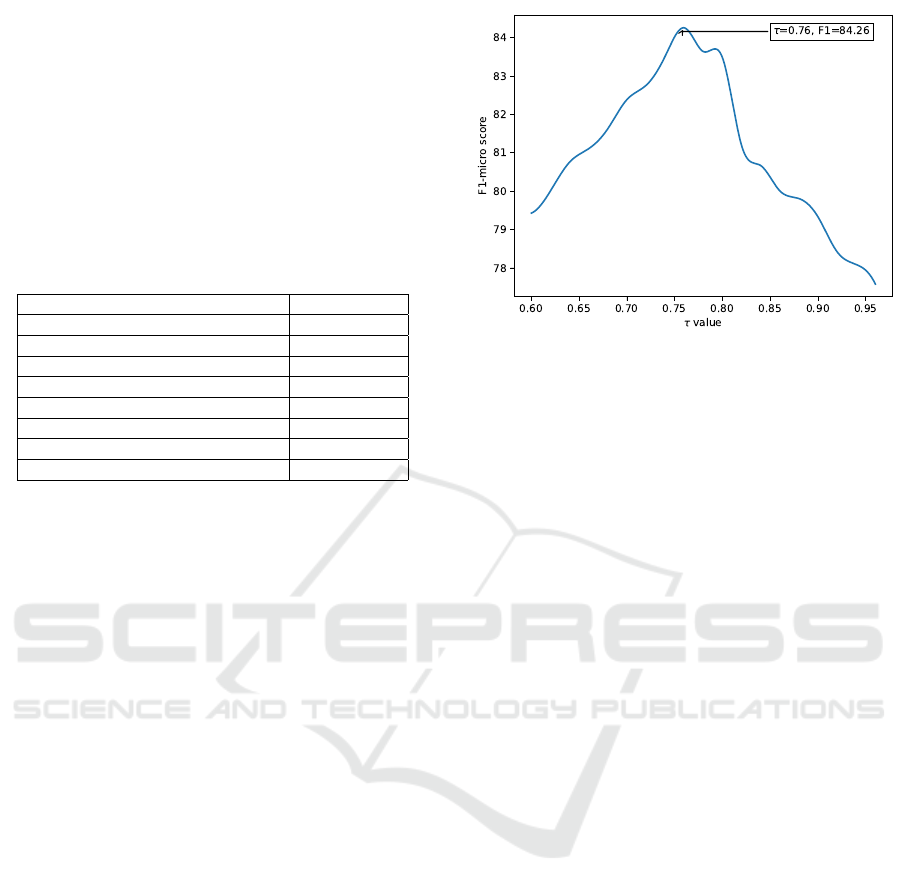
4.3.1 Optimal Similarity τ Ratio for Enhanced
ICD-10 Prediction
A grid search on a smaller dataset of 350,000 records
determined the optimal cosine similarity ratio. Ratios
from 0.6 to 0.96 were tested, with τ = 0.76 achieving
the best micro-F1 score of 84.26% (Figure 2).
4.3.2 Custom Loss Function Comparison
The proposed custom loss functions were compared
to binary cross-entropy (BCE) in ICD-10 classifi-
cation. As shown in Table 2, custom loss func-
tions significantly outperformed the baseline, achiev-
ing higher F1-micro and F1-weighted scores for train-
ing and testing.
WHML with α = 0.75 achieved the best F1-micro
score (96.83%) during training and 88.54% during
testing, demonstrating its robustness across classes. It
also converged faster (17 epochs) compared to BCE
(22 epochs), showing improved learning efficiency.
Figure 2: Micro-F1 scores for different cosine similarity ra-
tios, with the highest at τ = 0.76.
4.3.3 Specialty-Specific ICD-10 Prediction
Performance
Table 3 highlights the performance across medi-
cal specialties. WHML with α = 0.75 consistently
achieved the highest scores, particularly in Pediatrics
(97.51%), Ophthalmology (95.97%), and Dermatol-
ogy (94.26%), demonstrating its ability to handle
class imbalance and domain-specific nuances. BHML
also showed strong results, balancing recall and pre-
cision.
4.3.4 Performance on Challenging ICD-10
Codes
For complex ICD-10 codes prone to misclassification,
Table 4 shows that WHML significantly improved per-
formance. For example, ”K40.90” (inguinal hernia)
achieved 70.05%, a 10.19% improvement over BCE.
Similarly, codes like ”R10.4” (abdominal pain) ben-
efited from the semantic relationships leveraged by
custom loss functions.
