
CAMMA: A Deep Learning-Based Approach for Cascaded Multi-Task
Medical Vision Question Answering
Teodora-Alexandra Toader
a
, Alexandru Manole
b
and Gabriela Czibula
c
Department of Computer Science, Babes-Bolyai University, Mihai Kogalniceanu nr.1, Cluj-Napoca, Romania
Keywords:
Medical Visual Question Answering, Multi-Task Learning, Deep Learning.
Abstract:
Medical Visual Question Answering is a multi-modal problem which combines visual and language informa-
tion to address medical inquiries, offering potential benefits in computer-aided diagnosis and medical educa-
tion. Deep Learning has proven effective in this area, however the scarcity of data remains an issue for this
data-hungry approach. To tackle this, we propose CAMMA, a cascaded multi-task architecture for Medical Vi-
sual Question Answering, achieving state-of-the-art results on the OVQA dataset with 71.45% accuracy. The
model has all the advantages of a multi-task network, reducing overfitting and increasing data efficiency by
capitalizing on the additional output information for each input sample. To test the adaptability of our model,
we apply the same method on the VQA-Med 2019 dataset. We experiment with the choice of objectives in-
cluded in the multi-task framework and the weighting between them.
1 INTRODUCTION
Medical Visual Question Answering (MVQA) is
an emerging field within the multi-modal vision-
language domain with impressive applications in the
healthcare sector that can lead to increased accessi-
bility by providing second opinions to medical pro-
fessionals or assisting patients with their questions.
The aim of MVQA is to combine the inputs of tex-
tual and visual nature and understand them in order
to provide an informed and correct answer that takes
into account all the available information. The most
common approach in MVQA is using deep learning
(DL) models that can learn a joint representation of
the inputs to either generate or classify the answer.
Unlike the general task of Visual Question An-
swering (VQA) that has been more widely explored,
the MVQA task has the additional challenge of harder
to obtain data, given the domain, which leads to
smaller datasets. The smaller amounts of data can
generate issues such as overfitting and less generaliza-
tion when developing DL models for solving this task.
Moreover, simply applying the VQA state of the art
models on these sub-tasks is difficult as the best per-
forming models (Chen et al., 2023a) are jointly pre-
trained on very large amounts of data which does not
a
https://orcid.org/0009-0008-6447-5001
b
https://orcid.org/0009-0002-8728-5688
c
https://orcid.org/0000-0001-7852-681X
directly translate to medical data. However, transfer
learning has been successfully applied on the MVQA
task too. As many papers approach this problem as
a classification one and use a separate text and im-
age encoder in their methods, using pre-trained en-
coders and fusing the extracted features helped to ob-
tain good results. For text feature extraction most
literature use powerful transformers such as BERT,
however for image feature extraction the vision trans-
formers are not as explored, and Convolutional Neu-
ral Networks (CNNs) seem to be the most frequent
choice in this type of architecture.
Multi-task learning (MTL) (Caruana, 1997) is a
specific machine learning paradigm where the over-
all objective is formulated as a combination of two
or more task-specific loss functions, each correspond-
ing to a learning task. These tasks can be heteroge-
neous (i.e., combining classification with detection)
or homogeneous (i.e., combining multiple classifi-
cation tasks). In recent years, numerous such ap-
proaches were proposed, most of which can be de-
scribed as one of three categories: cascaded, paral-
lel and cross-talk. MTL is a paradigm with impres-
sive results in multiple domains, including the medi-
cal field (Zhao et al., 2023). The task of MVQA has
been only recently addressed with MTL.
In this paper, we propose a multi-task model
named CAMMA for the MVQA task that uses other an-
notations that most MVQA datasets have, such as
Toader, T.-A., Manole, A. and Czibula, G.
CAMMA: A Deep Learning-Based Approach for Cascaded Multi-Task Medical Vision Question Answering.
DOI: 10.5220/0013109100003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 193-200
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
193

question type, answer type, and organ type. The
model consists of a classic MVQA architecture in-
cluding a text encoder, an image encoder, for which
we experiment with pre-trained vision transformers
and obtain the best results using a Swin (Liu et al.,
2021) based transformer, and a fusion algorithm.
The experimental evaluation is performed on litera-
ture datasets, OVQA and VQA-Med 2019. First, the
CAMMA model is tested on the OVQA dataset on which
it achieved state-of-the-art results, then it is used on
the VQA-Med 2019 to assess its generalization ca-
pabilities. As far as we are aware, the CAMMA model
presented in this paper is new in the MVQA litera-
ture. In our research we aim to give conclusive an-
swers to the following research questions, answers
that can lead to the development of enhanced models
for solving the MVQA task: RQ1. Does multi-task
learning work as a method to improve generalization
and reduce overfitting for models developed for solv-
ing MVQA?; RQ2. Does the additional information
embedded in our multi-task approach lead to an en-
hanced performance of the model?; and RQ3. Would
symbiotic tasks for an MVQA multi-task approach be
useful for increasing model accuracy compared to the
single-task approach?
