
Petri Net Modeling of Root Hair Response to Phosphate Starvation in
Arabidopsis Thaliana
Amber H. B. Fijn*, Casper H. Stiekema*, Stijn Boere*, Marijan Vi
ˇ
si
´
c* and Lu Cao
a
Leiden Institute of Advanced Computer Science, Leiden University, Einsteinweg 55, Leiden, The Netherlands
{a.h.b.fijn, c.h.stiekema, s.boere.2, m.visic}@umail.leidenuniv.nl, l.cao@liacs.leidenuniv.nl
Keywords:
Petri Nets, Root Hair Elongation, Arabidopsis Thaliana.
Abstract:
Limited availability of inorganic phosphate (Pi) in soil is an important constraint to plant growth. In order to
understand better the underlying mechanism of plant response to Pi, the response to phosphate starvation in
Arabidopsis thaliana was investigated through use of Petri Nets, a formal language suitable for bio-modeling.
A. thaliana displays a range of responses to deal with Pi starvation, but special attention was paid to root hair
elongation in this study. A central player in the root hair pathway is the transcription factor ROOT HAIR
DEFECTIVE 6-LIKE 4 (RSL4), which has been found to be upregulated during the Pi stress. A Petri Net was
created which could simulate the gene regulatory networks responsible for the increase in root hair length, as
well as the resulting increase in root hair length. Notably, discrepancies between the model and the literature
suggested an important role for RSL2 in regulating RSL4. In the future, the net designed in the current study
could be used as a platform to develop hypotheses about the interaction between RSL2 and RSL4.
1 INTRODUCTION
Inorganic phosphate (Pi) is essential for plant growth
as plants incorporate it into their DNA and phospho-
lipids. In addition, plants require it for biological pro-
cesses such as photosynthesis (Crombez et al., 2019;
Kayoumu et al., 2023). Limited (bio)availability of Pi
in soil is an important constraint to plant growth and
reduces crop yields worldwide (Datta et al., 2014).
Common solutions to deal with this involve fertiliza-
tion. However, fertilization is associated with sustain-
ability issues as it can deteriorate soil ecology (Hop-
kins and Hansen, 2019). In this context, researchers
have focused efforts on understanding and elucidating
the molecular mechanisms underlying plant response
to Pi starvation. By doing this, researchers hope to
identify traits that can be manipulated to develop cul-
tivars with enhanced resilience to Pi starvation. It has
been well described that plants display a suit of re-
sponses to deal with limited Pi availability. For exam-
ple, to increase the bioavailability of Pi in soil, plants
secrete acid phosphatases and ribonucleases which re-
lease P esters bound to soil components (Abel et al.,
2000; Bariola et al., 1994).
However, the most distinct response can be found
a
https://orcid.org/0000-0002-1847-068X
∗
These authors contributed equally to this study.
in the roots. Plants display both an increased lateral
root formation as well as increased root hair elonga-
tion under Pi stress (Crombez et al., 2019). Increase
in root hair length and density has been reported to
account for 90% of the total Pi uptake during Pi stress
(F
¨
ohse et al., 1991). In this case study, we aimed to
gain further insight into the root hair response to phos-
phate (Pi) starvation in A. thaliana. As most research
focuses on single genes, we hoped that by incorpo-
rating these results into a single Petri Net further in-
sight could be gained into the importance and role of
the regulatory network components. We focused our
research on A. thaliana as the Pi response has been
well-studied in this plant model.
Petri Nets can be applied to model concurrent,
asynchronous, and stochastic systems (Murata, 1989).
Hence, Petri Nets have been used as a graphical and
mathematical tool for quantitative and/or qualitative
analyses of complex biological systems. Petri Nets
represent a directed, weighted, bipartite graph with
two types of nodes: places and transitions which are
connected by arcs. This framework has been success-
fully applied to various biological processes includ-
ing biochemical pathways, signal transduction as well
as epidemic and ecological models (Chaouiya, 2007;
Hardy and Robillard, 2004).
Fijn, A. H. B., Stiekema, C. H., Boere, S., Viši
´
c, M. and Cao, L.
Petri Net Modeling of Root Hair Response to Phosphate Starvation in Arabidopsis Thaliana.
DOI: 10.5220/0013102400003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 529-536
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
529

2 BIOLOGICAL BACKGROUND
Many genes are involved in plant response to root hair
elongation. Among these, the transcriptional regula-
tor ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4) is
recognized as a central player in the network as it acti-
vates many downstream targets essential for root mor-
phogenesis (Bhosale et al., 2018). In addition, it has
been found that RSL4 levels increase during Pi stress,
and knockout of RSL4 significantly impairs root hair
elongation. Besides RSL4, other transcription factors
such as ROOT HAIR DEFECTIVE 6 (RHD6) and
ROOT HAIR DEFECTIVE 6-LIKE 2 (RSL2) have
also been highlighted as key genes involved in root
hair development. Regulation of these transcription
factors occurs in complex networks that also involve
plant hormones. Auxin has been found to modulate
RSL4 levels (Yi et al., 2010). Auxin achieves this
by activating auxin response factors (ARFs) that regu-
late the expression of RSL2 and RSL4 (Bhosale et al.,
2018). Besides auxin, ethylene is also involved in the
root hair response to Pi stress (Song et al., 2016). In-
tracellular reception of ethylene increases the levels
of transcription factor ETHYLENE INSENSITIVE 3
(EIN3). Similar to the ARFs, EIN3 can regulate RSL4
expression and, in addition, has been found to share
gene targets with RSL4.