These results validate our relevancy-based ap-
proach, showing particular strength in handling am-
biguous and clinically similar codes.
5 DISCUSSION
The results of our study highlight the potential of in-
corporating semantic similarity and hierarchical rela-
tionships into the loss function for improving ICD-10
code prediction from clinical text. By moving beyond
strict equality matching and considering the clini-
cal relevance of the predicted codes, our proposed
approach demonstrates significant improvements in
both accuracy and efficiency.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
170
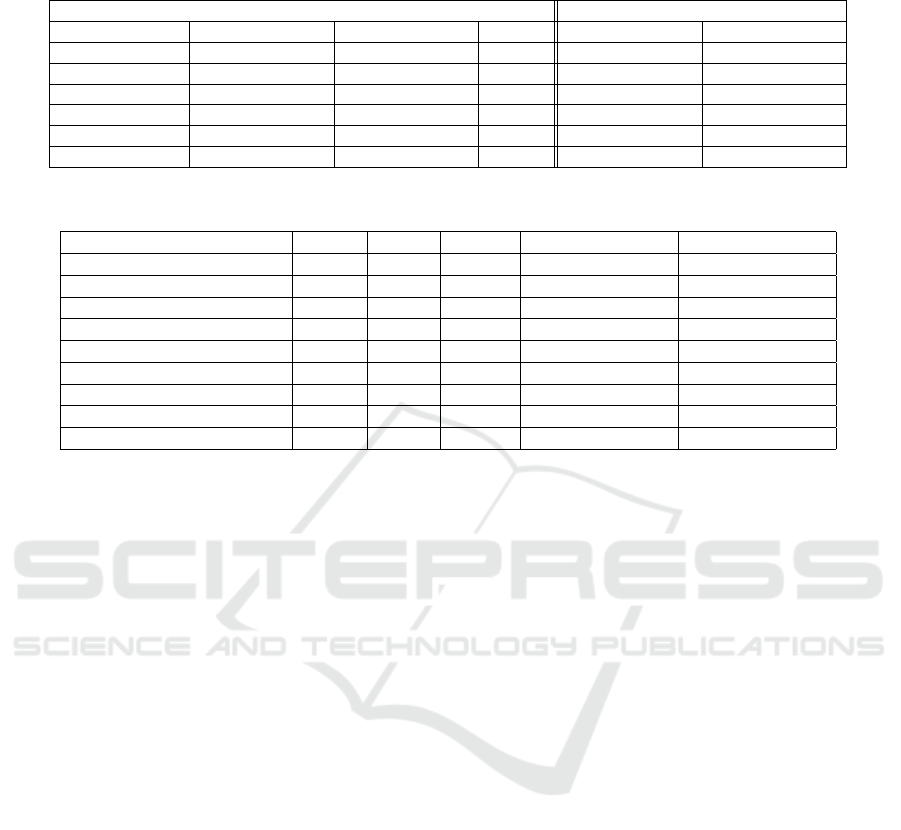
Table 2: Comparison between the baseline training and testing results for the custom loss functions TLCL, PLCL, and BHML
at τ = 0.76.
Training Results Testing Results
Experiment F1-micro F1-Weighted Epochs F1-micro F1-Weighted
EM 83.75 ± 5.81e-03 84.31 ± 6.52e-03 22 74.64 ± 2.28e-03 72.01 ± 2.20e-03
TLCL 94.18 ± 2.56e-03 92.78 ± 3.02e-03 17 85.72 ± 1.89e-03 83.61 ± 2.11e-03
PLCL 92.01 ± 3.19e-03 90.54 ± 4.12e-03 18 83.92 ± 2.37e-03 81.96 ± 3.08e-03
BHML 95.42 ± 2.32e-03 93.62 ± 3.51e-03 17 87.08 ± 1.95e-03 83.61 ± 2.34e-03
WHML α = 0.25 94.87 ± 3.41e-03 92.78 ± 3.88e-03 17 86.19 ± 2.25e-03 84.73 ± 2.48e-03
WHML α = 0.75 96.83 ± 3.01e-03 94.71 ± 3.89e-03 17 88.54 ± 2.58e-03 86.92 ± 2.99e-03
Table 3: Comparison of ICD-10 prediction F1-micro scores across various medical specialties, illustrating performance vari-
ations across metrics such as TLCL, PLCL, BHML, WHML α = 0.25, and WHML α = 0.75.