The structure of this paper is the following. Sec-
tion 2 presents the current state of the field by high-
lighting recent work. Section 3 will provide a more in
depth description of our proposed method while Sec-
tion 4 will cover the conducted experiments, their re-
sults and how they compare to other methods in the
literature. Section 5 will conclude the paper and also
propose some promising future research directions.
2 RELATED WORK
2.1 Medical Visual Question-Answering
MVQA is a task that combines both the Natural Lan-
guage Processing (NLP) domain and Computer Vi-
sion (CV) while having the added challenge of data
scarcity. The MVQA task has been tackled as both a
classification problem in which each possible answer
is a class or as a generation problem in which the re-
sponse is openly generated by the model.
Many architectures have been developed for
MVQA. One type of approach is developing image
classification models that integrate information from
question as well, but not by fusing the two. (Al-Sadi
et al., 2019) proposed creating multiple image classi-
fication models, one for each question category, and
then using pattern matching on the question to de-
duce the model that has to be used. (Liao et al., 2020)
proposed a multi-task image classification model that
first uses Skeleton-based Sentence Mapping (SSM) to
map similar questions into a unified backbone from
which certain information such as modality, existence
of abnormality, type of abnormality can be extracted.
(Gong et al., 2021b) focused only on the image fea-
ture extraction and transformed the task into an im-
age classification task as the dataset for the Image-
CLEF 2021 competition focuses on abnormality ques-
tions. They achieved the best results using a Mixup
(Huang et al., 2020) strategy for data augmentation,
label smoothing and curriculum learning.
Another base architecture that is used as a starting
point for most research approaching this task is using
a text encoder and an image encoder to extract fea-
tures from the two types of inputs independently and
then fusing these features using a fusion algorithm to
learn a shared representation that is then used as input
to a classifier for the final answer. The two highest-
ranking teams at the ImageCLEF 2019 competition
for the VQA-Med task combined features extracted
from image and text using a fusion algorithm. Yan et
al. (Yan et al., 2019) used a VGG-16 inspired network
combined with Global Average Pooling (GAP) for
image feature extraction and the basic BERT model
as the question encoder. The fusion of the two types
of features extracted was achieved by using multi-
modal factorized bilinear pooling with co-attention
(Yu et al., 2017). (Nguyen et al., 2019) aimed to over-
come the data limitation and proposed a Long-Short
Term Memory (LSTM) network to extract features
from the question and extracts the image features by
using a Mixture of Enhanced Visual Features (MEVF)
module. The features are combined using an atten-
tion based fusion method and the output is fed into the
classifier. The MEVF module makes use of two im-
portant components: Model-Agnostic Meta-Learning
(MAML) that helps to learn quickly adaptable meta-
weights and Convolutional Denoising Auto-Encoder
(CDAE) which is trained on a large amount of images
collected by the authors and thus is able to add the
learnt information into the model without the need of
extra annotations. (Do et al., 2021) introduced MMQ
model (Multiple Meta-model Quantifying) that uses a
special module for image feature extraction composed
of three sub-modules: meta-training, data refinement
and meta-quantifying.
A method that makes great use of transformer ca-
pabilities is proposed by (Khare et al., 2021), where
the authors propose Multimodal Medical BERT
(MMBERT), a BERT like architecture that is pre-
trained using self-supervised learning. The model is
pre-trained on medical images and their correspond-
ing captions using MLM (Masked Language Model-
ing). The image features are extracted and the cap-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
194

tions are modified by replacing medical terms with the
[MSK] token and then the embeddings are obtained
using BERT. The obtained embeddings are passed
through a BERT-like encoder and then a classifier
is used to predict the initially masked word. An-
other approach that provides a solution for MVQA by
using the transformer architecture and pre-trainig is
presented by (Chen et al., 2023b) where the authors
proposed the PTUnifier model. They introduced an
approach to unify two medical vision language pre-
training paradigms: learning the joint vision-language
representation and learning the visual representation
from text. They also introduced two prompt pools,
one for visual tokens and one for textual tokens in or-
der to make the model be able to perform text only,
image only and image-text tasks.
(Van Sonsbeek et al., 2023) stir away from the
classification based methods for MVQA and propose
a model tailored for open answers. Their approach
encodes the image into a set of learnable tokens and
adds it as a prefix to the question before using a lan-
guage model to obtain the answer. For image encod-
ing they use a pre-trained vision encoder to obtain the
features which are then passed through a small map-
ping network and transformed into the visual prefix.
They obtained the best results using the GPT-2 model.
2.2 Multi-Task MVQA
(Gong et al., 2021a) leveraged multi-task learning in
order to create a performant image encoder. The pro-
posed model combines image understanding, which
depending on the dataset can be either image type
classification or semantic segmentation, with a novel
task: image-question compatibility. For the latter, the
combined visual and natural language features, fused
through a newly introduced attention-based module,
are used to classify whether or not the given question
is related to the input image.