In the root epidermis, root hair development of
epidermal cells is tightly regulated (Montiel et al.,
2004). Epidermal cells receiving a positional sig-
nal from two cortical cells develop a root hair (tri-
choblasts), whereas cells receiving a positional sig-
nal from a single cortical cell remain hairless (atri-
choblasts). To commit to the trichoblast fate a process
of lateral inhibition is essential (Savage et al., 2013).
Differentiating atrichoblast cells express CAPRICE
(CPC). The CPC proteins, among other proteins,
move to adjacent trichoblast cells where they form
a complex with GLABRA3 (GL3) and TRANSPAR-
ENT TESTA GLABRA1 (TTG1). This complex al-
lows the differentiating trichoblast cell to remain in
the default, trichoblast pathway. In addition, CPC is
essential for the expression of the transcription fac-
tor RHD6 which is involved in the early stages of
root hair development (Salazar-Henao et al., 2016).
RSL4 is one of its targets and is necessary for the tran-
scription of genes required for root hair formation.
RHD6 was also found to control RSL2, which like
RSL4 controls downstream targets involved in root
hair morphogenesis, indicating some redundancy be-
tween the two (Mangano et al., 2018). Under Pi star-
vation, the plant hormones auxin and ethylene play
a vital role in enhanced root hair elongation (Vis-
senberg et al., 2020). The networks involved in this
hormone-dependent response to Pi stress will be dis-
cussed in the following sections and an overview can
be found in Figure 1. Together, these networks result
in a 1.7-fold increase in root hair length from Pi suf-
ficient to Pi deficient conditions (Datta et al., 2015).
2.1 Auxin-Dependent Pathway
It has been reported that auxin levels increase
2.5-fold in response to Pi stress (Bhosale et al.,
2018). Increased auxin biosynthesis occurs in the
root tip and is dependent on the gene TRYPTO-
PHAN AMINOTRANSFERASE OF ARABIDOP-
SIS1 (TAA1), which is upregulated upon Pi stress.
The auxin transporter AUXIN RESISTANT 1
(AUX1) is responsible for the subsequent transport of
auxin towards the root hair elongation zone. Once
auxin reaches the root hair zone, it causes enhanced
activity of the auxin-inducible transcription factors
ARF7 and ARF19. For ARF19 a 2-fold increase of
transcript levels has been described (Bhosale et al.,
2018). ARF19 can activate the expression of RSL4
and RSL2, whereas ARF7 can activate only RSL4.
Together with other transcription factors, this leads
to 1.8-fold and 2.3-fold changes of RSL4 and RSL2
mRNA, respectively, during Pi starvation.
2.2 Ethylene-Dependent Pathway
Changes in ethylene levels during Pi starvation are
less well described and no clear effect has been eluci-
dated (Roldan et al., 2013). Nonetheless, it has been
found that, during Pi stress, levels of the ethylene-
response factor EIN3 are increased 2-fold (Song et al.,
2016). Various functions in regulating root hair re-
sponse in low Pi have been ascribed to EIN3. Among
these, transcriptional regulation of various genes is in-
volved in root hair development. Many of these genes
are also regulated by RSL4, highlighting the redun-
dancy present in the gene regulatory network. In ad-
dition, EIN3 is involved in the regulation of RSL4 it-
self, through inhibition of MYB30 (Xiao et al., 2021).
MYB30 can bind to the promoter region of RSL4 and
inhibit its expression. Ethylene enhances the physical
association of EIN3 with MYB30 to form the EIN3-
MYB30 complex. This effectively reduces the associ-
ation of MYB30 with the RSL4 promoter and thus al-
lows for increased expression of RSL4. Finally, EIN3
also physically interacts with RHD6, and the tran-
scription factors cooperatively bind to the promoter
region of RSL4 with increased affinity compared to
the binding affinity when RHD6 binds by itself (Feng
et al., 2017).
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
530

Figure 1: Overview of regulatory networks towards root
hair elongation. Figure created in BioRender.
2.3 RSL4 Regulation
In addition to the hormone-mediated regulation of
RSL4 expression, several independent regulatory
loops have been found crucial for the fine-tuning of
timing and amount of total RSL4 product. RSL4
has been shown to bind to its own promoter and en-
hance transcription, forming a positive feedback loop
(Hwang et al., 2017). GT-2-LIKE-1 (GTL1) and its
less studied homolog DF1 form a negative feedback
loop with RSL4 on a transcriptional level (Shibata
et al., 2018). Expression of GTL1 is initiated by the
RSL4 protein, after which GTL1 binds to the pro-
moter of RSL4, causing repression of transcription.
GTL1 also represses gene expression downstream
from RSL4 and its expression. An additional negative
feedback loop, centered around LONG ROOT HAIR
(LRH), is present on the translational level (Cui et al.,
2024). RSL4 initiates the transcription of LRH, which
then binds to the eukaryotic TRANSLATION INITI-
ATION FACTOR 4E (eIF4E). eIF4E recognizes the
5’ cap of the mature mRNA molecules and helps with
the ribosome binding. However, by binding to LRH,
eIF4Es lose the affinity to associate with the mRNA of
RSL4, effectively inhibiting translation. Besides the
pathways mentioned here, RSL4 is regulated by many
other genes and complexes, including ZINC-FINGER
PROTEIN 1 (ZP1) (Han et al., 2019) and RALF1-
FERONIA complex (Zhu et al., 2020), but the details
about their impact on total expression are less clear.
The result of this complex regulation is a sequence of
events determining the final length of the hair (Datta
et al., 2015). RSL4 protein starts being synthesized
2 hours before the root hair growth initiation and
achieves its highest abundance 2 hours after the ini-
tiation. The expression of RSL4 mRNA is detectable
only before initiation of hair growth. The abundance
of RSL4 protein starts to decrease gradually over sev-
eral hours during the elongation phase due to degrada-
tion mediated by proteasome 26S (Datta et al., 2015).