Specialty TLCL PLCL BHML WHML α = 0.25 WHML α = 0.75
Cardiology 90.56 88.12 91.64 88.56 92.14
Dental 87.28 86.36 88.89 86.87 89.87
Dermatology 92.42 89.94 93.95 91.36 94.26
ENT 91.67 90.27 93.09 90.88 94.01
Internal Medicine 86.73 85.08 87.23 85.81 88.22
Obstetrics and Gynaecology 92.38 90.46 93.89 90.79 94.31
Orthopedics 93.80 92.18 94.73 93.53 94.98
Pediatrics 94.51 93.96 97.08 94.12 97.51
Emergency 74.26 73.85 76.34 74.02 77.09
5.1 Cost-Effectiveness and
Computational Complexity
While the performance improvements of our cus-
tom loss functions are clear, it is also important to
consider the computational complexity and runtime
efficiency of the proposed methods. All four loss
functions (TLCL, PLCL, BHML, and WHML) involve
calculating cosine similarities between vector repre-
sentations of the ICD-10 codes, which adds addi-
tional overhead compared to traditional binary cross-
entropy loss (EM).
The complexity of calculating cosine similarity
for each label in a multi-label classification setting is
O(d), where d is the dimensionality of the vector rep-
resentations. Given that we compute this for every la-
bel, the complexity of each loss function for a single
sample is O(|y| · d), where |y| is the number of pre-
dicted or true labels. For the entire dataset of n sam-
ples, the total complexity becomes O(n · |y| · d). This
makes the proposed loss functions computationally
more expensive than traditional binary cross-entropy,
but the improved accuracy and recall justify this over-
head for large, complex datasets like ours.
In terms of runtime, our experiments showed that
the models trained with WHML and BHML required
fewer epochs to converge (17 compared to 22 epochs
for the traditional method), indicating greater effi-
ciency in training. This reduction in epochs helps
offset the higher per-iteration cost of the custom loss
functions.
5.2 Limitations of the Proposed Method
Despite the promising results, there are several lim-
itations to our approach. Firstly, while we studied
a very large number of labels (3,100 unique ICD-10
codes), the number of relevant labels varies signifi-
cantly across different healthcare facilities. For ex-
ample, certain rare codes, such as W58 - Bitten or
struck by crocodile or alligator, were absent from our
dataset, which was collected from a hospital in Saudi
Arabia. The absence of such rare codes limits the gen-
eralizability of the model to other regions or facilities
that may encounter different medical conditions.
Additionally, the dataset used in this study is in-
herently imbalanced, with certain medical special-
ties and ICD-10 codes being much more frequent
than others. This imbalance may have impacted
the model’s ability to generalize to underrepresented
classes, potentially leading to suboptimal perfor-
mance in these areas. Future work could involve ex-
ploring advanced techniques such as resampling or
class weighting to mitigate the effects of data imbal-
ance and improve the model’s robustness across all
classes.
While our model showed strong performance
across most specialties, some specialties did not
show as much improvement. Further investigation is
needed to understand the reasons behind the weaker
performance in these areas, and whether specific
characteristics of the specialties or the corresponding
ICD-10 codes contributed to this outcome. Address-
Beyond Equality Matching: Custom Loss Functions for Semantics-Aware ICD-10 Coding
171
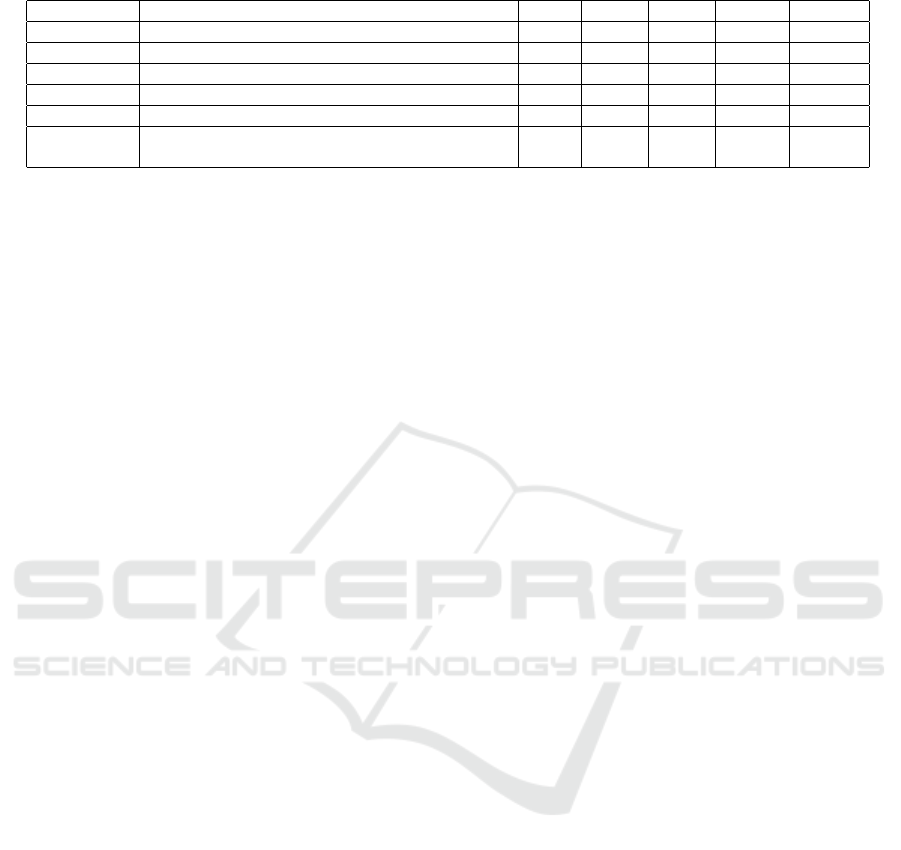