The visual features are obtained through the con-
catenation of three feature maps, each obtained with
a ResNet-34 architecture, pre-trained on an external
dataset with the weights obtained from the image un-
derstanding task. The question is encoded through the
use of an LSTM. Both visual and language features
are fed into the cross-modal self-attention module in
order to perform the compatibility measurement, in-
troduced to embed a better understanding regarding
the relation between image and question in the model.
The resulting image encoder was used to improve the
performance of VQA on the VQA-RAD (Lau et al.,
2018) dataset by around 2% accuracy. (Cong et al.,
2022) proposed a complex multi-task framework for
VQA on the same dataset. In order to fully under-
stand the visual information, a captioning component
is added to the network. The extracted image fea-
tures, the caption features, and the attention maps
used to obtain the caption from the image are com-
bined through a cross-module attention-based block.
These resulting dense visual features are combined
with BERT extracted question features in order to ob-
tain the answer to the given question.
3 APPROACH
This section presents our proposed model for MVQA
named CAMMA (CAscaded Multi-Task Medical visual
question Answering). As stated before, we train and
evaluate the method on two datasets. We develop the
architecture using the OVQA dataset and then take
the approach and replicate it on the other dataset with
small changes to the multi-task approach, namely the
selected tasks due to the nature of the dataset and the
weighting between the losses.
The tasks addressed in this paper are classification
tasks. For example, for the OVQA dataset (Huang
et al., 2022), we obtained the best results using four
tasks, one being the answer classification for the final
response and the others answer type, question type
and image organ classifications. A simplified illustra-
tion of the MVQA task and our multi-task formula-
tion of the problem can be observed in Figure 1.
Figure 1: Overview of the MVQA task in the normal and
multi-task formulation.
Denoting by I a set of medical images, by Q a set
of questions/texts in natural language and by C a set
of classes corresponding to the considered tasks (in
our case the main answer, the image organ type, the
the answer type and the question type), the MVQA
task in a MT formulation can be formalized as the
problem of learning a mapping Φ : I xQ → C .
CAMMA: A Deep Learning-Based Approach for Cascaded Multi-Task Medical Vision Question Answering
195

3.1 Datasets
OVQA Dataset. The OVQA dataset, created by
(Huang et al., 2022), is a semi-automatically gener-
ated collection of orthopedic medical images and re-
lated QA pairs. It contains 2001 images and 19020
QA pairs, averaging 9.5 questions per image. The
dataset is split into training (2000 images, 15216 QA
pairs), validation (1235 images, 1902 QA pairs), and
testing (1234 images, 1902 QA pairs). The data cov-
ers various orthopedic body parts (hands, legs, etc).
The questions are divided into six categories:
abnormality, attribute other, conditions
presence, modality, organ system and plane.
The plane category includes both open and closed
questions relating to ten different plans. The closed
questions can be in the form of questions with yes
or no answer or given as questions with multiple
plane options mentioned. The organ system type
questions contains 129 possible answers in the whole
dataset, including closed and open type answers that
can refer to one or multiple organs.
The modality category questions inquire about
three main types of modalities: CT, MRI and X-Ray.
The closed questions have yes or no answers with the
open ones having two possible answers throughout
the dataset: CT and X-Ray. The attribute other
questions are an open question category with 377 pos-
sible answers in the dataset. In contrast, the condi-
tion category, which inquires about possible condi-
tions/diseases, is a closed questions category. The an-
swers can be in the form of yes/no or they can rep-
resent a disease that is mentioned in the question text
following a certain template.
The abnormality category includes both closed
and open questions that enquire about the normality
of the medical image. It is a complex category with
308 possible answers for the questions in this dataset.
VQA-Med-2019 Dataset. The VQA-Med-2019
dataset, introduced at ImageCLEF 2019 for the VQA
task (Abacha et al., 2019), includes 4200 images from
the MedPix database and 15,292 questions and an-
swers. It is split into training (3200 images, 12792
QA pairs), validation (500 images, 2000 QA pairs),
and testing (500 images and QA pairs).
The questions were divided into four different cat-
egories: organ, plane, modality and abnormality.
The plane category includes images in 16 different
planes. The organ category has the smallest num-
ber of classes, the possible answers to all the ques-
tions belonging to a set of ten organs and organ sys-
tems. The modality category is slightly more com-
plex than the previous two. There are 36 modali-
ties, and the question can refer to the type of modal-
ity used, either what or yes/no questions. There are
also questions related to contrast in the image, what
type of contrast is used, and specifics of MRIs. In
total, there are 44 possible answers for all modality
questions. The abnormality category includes both
closed questions that inquire about the state of the im-
age; if it is normal/abnormal, and open questions that
inquire about the abnormality shown in the picture.
The latter is the most complex, with 1485 possible
answers in the training set.