As long as the RSL4 protein is present in the tri-
choblasts, the hair keeps growing at a steady rate of
1 micrometer per minute (Datta et al., 2015; Yi et al.,
2010).
3 PETRI NETS MODELING
Petri Nets are modeling structures based on math-
ematical formalisms and are suitable for describ-
ing complex systems with multiple co-occurring and
co-dependent processes. A Qualitative Petri Net
(QPN) is defined by the following quadruple N =
(P,T, f , m
0
) (Heiner et al., 2008). Here, P refers to a
set of places (represented by circles), and T to a set of
transitions (represented by rectangles). f defines the
set of directed arcs that connect transitions and places,
which can be weighted by non-negative integers, and
finally m
0
which gives the initial marking. Places are
typically used to model passive system components,
which in a biological context can be used to repre-
sent species, proteins, or molecules. The available
amount of these compounds or species is represented
by tokens, shown as black dots or non-negative in-
tegers. Transitions are active components represent-
ing processes such as chemical reactions or interac-
tions. Arcs connect places and transitions (Heiner
et al., 2008; Bl
¨
atke et al., 2011). The tokens assigned
to the places before the first firing is the initial mark-
ing. Tokens move through the net by firing transitions
(Bl
¨
atke et al., 2011). When a transition is fired, the
tokens are removed from the pre-place(s) and added
to the post-place(s). The number of moving tokens is
determined by the weights of the ingoing and outgo-
ing arcs of a transition. Firing happens atomically and
consumes no time in the QPN (Heiner et al., 2008).
In this model, we used a Stochastic Petri Net,
where a firing delay of a transition is randomly com-
puted based on the probability distribution with a fir-
ing rate. This allows the incorporation of time prop-
erty and randomness effect. They are valuable for
modeling biological processes. Most functions in our
model are based on mass action kinetics, where the
firing rate of a transition depends on the number of
tokens in its pre-place(s), multiplied by a constant
that was determined empirically for every transition.
The use of alternative functions, namely logarithmic
and minimum functions, is motivated in subsequent
Petri Net Modeling of Root Hair Response to Phosphate Starvation in Arabidopsis Thaliana
531
sections. Besides stochastic transitions, there are de-
terministic transitions that fire without delay and are
only active during certain simulation periods.
Tokens in the net represent both information and
actual amount. For example, the number of tokens
of Pi represents the concentration (µM) of Pi in suffi-
cient and deficient conditions. For transcription fac-
tor activation, tokens represent information, as infor-
mation from the transcription factor is transferred to
its activated gene. In order to allow the Pi signal
to directly affect the abundance of proteins and the
eventual root hair length, most places had zero ini-
tial marking. Only MYB30 and eIF4E were given
non-zero initial marking since both components are
already present in the cell. The model can be inter-
preted as the intracellular processes in response to a
specific Pi concentration. Two Pi concentrations will
be investigated, 3 and 300 µM. These were concen-
trations used to represent Pi deficient and sufficient
conditions.
In a Petri Net several additional types of arcs can
be defined (Marwan et al., 2011; Bl
¨
atke et al., 2011).
With read arcs, the transition fires only when its pre-
place is populated with an equal or larger number of
tokens as its associated arc weight. Tokens are not
consumed from the pre-place. Inhibitor arcs prevent