Table 4: F1-scores for challenging ICD-10 codes across different loss functions.
ICD-10-AM Description EM TLCL PLCL BHML WHML
J18.9 Pneumonia, unspecified 63.59 75.12 74.87 76.05 76.18
F41.9 Anxiety disorder, unspecified 53.17 56.94 56.32 57.21 57.29
M54.5 Low back pain 57.43 60.09 61.14 63.67 64.20
R10.4 Other and unspecified abdominal pain 50.38 58.40 57.78 59.17 59.64
G93.9 Disorder of brain, unspecified 49.08 50.21 50.14 51.02 50.83
K40.90
Unilateral or unspecified inguinal hernia without
obstruction or gangrene, not specified as recurrent
59.86 67.22 65.47 69.13 70.05
ing this limitation will require further tuning of the
model and potentially incorporating domain-specific
knowledge into the training process.
Moreover, this study focused on optimizing the
loss functions rather than customizing the underly-
ing model architecture or the optimizer. Future work
could explore integrating customized optimizers and
classifiers to further enhance the model’s predictive
power. This would allow us to tailor both the learning
process and the architecture more closely to the needs
of ICD-10 classification tasks, potentially unlocking
even greater improvements.
5.3 Real-World Implementation and
Future Work
One strength of our study is that the trained model
has been implemented in a real-world hospital set-
ting, where it is currently undergoing pilot testing.
This provides valuable practical insights and demon-
strates the feasibility of applying the proposed method
in healthcare environments. Initial feedback from the
pilot testing has been positive, though challenges have
emerged, such as integrating the model into existing
hospital workflows and ensuring compatibility with
the hospital’s electronic health record (EHR) systems.
Additionally, the model’s performance in handling
ambiguous or incomplete clinical notes during real-
time use is another area that requires further refine-
ment.
Looking ahead, there are several avenues for fu-
ture research. While we already utilize a tool that
improves the quality of clinical textual data by unify-
ing medical terms, expanding abbreviations, and nor-
malizing investigational values, further refinements in
data preprocessing techniques could enhance model
performance even more. For instance, additional
techniques such as advanced semantic normalization
and entity resolution may help in handling even more
nuanced and noisy clinical texts, especially in diverse
medical contexts.
Additionally, exploring different configurations of
the α parameter for the WHML loss function across
various specialties could lead to further optimizations.
This would allow the model to be fine-tuned to the
specific characteristics of each medical specialty.
Finally, customizing the optimizer and classifier
will be important next steps to maximize the effec-
tiveness of our approach and ensure it is adaptable
to various healthcare contexts. Integrating more ad-
vanced learning techniques and architecture optimiza-
tions could lead to even greater improvements in ICD-
10 code prediction accuracy and efficiency.
6 CONCLUSION
This study introduces semantics-aware loss functions
for ICD-10 code prediction that incorporate clinical
relevance and hierarchical relationships through vec-
tor representations. Our approach significantly out-
performed traditional methods, achieving an 88.54%
test set F1-micro score compared to 74.64% with bi-
nary cross-entropy. The Weighted Harmonic Mean
Loss (WHML) demonstrated particularly robust per-
formance across medical specialties.