3.2 General Architecture
An overview of the architecture can be seen in Figure
2. We develop the model starting from a well estab-
lished architecture category for MVQA, namely using
two feature extractors, for text and image, and joining
the features using a fusion algorithm. We developed
the model on the OVQA dataset and then used the
same architecture to assess the generality on another
dataset, namely, VQA-Med 2019.
For the VQA-Med-2019 dataset, the general
CAMMA architecture was adapted as the dataset con-
tains no organ information and thus the classification
head responsible for that objective was removed.
The first elements of the model are therefore the
text and image encoders. In order to extract the fea-
tures from text we choose the BERT model since the
literature shows it as a very powerful choice. We use
the base uncased version of the model. For text pre-
processing we take a minimal approach by just re-
moving unnecessary space characters which results in
the question that will then be tokenized with the cor-
responding BERT tokenizer.
For the image feature extractor we experimented
with three types of models for extracting the features,
one CNN based, namely the VGG19 model and two
transformer based models: the Swin Transformer and
the Vision Transformer (ViT). Based on the obtained
results we landed on the Swin Transformer, more
specifically the base version. A more detailed descrip-
tion of the results on which we based our decision will
be presented in the next section. The pre-processing
step consists of a simple resize of the image to the
dimension required by the vision models.
The features from the question and the image
are extracted independently using these two encod-
ing models and then fused in order to obtain the
joint representation. For the fusion algorithms, we
experimented with Multi-modal Factorized Bilinear
pooling (MFB) as well as with its extended version
Multi-modal Factorized High-order pooling (MFH)
(Yu et al., 2018) with two MFB blocks (the latter is
selected for our model). After passing through the
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
196
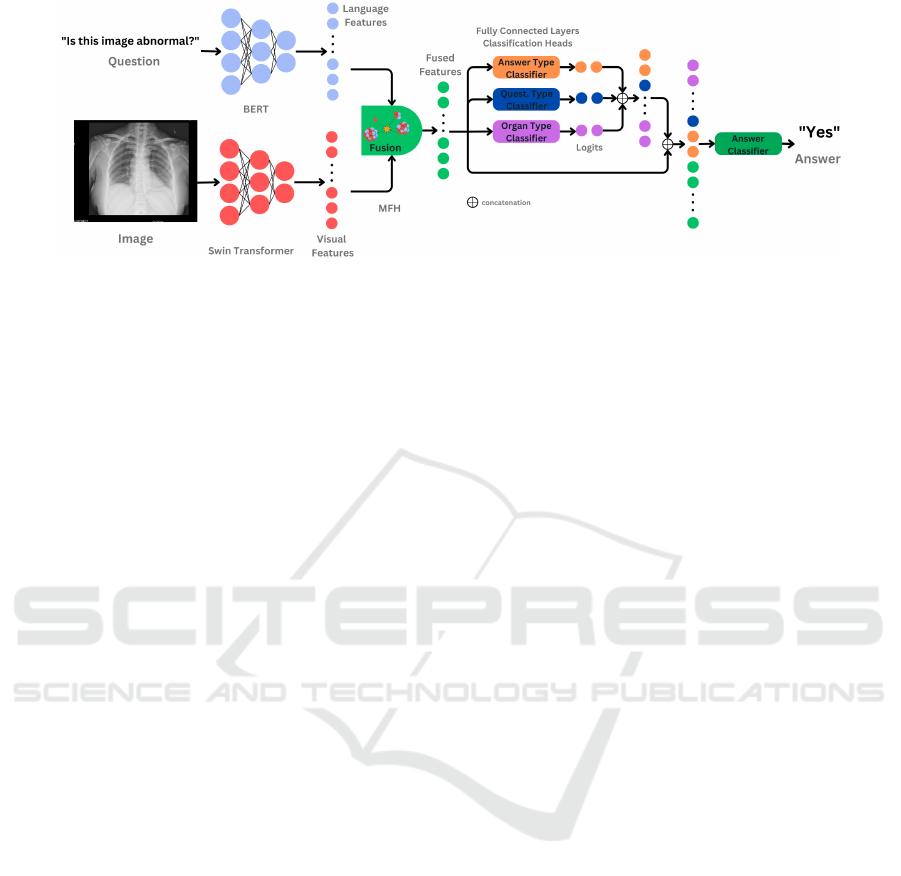
Figure 2: Overview of the proposed CAMMA model. Each component of the multi-modal input is fed into a specific encoder
(BERT for the question, respectively Swin Transformer for the image) in order to extract features. The resulting information
is combined through the use of the MFH feature fusion module. Based on the fused features three classification heads, in the
form of a fully connected dense layers, perform: answer-type, question type and organ type classification. The logits obtained
through this process are concatenated to the MFH fused features prior to the main classification task: answer classification.
MFH fusion module a joint image-text representation
is obtained that is used as input for our classifiers.
We have a total of three classifiers for the VQA-Med
2019 dataset and four for the OVQA dataset depend-
ing on the number of tasks. For all tasks except the
answer classification one, a classification head takes
the joint image-text representation in order to obtain
the correct class. For the main task we use a cascading
multi-task approach in which the output of the addi-
tional tasks is concatenated to the joint representation
in order to create the input for the classification head.