a transition as long as its pre-place holds an equal or
larger number of tokens to the arc weight. Modifier
arcs are used with stochastic transitions and attribute
to a token-dependent modification of the firing rate.
No tokens are consumed from the pre-place.
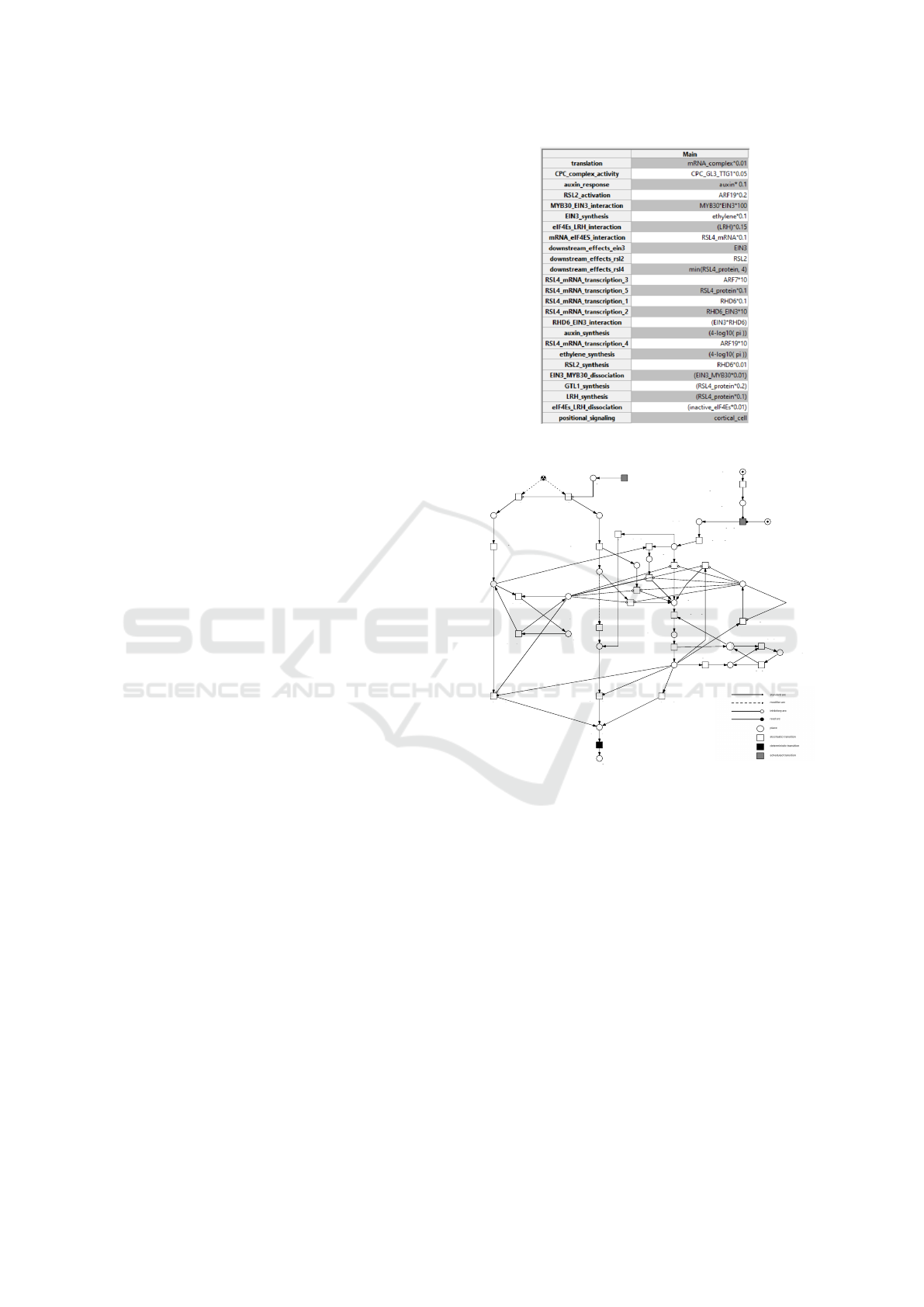
To create the Petri Net, the Snoopy software was
employed (Heiner et al., 2012). The complete Petri
Net can be seen in Figure 3, with the initial mark-
ing representing low Pi configurations. In the model,
custom functions were ascribed to each transition and
designed to be in line with the biological system. In
Figure 2 the functions used for each transition are dis-
played.
The epidermal cell resolution based on positional
signaling was modeled by including the components
necessary for determining trichoblast cell fate. Posi-
tional signaling from cortical cells and CPC originat-
ing from adjacent atrichoblasts were required for the
production of RHD6 and for the cell to commit to tri-
choblast cell fate.
3.1 Modeling Auxin-Dependent
Pathway
Pi influences auxin levels through a modifier arc with
the function (4 − log10(pi)) which causes higher lev-
els of Pi to be associated with a lower firing rate.
The log function was selected as a log relation has
Figure 2: The functions in Petri Net transitions.
ethylene
EIN3
auxin
ARF19
RSL2
downstream growth factors
RSL4 protein
RSL4 mRNA
RHD6
CPC GL3 TTG1
cortical cell
GL3 TTG1
CPC
MYB30
5
EIN3 MYB30
eIF4Es
50
inactive eIF4Es
GTL1
mRNA complex
RHD6 EIN3
ARF7
pi
Hair length
LRH
hormone stopper
translation
CPC complex activity
auxin response
RSL2 activation
MYB30 EIN3 interaction
EIN3 synthesis
eIF4Es LRH interaction
mRNA eIF4ES interaction
downstream effects ein3
downstream effects rsl2
downstream effects rsl4
RHD6 EIN3 interaction
auxin synthesisethylene synthesis
RSL2 synthesis
EIN3 MYB30 dissociation
GTL1 synthesis
LRH synthesis
eIF4Es LRH dissociation
positional signaling
<0.004>
stopper
root hair formation
4
10
80
3
2
10
10
5
5
5
5
5
5
Figure 3: The Petri Net model of root hair elongation in
response to inorganic phosphate (Pi).
been described between Pi concentration and root hair
length (Bates and Lynch, 2000). Since experimental
Pi levels in literature do not go above 1000 µM, the
upper log-scaled limit was set to one order of magni-
tude more, which is 4. The modifier arc ensures that
tokens are not consumed, resulting in the hair elon-
gation being limited only by the marking-dependent
firing rate. This was a simplified method to repre-
sent the Pi concentration which is unlikely to change
much during root hair elongation. Auxin response
was modeled through ARF7 and ARF19, which af-
fect RSL2 and RSL4. The firing rate functions for
these transitions differ, with the coefficients deter-
mined by empirical evaluation to keep the ratios be-
tween the downstream changes caused by these pro-
teins in alignment with the literature.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
532

3.2 Modeling Ethylene-Dependent
Pathway
The ethylene-dependent pathway begins with a mod-
ifier arc originating from Pi and includes a transition
that affects ethylene levels in the same manner as it
does for auxin. The effect of ethylene is mediated
by EIN3. EIN3 binds to RHD6 and increases RSL4
transcription. As can be seen from its function, EIN3-
RHD6 increases the RSL4 mRNA level more strongly
than RHD6 alone. In addition, EIN3 can bind to
MYB30. As this transition consumes tokens from
MYB30, it can disrupt the inhibition of MYB30 on
RSL4 transcription. As proteins are dynamic and are
not infinite in a complex once bound, a loop was cre-
ated to display the dissociation of the EIN3-MYB30
complex.