While cosine similarity calculations added com-
putational overhead, faster convergence partially off-
set this cost. Pilot testing validates our approach’s
feasibility, though challenges remain in workflow
integration and real-time processing. Future work
will address ICD-10 code distribution variability,
dataset imbalances, and specialty-specific optimiza-
tion through refined WHML configurations and en-
hanced preprocessing techniques.
By improving automated medical coding accu-
racy and efficiency, our approach has the potential to
streamline healthcare operations and support more in-
formed clinical decision-making. With further refine-
ments in model architecture and optimization strate-
gies, these methods promise to advance both medical
coding automation and healthcare analytics.
ACKNOWLEDGMENT
The authors would like to express their deepest grat-
itude to the Medical Coding Department at Special-
ized Medical Center Hospital in Saudi Arabia. Their
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
172

unwavering support and dedication have been instru-
mental in the success of this research. A special note
of thanks is extended to the medical coders who have
shown exceptional commitment and diligence. Their
tireless efforts and invaluable support have signifi-
cantly enriched our work.
This work has been achieved in the frame of the
EIPHI Graduate school (contract ”ANR-17-EURE-
0002”).
CONFLICT OF INTEREST
The authors declare that they have no known com-
peting financial interests or personal relationships that
could have appeared to influence the work reported in
this paper.
REFERENCES
Al-Anzi, F. and AbuZeina, D. (2020). Enhanced latent se-
mantic indexing using cosine similarity measures for
medical application. The International Arab Journal
of Information Technology, 17(5):742–749.
Al-Bashabsheh, E., Alaiad, A., Al-Ayyoub, M., Beni-Yonis,
O., Zitar, R. A., and Abualigah, L. (2023). Improving
clinical documentation: automatic inference of icd-10
codes from patient notes using bert model. The Jour-
nal of Supercomputing, 79(11):12766–12790.
Alsentzer, E., Murphy, J., Boag, W., Weng, W.-H., Jin, D.,
Naumann, T., and McDermott, M. (2019). Publicly
available clinical BERT embeddings. In Proceed-
ings of the 2nd Clinical Natural Language Process-
ing Workshop, pages 72–78. Association for Compu-
tational Linguistics.
Boldini, D., Friedrich, L., Kuhn, D., and Sieber, S. A.
(2022). Tuning gradient boosting for imbalanced
bioassay modelling with custom loss functions. Jour-
nal of Cheminformatics, 14(1).
del Barrio, E., Gordaliza, P., and Loubes, J.-M. (2020). Re-
view of mathematical frameworks for fairness in ma-
chine learning. ArXiv.
Devlin, J., Chang, M., Lee, K., and Toutanova, K. (2019).
BERT: pre-training of deep bidirectional transformers
for language understanding. In NAACL-HLT 2019,
Minneapolis, MN, USA, June 2-7, Volume 1, pages
4171–4186. Association for Computational Linguis-
tics.
Dinkel, H., Wu, M., and Yu, K. (2019). Text-based depres-
sion detection on sparse data. arXiv.
Esteva, A., Robicquet, A., Ramsundar, B., Kuleshov, V.,
DePristo, M., Chou, K., Cui, C., Corrado, G., Thrun,
S., and Dean, J. (2019). A guide to deep learning in
healthcare. Nature Medicine, 25(1):24–29.
Ferger, W. F. (1931). The nature and use of the harmonic
mean. Journal of the American Statistical Association,
26(173):36–40.
Giyahchi, T., Singh, S., Harris, I., and Pechmann, C. (2022).
Customized training of pretrained language models to
detect post intents in online health support groups.
Multimodal AI in Healthcare, pages 59–75.
Grabner, C., Safont-Andreu, A., Burmer, C., and Schekoti-
hin, K. (2022). A bert-based report classification for
semiconductor failure analysis. International Sympo-
sium for Testing and Failure Analysis.
Hatoum, M., Charr, J.-C., Guyeux, C., Laiymani, D., and
Ghaddar, A. (2023). Emte: An enhanced medical
terms extractor using pattern matching rules. 15th In-
ternational Conference on Agents and Artificial Intel-
ligence, pages 301–311.
Hatoum, M. B., Charr, J. C., Ghaddar, A., Guyeux, C., and
Laiymani, D. (2024a). Nnbsvr: Neural network-based
semantic vector representations of icd-10 codes. Un-
der revision.