3.3 Multi-Task Learning
Our multi-task approach is based on a combination
of parallel and cascading multi-task. The chosen
tasks are the answer classification (main task) and
two or three additional tasks: question type classifica-
tion (into the categories given in each dataset), answer
type classification (referring to whether the answer is
an open or closed question). For the OVQA dataset
image organ classification is also added, as this extra
annotation is available in this dataset. We use the par-
allel MTL approach for all tasks except the main one
as all predictions are made based on the joint repre-
sentation input. For the main task we use cascading
MTL by concatenating the image-text representation
with the output of the other classifiers in order to cre-
ate the classifier input.
During training we experimented with different
methods of combining the loss functions. Our exper-
iments lead to two slightly different methods depend-
ing on the dataset that is used for training. Therefore,
for OVQA data we obtained the best results by sim-
ply summing the losses of the four tasks, while for
the VQA-Med 2019 dataset, a weighted approach in
which the main task has a higher weight proved to
be most effective, specifically a weight of 0.7 for the
answer classification and a 0.15 weight for both ques-
tion type classification and answer type classification
when using all three tasks. The weights were selected
empirically. However, the best result on VQA-Med
2019 was attained using only the main task and the
answer classification task with corresponding weights
of 0.7 and 0.3 respectively. We computed each indi-
vidual loss using Cross Entropy Loss.
4 EXPERIMENTAL EVALUATION
4.1 Experimental Setup
We evaluated our model using the two metrics that
are mostly used in the literature, namely accuracy
and BLEU score (Papineni et al., 2002). Accuracy
or overall accuracy as it appears in most evaluations
is computed as the number of correct answers over the
number of total predictions. An answer is considered
correct if it is an exact match with the ground truth.
The BLEU score is computed by counting matching
n-grams between the two sentences while taking into
account the occurrence of the words in the ground
truth text, consideration assured by the n-gram pre-
cision. In our evaluation we use overall accuracy and
BLEU1 as the metrics for our model.
Each of our models is trained using the Nvidia
T4 GPU integrated into the Google Colaboratory en-
vironment over 200 epochs on OVQA and 100 on
VQA-Med 2019, using the Adam optimizer with an
initial learning rate of 1e
−3
with a cycle scheduler to
adjust the learning rate over time during training. This
allows the model to explore the search space through
the use of a large learning rate, which is then reduced
in order to better identify local minima.
CAMMA: A Deep Learning-Based Approach for Cascaded Multi-Task Medical Vision Question Answering
197
4.2 Results and Discussion
We performed multiple experiments on the OVQA
dataset in order to decide on the best image encoder
and fusion strategy and then experimented with the
obtained architecture on both datasets to see if adding
the multitask approach provides an improvement. The
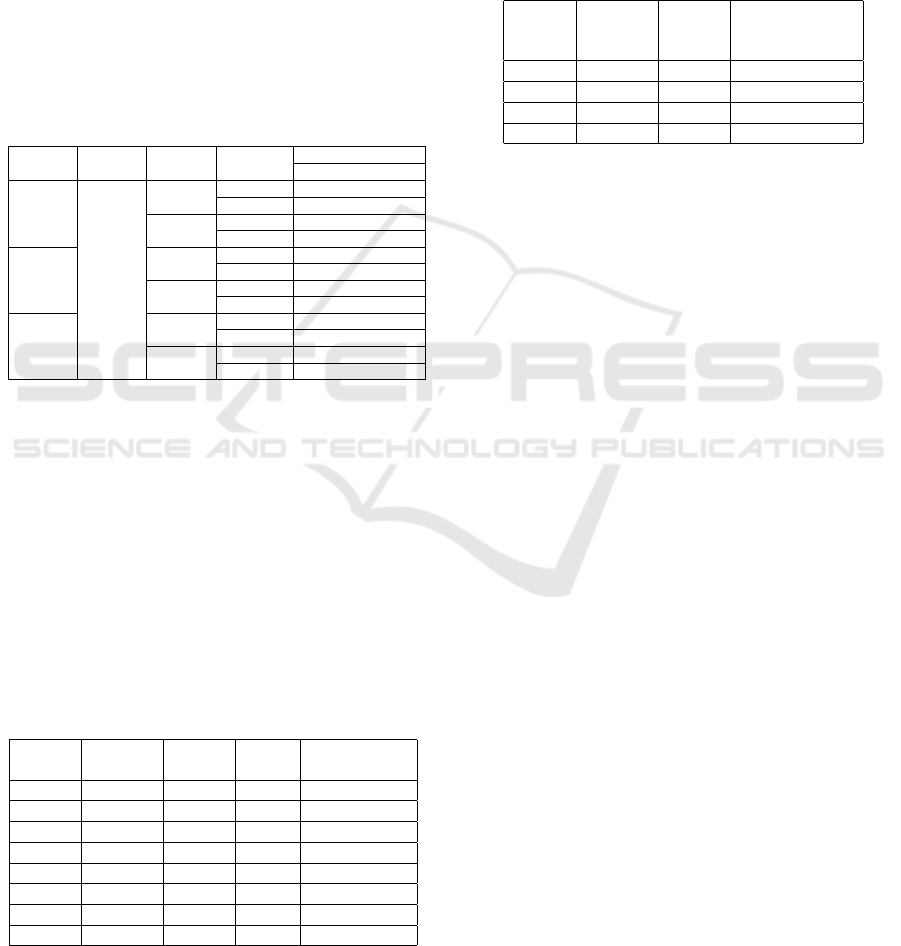
results of these experiments can be seen in Table 1.