3.3 Modeling RSL4 Regulation
RSL4 is produced in a pulse during root hair elonga-
tion. Two feedback loops responsible for the pulse
are included in the net. The first feedback loop is the
GTL1 loop. Production of the RSL4 protein leads
to GTL1 production which inhibits transcription of
RSL4 mRNA. This inhibitory loop only occurs when
sufficient RSL4 has been produced. Moreover, since
GTL1 also inhibits itself, a self-inhibitory arc was
added. The second is LRH-mediated negative feed-
back loop. In the net, eIF4Es have a marking of
50, meaning that only when sufficient RSL4 has been
produced, translation is fully inhibited. Dissociation
of eIF4E and LRH can make eIF4E available again,
reproducing the dynamic process of protein interac-
tions in the cell. A simplification was made by mark-
ing downstream growth factors as a single place be-
cause the population of downstream growth factors
was too large to model and limited information is
known. Since RSL4 has been highlighted as a nec-
essary transcription factor for root hair elongation,
read arcs are included from RSL4 to transitions com-
ing from EIN3 and RSL2. This ensures that, without
RSL4, root hair elongation is not possible. To de-
crease the effect of the abundance of RSL4 on hair
growth rate, the coefficient for firing rate was lim-
ited to 4 using the minimum function. From RSL4,
RSL2, and EIN3, downstream growth factors are ac-
tivated. These are simplified as a single place. From
downstream growth factors, a deterministic transition
is modeled towards the final hair length so that the
elongation period becomes dependent on the number
of tokens in pre-place with the rate held constant.
4 RESULTS
4.1 Snoopy Simulation
Using Snoopy simulation, a comparison was made for
the root hair length, as well as protein and hormone
abundances under low and high Pi concentrations. All
graphs obtained from simulations are an averaged re-
sult of 100 separate runs.
(a) Auxin. (b) RSL2.
(c) ARF7. (d) ARF19.
(e) EIN3. (f) MYB30.
(g) RSL4 mRNA. (h) RSL4 protein.
Figure 4: Auxin (a), RSL2 (b), ARF7 (c), ARF19 (d), EIN3
(e), MYB30 (f), RSL4 mRNA (g) and RSL4 protein (h)
given a low (Pi-) and a high (Pi+) concentration of Pi.
A decrease in Pi concentration causes increased
auxin biosynthesis. Figure 4a shows the presence of
auxin in both low and high Pi concentrations. Auxin
increases activity of ARF7 and ARF19 as shown in
Figure 4c and 4d. The abundance of RSL2 in low
and high Pi concentrations is shown in Figure 4b.
Petri Net Modeling of Root Hair Response to Phosphate Starvation in Arabidopsis Thaliana
533
Under the influence of ARF19, RSL2 increases the
transcription of downstream growth factors. How-
ever, this transcription also requires the presence of
RSL4. Since the transcription of RSL2 in the auxin-
dependent pathway occurs faster than the translation
of RSL4, many tokens accumulate in RSL2 before
RSL4 allowing them to flux towards the transcrip-
tion of downstream growth factors. This illustrates
why a peak can be observed for RSL2 abundance over
time. The peak is lower in low Pi concentration due
to higher RSL4 protein production, which leads to an
earlier flux of RSL2 towards downstream growth fac-
tors.
The abundance of ethylene follows the same trend
as auxin. Lower Pi leads to higher EIN3 levels as
shown in Figure 4e. EIN3 can physically interact with
RSL4-inhibitor MYB30, and thus effectively release
RSL4 from this inhibition. Figure 4f shows the pres-
ence of MYB30 given both low and high Pi concen-
trations. The abundance of MYB30 quickly drops for
both Pi concentrations, followed by a gradual increase
and leveling out. This is due to the termination ef-
fect exerted on the ethylene upstream, which causes
EIN3 to decline and allows MYB30 to become avail-
able again.
As evident from the Figure 4g, lower Pi concen-
tration leads to a higher accumulation of RSL4. This
is due to the culmination of activities from various
proteins and hormones that are influenced by Pi. The
general curve-shape of RSL4 abundance can be at-
tributed to the inhibitory effect from GTL1, which
inhibits transcription of RSL4 after some time, and
subsequent RSL4 translation as shown in Figure 4h.
The fluctuating levels observed for RSL4 protein are
caused by eIF4E, which is required for translation of
RSL4 mRNA. After a large amount of RSL4 is trans-
lated, eIF4E is shortly disabled which causes a dip
in the abundance of RSL4 protein, as the protein is
still fluxed towards root hair elongation. Once eIF4E
is enabled, an increase in RSL4 protein occurs, once
again disabling eIF4E. Although the peak amount of
tokens in the place representing RSL4 protein is simi-
lar in both Pi conditions, this should not be interpreted
as RSL4 being produced in equal amounts throughout
the simulation because it is constantly used by down-
stream processes. The slower leveling out in lower Pi
conditions shows that RSL4 persists for a longer time
in the cell, resulting in longer hairs.