Hatoum, M. B., Charr, J. C., Ghaddar, A., Guyeux, C.,
and Laiymani, D. (2024b). Utp: A unified term pre-
sentation tool for clinical textual data using pattern-
matching rules and dictionary-based ontologies. Lec-
ture Notes in Computer Science, pages 353–369.
Kulkarni, D., Ghosh, A., Girdhari, A., Liu, S., Vance,
L. A., Unruh, M., and Sarkar, J. (2024). Enhancing
pre-trained contextual embeddings with triplet loss as
an effective fine-tuning method for extracting clinical
features from electronic health record derived mental
health clinical notes. Natural Language Processing
Journal, 6:100045.
Kumari, S. and Pushphavati, T. (2022). Question answer-
ing and text generation using bert and gpt-2 model. In
Computational Methods and Data Engineering: Pro-
ceedings of ICCMDE 2021, pages 93–110. Springer.
Kurokawa, R., Ohizumi, Y., Kanzawa, J., Kurokawa, M.,
Kiguchi, T., Gonoi, W., and Abe, O. (2024). Diagnos-
tic performance of claude 3 from patient history and
key images in diagnosis please cases. medRxiv.
Long, R. (2021). Fairness in machine learning: Against
false positive rate equality as a measure of fairness.
Journal of Moral Philosophy, 19(1):49–78.
Mihalache, A., Grad, J., Patil, N. S., Huang, R. S., Popovic,
M. M., Mallipatna, A., Kertes, P. J., and Muni, R. H.
(2024). Google gemini and bard artificial intelligence
chatbot performance in ophthalmology knowledge as-
sessment. Eye, pages 2530–2535.
Mittelstadt, B., Wachter, S., and Russell, C. (2023). The un-
fairness of fair machine learning: Levelling down and
strict egalitarianism by default. Michigan Technology
Law Review.
Mohammadi, S. and Chapon, M. (2020). Investigating the
performance of fine-tuned text classification models
based-on bert. In 2020 IEEE 22nd International Con-
ference on High Performance Computing and Com-
munications, pages 1252–1257.
Mou, C., Ye, X., Wu, J., and Dai, W. (2023). Automated
icd coding based on neural machine translation. In
2023 8th International Conference on Cloud Comput-
ing and Big Data Analytics (ICCCBDA), pages 495–
500. IEEE.
Beyond Equality Matching: Custom Loss Functions for Semantics-Aware ICD-10 Coding
173

Nayyar, A., Gadhavi, L., and Zaman, N. (2021). Machine
learning in healthcare: review, opportunities and chal-
lenges. Machine Learning and the Internet of Medical
Things in Healthcare, pages 23–45.
Otero Varela, L., Doktorchik, C., Wiebe, N., Quan, H., and
Eastwood, C. (2021). Exploring the differences in icd
and hospital morbidity data collection features across
countries: an international survey. BMC Health Ser-
vices Research, 21(1).
Silva, H., Duque, V., Macedo, M., and Mendes, M. (2024).
Aiding icd-10 encoding of clinical health records us-
ing improved text cosine similarity and plm-icd. Al-
gorithms, 17(4).
Wang, Y., Sohn, S., Liu, S., Shen, F., Wang, L., Atkinson,
E. J., Amin, S., and Liu, H. (2019). A clinical text
classification paradigm using weak supervision and
deep representation. BMC Medical Informatics and
Decision Making, 19(1).
Wu, T., He, S., Liu, J., Sun, S., Liu, K., Han, Q.-L., and
Tang, Y. (2023). A brief overview of chatgpt: The
history, status quo and potential future development.
IEEE/CAA Journal of Automatica Sinica, 10(5):1122–
1136.
Yu, Y., Qiu, T., Duan, J., and Wang, J. (2023). Multigran-
ularity label prediction model for automatic interna-
tional classification of diseases coding in clinical text.
Journal of Computational Biology, 30(8):900–911.
Zhang, Z. and Sabuncu, M. R. (2018). Generalized cross en-
tropy loss for training deep neural networks with noisy
labels. In proceedings 32nd International Conference
on Neural Information Processing Systems, NIPS’18,
pages 8792–8802, NY, USA. Curran Associates Inc.
Zhou, L., Cheng, C., Ou, D., and Huang, H. (2020).
Construction of a semi-automatic icd-10 coding sys-
tem. BMC Medical Informatics and Decision Making,
20(1).
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
174