As we can observe the best model is obtained while
using the SWIN model as an image encoder with an
MFH fusion and while integrating out multitask ap-
proach. We can also observe that between each two
models that use the same building blocks, adding the
multitask learning helps achieve an improvement.
Table 1: Performance of CAMMA based on the choice of im-
age encoder, fusion module and use of MTL.
Image Text Fusion Multitask OVQA
Encoder Encoder Strategy Accuracy BLEU
SWIN
BERT
MFH
X 0.6230 0.6784
✓ 0.7145 0.7559
MFB
X 0.5846 0.6398
✓ 0.674 0.7194
VGG19
MFH
X 0.5962 0.6543
✓ 0.6451 0.6979
MFB
X 0.5588 0.6143
✓ 0.6161 0.6731
ViT
MFH
X 0.6119 0.6683
✓ 0.6803 0.7305
MFB
X 0.5799 0.636
✓ 0.6424 0.7012
The results presented in Table 1 are for a mul-
titask approach that uses all the tasks mentioned in
Section 3. However, we would like to further discuss
the decision of choosing these tasks and see if the ob-
tained combinations lead indeed to the best informa-
tion transfer to the model. Therefore, Table 2 shows
different results obtained on the OVQA dataset with
different task combinations using the model selected
based on Table 1 results. For this case we can observe
that using all three additional tasks provides the best
result, with a quite significant difference between the
accuracy of the model without and with multi-task.
Table 2: Results on the OVQA dataset using different clas-
sification tasks selection.
main question answer image OVQA
answer type type organ Test accuracy
✓ 0.623
✓ ✓ 0.6524
✓ ✓ 0.6335
✓ ✓ 0.6482
✓ ✓ ✓ 0.6766
✓ ✓ ✓ 0.6992
✓ ✓ ✓ 0.6824
✓ ✓ ✓ ✓ 0.7145
The best architecture obtained on OVQA was ap-
plied on the VQA-Med 2019 dataset. The results of
the approach with different task combinations can be
seen on Table 3. The best result was obtained for a
multi-task approach consisting of the main task and
the answer classification task. When comparing all
multitask results with the base method we observe
an improvement for each individual task combination.
When using summing of the losses, as on OVQA, we
obtained better results for the combination of all tasks.
Table 3: Results on the VQA-Med 2019 dataset using the
same notations as in Table 2.
main
answer
question
type
answer
type
VQA-Med-2019
Test
Accuracy
✓ 0.552
✓ ✓ 0.558
✓ ✓ 0.568
✓ ✓ ✓ 0.562
The difference in results between the two datasets
from an overall improvement perspective and differ-
ent responses to the loss combination methods may be
due to the differences in how the answer type annota-
tion is done. For VQA-Med 2019, answer type refers
strictly to an affirmative or negative answer, while on
OVQA the interpretation is different since these ques-
tions can have answers other than “yes” or “no”. The
question type category also differs between the two
datasets, with OVQA having two additional classes,
particularity that might have led to a more powerful
addition in the final input for the answer classifier.
We also compared our model with other ap-
proaches from the literature on the OVQA dataset.
The approaches chosen for comparison were selected
based on performance and relevance to the field and
are all single-task approaches. Therefore, we chose
to compare with a diverse suite of models that illus-
trate the developments in solving the MVQA prob-
lem. These approaches span from feature extraction
from images and text using separate encoders, with
a focus on the image feature extraction module, like
MEVF and MMQ to methods that leverage transform-
ers, such as MMBERT and PTUnifier, which use pre-
training on multimodal data and even generative pro-
posals that include generating open-ended answers
by combining visual features with language models
like GPT-2. All of the mentioned methods find inge-
nious ways to overcome data limitations, however we
believe that the extra annotations provided can con-
tribute even more to reduce the challenges of data
scarcity, especially when leveraged in a MTL ap-
proach. Given the advanced current state of largely
available multimodal models we also considered of
interest to compare our work with one of these gen-
eral models, specifically GPT-4o. We obtained the
answers by using the OpenAI API and providing the
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
198

image and the question preceded by a prompt. We ob-
tained the best results with the following prompt: “I
am working on visual question answering using medi-
cal images. Please provide an answer to the following
question. If the question requires a yes/no answer, re-
spond with a single word only, without punctuation.