The transcription of the downstream growth fac-
tors is influenced by three factors: RSL4 protein
which is influenced by RSL4 regulation as well as the
ethylene- and auxin-dependent pathway, RSL2 which
is produced through the auxin-dependent pathway,
and finally EIN3 which is influenced by the ethylene-
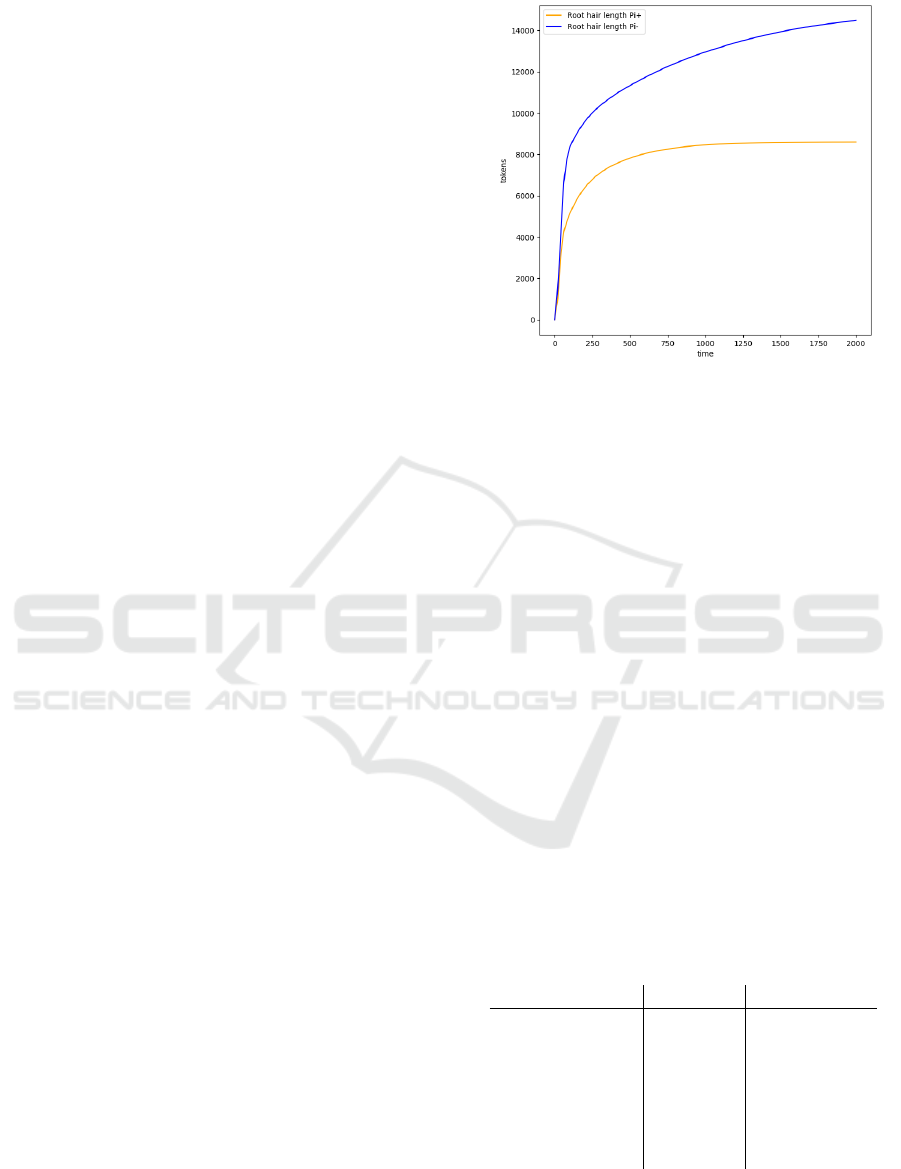
Figure 5: Root hair length given a low and a high concen-
tration of Pi.
dependent pathway. Figure 5 shows the hair length
given a low and a high concentration of Pi. While
Pi cannot directly influence hair length, the previous
figures indicated the effect that Pi has on the proteins
and hormones, which eventually determine the root
hair length. The hair growth does not follow a steady
rate as described in the literature. Instead, a high ini-
tial growth rate can be observed, likely due to the
high number of tokens accumulated in RSL2, and to a
lesser extent EIN3, that cannot immediately flow to-
wards downstream growth factors because RSL4 pro-
tein is required through the read arcs.
4.2 Validation
For various hormones and proteins, literature has re-
ported fold changes in their abundance given a lower
or higher concentration of Pi. To validate our net-
work we compare the results between low and high
Pi concentration simulations to those obtained from
literature.
Table 1: The relative abundance of hormones and proteins
in low Pi conditions compared to a high Pi conditions.
Literature Our Petri Net
RSL4 mRNA 1.8 2.3
RSL4 protein 2.5 2.0
RSL2 2.3 1.5
ARF19 2.0 2.3
Auxin 2.5 2.3
EIN3 2.0 2.3
Root hair length 1.7 1.7
Table 1 shows that our model achieves compara-
ble results to some protein and hormone values ob-
tained from the literature (e.g., ARF19, EIN3) yet
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
534

lacks in accuracy when compared to others. For ex-
ample, the result obtained for RSL2 appears biolog-
ically out of bounds when considering the respective
literature (Bhosale et al., 2018). Quantitative inaccu-
racies are due to incapable of capturing the intricate
interplay between these proteins and hormones from
limited available knowledge in general. Moreover, the
validation set was not obtained from a single study,
which could have affected the compatibility of simu-
lating these in a single model.
5 CONCLUSION
We developed a Petri Net to simulate the regulatory
network governing root hair length in A. thaliana,
with specific attention to soil Pi availability. Our
model, based on stochastic properties of biological
processes, incorporates key regulatory components of
the system, resulting in outputs that closely align with
experimental data reported in the literature. This in-
cludes the relative size of root hair, the amount of
hormones and their response factors in the cell, and
the pulse-like expression of the key regulator RSL4,
including both its mRNA and protein. Given the
scarcity of review articles on this topic, our model
advances the current understanding of this biological
process. It also highlights critical gaps in the exist-
ing knowledge. Further research should focus on the
regulation of RSL2, as understanding RSL4 regula-
tion alone is insufficient to fully replicate all aspects
of root hair growth. Obtaining these data could aid in
addressing some of the quantitative inaccuracies. Ad-
ditionally, the permanent transcriptional repression of
RSL4 should be further investigated, and it may be
linked to epigenetic modifications initiated by down-
stream genes. Due to the lack of this information,
we had to opt for a simplistic approach of using a
scheduled transition to prevent infinite Pi response.