Here is the question:”
The comparative results from Table 4 show that
our model achieves state-of-the-art results, highlight-
ing that the cascading multitask addition can provide a
notable improvement. The improvement in accuracy
achieved by CAMMA with respect to the related work
from Table 4 is significant at a significance level 0.01,
as confirmed by a one-tailed paired Wilcoxon signed-
rank test.
Table 4: Comparison to related work on OVQA dataset.
The accuracy values for MEVF-SAN, MEVF-BAN and PT-
Unifier models are taken from (Hong et al., 2024), while
for MMQ-SAN, MMQ-BAN, MMBERT are taken from
(Van Sonsbeek et al., 2023).
Model Accuracy
Our CAMMA model 0.7145
MEVF-SAN (Nguyen et al., 2019) 0.6190
MEVF-BAN (Nguyen et al., 2019) 0.6100
MMQ-SAN (Do et al., 2021) 0.6850
MMQ-BAN (Do et al., 2021) 0.650
MMBERT (Khare et al., 2021) 0.6330
PTUnifier (Chen et al., 2023b) 0.7130
Generative LLM (Van Sonsbeek et al., 2023) 0.7100
OpenAI’s GPT-4o 0.3123
The comparative results are included just for the
OVQA dataset as in this paper we aimed to intro-
duce the cascaded multi-task approach for MVQA as
a proof of concept and thus, the base model that cre-
ates the joint image-text features was tailored for this
dataset. Different architectures attached to the cas-
caded multi-task module will be further investigated
to allow a proper comparison to related work on the
VQA-Med-2019 dataset.
As previously shown, our proposed strategy ob-
tains advanced results on the OVQA dataset and on
both datasets we can see that the cascading multi-task
learning addition generates better results. In this sub-
section we will discuss these results in more depth.
Figure 3 illustrates, for the answer classification
task, the accuracy by certain categories on the OVQA
dataset. For different image organ classes the perfor-
mance is quite stable with a slightly smaller accuracy
for questions showing legs and an increase for chest
questions. While comparing closed and open ended
questions accuracy we can observe that the model is
better suited for answering closed ended questions
which is expected since these questions have either
a smaller pool of answers such as affirmative or neg-
ative or they contain the answer and therefore this in-
Figure 3: Performance comparison based on question cate-
gory, body part and answer type on OVQA Dataset.
formation will be present in the classifiers input.
Open ended questions are a more difficult class
of questions due to the large number of possible an-
swers and we plan to improve the performance on
this task in the future. For different question cate-
gories the worst results are for the plane, organ sys-
tem and attribute other categories which correspond
to the least represented categories in the dataset. In
order to better understand the shortcomings of the
model we analyzed the most frequent miss-predicted
answers. We observed that for the organ system cate-
gories some of these answers are correct predictions,
but the way in which punctuation is used in the train-
ing and test classes does not create an exact match.
For example, our model predicts: “ulna,ulnar,and
distal radius” while the answer in the test set
is “ulna,ulnar and distal radius” which dif-
fers just by the use of a comma, and this is not an
isolated case. We did not alter the text in any way as
an exact match is used, and we do not want to create
an unfair advantage to other works that might have
been using the exact string in the datasets. However,
we computed the accuracy on the cleaned strings and
we observed that it increased to 0.7297.
In Figure 4 we can see the broken down results
for the VQA-Med 2019 dataset. As we can observe
the worst performing category is the abnormality one
which is the case for most papers using this dataset.
The large number of possible answers in addition to
the possible answers in the test dataset that are not
present in the training data contribute to this short-
coming of the model. However, steps can be made to
create a better model such as using augmentations.
To conclude, the research questions formulated
in Section 1 have been answered. As an answer to
RQ1, through the experimental evaluation of our pro-
Figure 4: Performance comparison based on question cate-
gory and answer type VQA-Med-2019.
CAMMA: A Deep Learning-Based Approach for Cascaded Multi-Task Medical Vision Question Answering
199

posed CAMMA model we highlighted that multi-task
learning is useful for improving generalization and
reduce overfitting for models developed for solving
the MVQA task. Through the performed experiments
on the OVQA and MVQA datasets it was empirically
proven, in response to RQ2, that embedding addi-
tional information into the model through the use of
multiple classification heads is beneficial for improv-
ing the model performance. In what concerns RQ3,
the experimental results highlighted that using addi-
tional tasks lead to a significant improvement in the
model accuracy compared to the single-task model.
5 CONCLUSIONS
In this paper we presented CAMMA, a cascading multi-
task architecture created for Medical Visual Question
answering that obtained state-of-the-art results on the
OVQA dataset. In our experimental set-up, multi-
task learning showed its prowess in the MVQA task
leading to improved performance and reduced over-
fitting. Although our choice of tasks is limited to
the categories for which we have annotations in the
OVQA and VQA-Med 2019 datasets, embedding ad-
ditional information into the model through the use of
multiple classification heads is a useful technique that
allows us to deal with data scarcity. A clear constant
we observe, however, is that for this task, a cascaded
approach results in increased performance suggesting
that answer classification is enhanced by knowledge
regarding question type and answer type.