This data could additionally provide the information
needed for better timing of the net, as we were not
able to fully replicate the sequence of the events in
time. Additional data needed to circumvent these
discrepancies includes the rate of proteasomal degra-
dation of RSL4, relative binding affinities of RSL4
to different components, and the stability of RSL4
mRNA. Despite this, the key features of the process
are evident. This is particularly true for the expres-
sion of RSL4 mRNA in the initial stage, while RSL4
protein is synthesized over longer period of time but
with gradual decrease in abundance.
Our model represents an initial step towards con-
structing a comprehensive plant root system model.
Future enhancements should incorporate missing ge-
netic data and account for responses to other soil nu-
trients, such as iron and bioavailable nitrogen, given
their interdependent effects (Crombez et al., 2019;
Liu et al., 2019). Integrating this genetic model with
morphological root models, such as SimRoot (Postma
et al., 2017), could yield a robust exploratory tool for
simulating plant responses to diverse ecological vari-
ables. Considering the conserved nature of Pi sens-
ing mechanisms between the model plant A. thaliana
and other plants, including crops like rice and maize
(Hwang et al., 2017; Ren et al., 2023), comprehensive
models could become perfect tools for accelerating
the development of plants with optimized agronomic
traits.
REFERENCES
Abel, S., N”urnberger, T., Ahnert, V., Krauss, G.-J., and
Glund, K. (2000). Induction of an extracellular cyclic
nucleotide phosphodiesterase as an accessory ribonu-
cleolytic activity during phosphate starvation of cul-
tured tomato cells. Plant Physiology, 122(2):543–552.
Bariola, P. A., Howard, C. J., Taylor, C. B., Verburg, M. T.,
Jaglan, V. D., and Green, P. J. (1994). The Arabidop-
sis ribonuclease gene RNS1 is tightly controlled in re-
sponse to phosphate limitation. The Plant Journal,
6(5):673–685.
Bates, T. R. and Lynch, J. P. (2000). Plant growth and
phosphorus accumulation of wild type and two root
hair mutants of Arabidopsis thaliana (Brassicaceae).
American Journal of Botany, 87(7):958–963.
Bhosale, R., Giri, J., Pandey, B. K., Giehl, R. F. H., Hart-
mann, A., Traini, R., Truskina, J., Leftley, N., Han-
lon, M., Swarup, K., Rashed, A., Voß, U., Alonso,
J., Stepanova, A., Yun, J., Ljung, K., Brown, K. M.,
Lynch, J. P., Dolan, L., Vernoux, T., Bishopp, A.,
Wells, D., von Wir
´
en, N., Bennett, M. J., and Swarup,
R. (2018). A mechanistic framework for auxin depen-
dent Arabidopsis root hair elongation to low external
phosphate. Nature Communications, 9(1).
Bl
¨
atke, M. A., Heiner, M., and Marwan, W. (2011). Tutorial
- Petri nets in systems biology. Technical report.
Chaouiya, C. (2007). Petri net modelling of biological net-
works. Briefings in Bioinformatics, 8(4):210–219.
Crombez, H., Motte, H., and Beeckman, T. (2019). Tackling
plant phosphate starvation by the roots. Developmen-
tal Cell, 48(5):599–615.
Cui, M. Q., Xu, C., Wang, T., Zhao, L. H., Wang, Y. X., Li,
G. X., Yan, J. Y., Xu, J. M., Liu, R., Wang, Z. Y., Har-
berd, N. P., Zheng, S. J., and Ding, Z. J. (2024). An
LRH-RSL4 feedback regulatory loop controls the de-
terminate growth of root hairs in Arabidopsis. Current
Biology, 34(2):313–326.e7.
Datta, A., Shrestha, S., Ferdous, Z., and Win, C. C. (2014).
Strategies for Enhancing Phosphorus Efficiency in
Crop Production Systems, page 59–71. Springer In-
dia.
Petri Net Modeling of Root Hair Response to Phosphate Starvation in Arabidopsis Thaliana
535

Datta, S., Prescott, H., and Dolan, L. (2015). Intensity of
a pulse of RSL4 transcription factor synthesis deter-
mines Arabidopsis root hair cell size. Nature Plants,
1(10).
Feng, Y., Xu, P., Li, B., Li, P., Wen, X., An, F., Gong,
Y., Xin, Y., Zhu, Z., Wang, Y., and Guo, H. (2017).
Ethylene promotes root hair growth through coordi-
nated EIN3/EIL1 and RHD6/RSL1 activity in Ara-
bidopsis. Proceedings of the National Academy of Sci-
ences, 114(52):13834–13839.
F
¨
ohse, D., Claassen, N., and Jungk, A. (1991). Phospho-
rus efficiency of plants: Ii. significance of root ra-
dius, root hairs and cation-anion balance for phos-
phorus influx in seven plant species. Plant and Soil,
132(2):261–272.
Han, G., Wei, X., Dong, X., Wang, C., Sui, N., Guo, J.,
Yuan, F., Gong, Z., Li, X., Zhang, Y., Meng, Z.,
Chen, Z., Zhao, D., and Wang, B. (2019). Arabidop-
sis ZINC FINGER PROTEIN1 acts downstream of
GL2 to repress root hair initiation and elongation by
directly suppressing bHLH genes. The Plant Cell,
32(1):206–225.
Hardy, S. and Robillard, P. N. (2004). Modeling and
simulation of molecular biology systems using Petri
nets: Modeling goals of various approaches. Jour-
nal of Bioinformatics and Computational Biology,
02(04):619–637.
Heiner, M., Gilbert, D., and Donaldson, R. (2008).
Petri Nets for Systems and Synthetic Biology, page
215–264. Springer Berlin Heidelberg.
Heiner, M., Herajy, M., Liu, F., Rohr, C., and Schwarick,
M. (2012). Snoopy – A Unifying Petri Net Tool, page
398–407. Springer Berlin Heidelberg.
Hopkins, B. G. and Hansen, N. C. (2019). Phosphorus man-
agement in high-yield systems. Journal of Environ-
mental Quality, 48(5):1265–1280.
Hwang, Y., Choi, H.-S., Cho, H.-M., and Cho, H.-T. (2017).
Tracheophytes contain conserved orthologs of a ba-
sic helix-loop-helix transcription factor that modulate
root hair specific genes. The Plant Cell, 29(1):39–53.
Kayoumu, M., Iqbal, A., Muhammad, N., Li, X., Li, L.,
Wang, X., Gui, H., Qi, Q., Ruan, S., Guo, R., Zhang,
X., Song, M., and Dong, Q. (2023). Phosphorus avail-
ability affects the photosynthesis and antioxidant sys-
tem of contrasting low-P-tolerant cotton genotypes.
Antioxidants, 12(2):466.
Liu, B., Wu, J., Yang, S., Schiefelbein, J., and Gan, Y.
(2019). Nitrate regulation of lateral root and root
hair development in plants. Journal of Experimental
Botany, 71(15):4405–4414.
Mangano, S., Denita-Juarez, S. P., Marzol, E., Borassi,
C., and Estevez, J. M. (2018). High auxin and
high phosphate impact on RSL2 expression and ROS-
homeostasis linked to root hair growth in Arabidopsis
thaliana. Frontiers in Plant Science, 9.
Marwan, W., Rohr, C., and Heiner, M. (2011). Petri Nets
in Snoopy: A Unifying Framework for the Graphi-
cal Display, Computational Modelling, and Simula-
tion of Bacterial Regulatory Networks, page 409–437.
Springer New York.
Montiel, G., Gantet, P., Jay-Allemand, C., and Breton, C.
(2004). Transcription factor networks. Pathways to
the knowledge of root development. Plant Physiology,
136(3):3478–3485.
Murata, T. (1989). Petri nets: Properties, analysis and ap-
plications. Proceedings of the IEEE, 77(4):541–580.
Postma, J. A., Kuppe, C., Owen, M. R., Mellor, N., Grif-
fiths, M., Bennett, M. J., Lynch, J. P., and Watt, M.
(2017). OpenSimRoot: widening the scope and appli-
cation of root architectural models. New Phytologist,
215(3):1274–1286.
Ren, M., Li, Y., Zhu, J., Zhao, K., Wu, Z., and Mao, C.
(2023). Phenotypes and molecular mechanisms un-
derlying the root response to phosphate deprivation in
plants. International Journal of Molecular Sciences,
24(6):5107.
Roldan, M., Dinh, P., Leung, S., and McManus, M. T.
(2013). Ethylene and the responses of plants to phos-
phate deficiency. AoB Plants, 5(0):plt013–plt013.
Salazar-Henao, J. E., V
´
elez-Berm
´
udez, I. C., and Schmidt,
W. (2016). The regulation and plasticity of root
hair patterning and morphogenesis. Development,
143(11):1848–1858.
Savage, N., Yang, T. J. W., Chen, C. Y., Lin, K.-L.,
Monk, N. A. M., and Schmidt, W. (2013). Posi-
tional signaling and expression of ENHANCER OF
TRY AND CPC1 are tuned to increase root hair den-
sity in response to phosphate deficiency in Arabidop-
sis thaliana. PLoS ONE, 8(10):e75452.
Shibata, M., Breuer, C., Kawamura, A., Clark, N. M., Ry-
men, B., Braidwood, L., Morohashi, K., Busch, W.,
Benfey, P. N., Sozzani, R., and Sugimoto, K. (2018).
GTL1 and DF1 regulate root hair growth through tran-
scriptional repression of ROOT HAIR DEFECTIVE
6-LIKE 4 in Arabidopsis. Development, 145(3).
Song, L., Yu, H., Dong, J., Che, X., Jiao, Y., and Liu,
D. (2016). The molecular mechanism of ethylene-
mediated root hair development induced by phosphate
starvation. PLOS Genetics, 12(7):e1006194.
Vissenberg, K., Claeijs, N., Balcerowicz, D., and Schoe-
naers, S. (2020). Hormonal regulation of root
hair growth and responses to the environment in
Arabidopsis. Journal of Experimental Botany,
71(8):2412–2427.
Xiao, F., Gong, Q., Zhao, S., Lin, H., and Zhou, H. (2021).
MYB30 and ETHYLENE INSENSITIVE3 antagonis-
tically modulate root hair growth in Arabidopsis. The
Plant Journal, 106(2):480–492.
Yi, K., Menand, B., Bell, E., and Dolan, L. (2010). A ba-
sic helix-loop-helix transcription factor controls cell
growth and size in root hairs. Nature Genetics,
42(3):264–267.
Zhu, S., Est
´
evez, J. M., Liao, H., Zhu, Y., Yang, T., Li,
C., Wang, Y., Li, L., Liu, X., Pacheco, J. M., Guo,
H., and Yu, F. (2020). The RALF1–FERONIA com-
plex phosphorylates eIF4E1 to promote protein syn-
thesis and polar root hair growth. Molecular Plant,
13(5):698–716.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
536