Although we achieved impressive results, the
complexity of the problem allows for further improve-
ments. A future work would be to use task weights as
hyperparameters, in order to allow the model to learn
the best balance between the tasks. Additionally, we
may consider and experiment on new tasks which
could be added to the framework in order to mea-
sure their impact on the proposed approach. Since
extra classification task annotations are not available,
a self-supervised candidate such as image or question
reconstruction could be a interesting approach.
REFERENCES
Abacha, A. B., Hasan, S. A., et al. (2019). VQA-Med:
Overview of the Medical VQA Task at ImageCLEF
2019. In Working Notes of CLEF 2019, volume 2380.
Al-Sadi, A., Talafha, B., et al. (2019). JUST at ImageCLEF
2019 Visual Question Answering in the Medical Do-
main. In Working Notes of CLEF 2019, volume 2380.
Caruana, R. (1997). Multitask learning. Machine Learning,
28:41–75.
Chen, X., Wang, X., et al. (2023a). PaLI: A Jointly-Scaled
Multilingual Language-Image Model. In Proceedings
of ICLR 2023, pages 1–33.
Chen, Z., Diao, S., et al. (2023b). Towards unifying medical
vision-and-language pre-training via soft prompts. In
Proceedings of ICCV’23, pages 23403–23413.
Cong, F., Xu, S., et al. (2022). Caption-aware medical VQA
via semantic focusing and progressive cross-modality
comprehension. In MM’22, pages 3569–3577.
Do, T., Nguyen, B. X., et al. (2021). Multiple meta-model
quantifying for medical visual question answering. In
MICCAI 2021: Part V 24, pages 64–74. Springer.
Gong, H., Chen, G., et al. (2021a). Cross-modal self-
attention with multi-task pre-training for MVQA. In
Proceedings of ICMR 2021, pages 456–460.
Gong, H., Huang, R., et al. (2021b). SYSU-HCP at VQA-
Med 2021: A Data-centric Model with Efficient Train-
ing Methodology for Medical Visual Question An-
swering. In CLEF (Working Notes), pages 1218–1228.
Hong, X., Song, Z., et al. (2024). BESTMVQA: A Bench-
mark Evaluation System for Medical Visual Question
Answering. In ECML–PKDD, pages 435–451.
Huang, L., Zhang, C., and Zhang, H. (2020). Self-
adaptive training: beyond empirical risk minimiza-
tion. Advances in neural information processing sys-
tems, 33:19365–19376.
Huang, Y., Wang, X., Liu, F., and Huang, G. (2022).
OVQA: A clinically generated visual question an-
swering dataset. In SIGIR 2022, pages 2924–2938.
Khare, Y., Bagal, V., et al. (2021). Mmbert: Multimodal
bert pretraining for improved medical VQA. In Pro-
ceedings of ISBI 2021, pages 1033–1036. IEEE.
Lau, J. J., Gayen, S., et al. (2018). A dataset of clinically
generated visual questions and answers about radiol-
ogy images. Scientific data, 5(1):1–10.
Liao, Z. et al. (2020). AIML at VQA-Med 2020: Knowl-
edge inference via a skeleton-based sentence mapping
approach for MVQA. In CLEF, pages 1–14.
Liu, Z., Lin, Y., et al. (2021). Swin transformer: Hierar-
chical vision transformer using shifted windows. In
Proceedings of the IEEE/CVF, pages 10012–10022.
Nguyen, B. D. et al. (2019). Overcoming data limitation in
medical visual question answering. In Proceedings of
MICCAI 2019, Part IV 22, pages 522–530.
Papineni, K., Roukos, S., and othrs (2002). Bleu: a Method
for Automatic Evaluation of Machine Translation. In
The 40th Annual meeting of the ACL, pages 311–318.
Van Sonsbeek, T., Derakhshani, M. M., et al. (2023). Open-
ended MVQA through prefix tuning of language mod-
els. In Proceedings of MICCAI 2023, pages 726–736.
Yan, X. et al. (2019). Zhejiang University at ImageCLEF.
In CLEF (Working Notes), volume 2380, pages 1–9.
Yu, Z. et al. (2017). Multi-modal factorized bilinear pooling
with co-attention learning for VQA. In Proceedings of
ICCV’17, pages 1821–1830.
Yu, Z. et al. (2018). Beyond bilinear: Generalized multi-
modal factorized high-order pooling for VQA. IEEE
Tran. Neural Netw. Learn. Syst., 29(12):5947–5959.
Zhao, Y., Wang, X., et al. (2023). Multi-task deep learning
for medical image computing and analysis: A review.
Computers in Biology and Medicine, 153:106496.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
200
