
Cross-Domain Transfer Learning for Domain Adaptation in Autism
Spectrum Disorder Diagnosis
Kush Gupta
a
, Amir Aly
b
and Emmanuel Ifeachor
c
University of Plymouth, Plymouth, U.K.
Keywords:
Cross-Domain Transfer Learning, Autism Diagnosis, Vision Transformers.
Abstract:
A cross-domain transfer learning approach is introduced to address the challenges of diagnosing individu-
als with Autism Spectrum Disorder (ASD) using small-scale fMRI datasets. Vision Transformer (ViT) and
TinyViT models pre-trained on the ImageNet, were employed to transfer knowledge from the natural image
domain to the brain imaging domain. The models were fine-tuned on ABIDE and CMI-HBN, using a teacher-
student framework with knowledge distillation loss. Experimental results demonstrated that our method out-
performed previous studies, ViT models, and CNN-based models. Our approach achieved competitive per-
formance (F-1 score 78.72%) with a much smaller parameter size. This study highlights the effectiveness
of cross-domain transfer learning in medical applications, particularly for scenarios with small datasets. It
suggests that pre-trained models can be leveraged to improve diagnostic accuracy for neuro-developmental
disorders such as ASD. The findings indicate that the features learned from natural images can be adapted to
fMRI data using the proposed method, potentially providing a reliable and efficient approach to diagnosing
autism.
1 INTRODUCTION
Autism is a neuro-developmental condition that af-
fects brain development and can typically be identi-
fied as early as 16 months of age (Ali, 2020). In the
current era, the global incidence of autism is on the
rise. Statistics indicate that 1 in 68 children are diag-
nosed with autism, with boys at higher risk, showing
a prevalence rate of four boys for every girl (Lahiri
et al., 2011). However, detecting autism is chal-
lenging because autistic children do not have distinct
physical characteristics. Typically, doctors utilize a
screening tool to assess the likelihood of autism in
children between 16 and 30 months of age (Sharda
et al., 2016); (Jennings Dunlap, 2019).
Inaccurate diagnoses of children with ASD have
often resulted from insufficient experience and train-
ing among doctors (Manaswi et al., 2018). Traditional
diagnostic methods are based on subjective behav-
ioral assessments (ADOS)
1
, which can lead to errors
a
https://orcid.org/0009-0008-9930-6435
b
https://orcid.org/0000-0001-5169-0679
c
https://orcid.org/0000-0001-8362-6292
1
The ADOS is a partially systematic diagnosis tool de-
signed by (Lord et al., 2000). ADOS score is used to deter-
mine the severity of autism.
in the identification of neuro-developmental disorders
such as ASD. These challenges can hinder pediatric
screening efforts, as no straightforward method cur-
rently exists for diagnosing ASD. An accurate diag-
nosis requires frequent follow-up with each patient
with ASD to ensure reliable results. Measurable
approaches, such as functional Magnetic Resonance
Imaging (fMRI), have become a focus of research,
with fMRI being recognized as the leading method
for identifying ASD (Klin, 2018). fMRI can pro-
vide objective, measurable biomarkers of brain activ-
ity (Traut et al., 2022), thus reducing the reliance on
subjective observations.
The scientific and clinical usefulness of computer-
assisted diagnosis (CAD) powered by machine learn-
ing has gained increasing recognition over the past
two decades. Deep neural networks (DNNs) are used
to extract compact, fixed-dimensional feature repre-
sentations from large-scale public datasets. Through
transfer learning, these representations are then em-
ployed to fine-tune models across various research
areas, offering improved generalizability across do-
mains. Recent studies have shown that neural net-
works and transfer learning can serve as effective
clinical tools for the prevention of mental illness
(Durstewitz et al., 2019). However, the application
Gupta, K., Aly, A. and Ifecahor, E.
Cross-Domain Transfer Learning for Domain Adaptation in Autism Spectrum Disorder Diagnosis.
DOI: 10.5220/0013113000003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 53-64
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
53

of transfer learning techniques to the investigation
of autism spectrum disorder (ASD) has seen sparse
progress. This limitation is partly because ASD is
a diverse neuro-developmental condition character-
ized by complex cognitive features (Cao and Cao,
2023). Consequently, there are significant challenges
in gathering data on individuals with ASD and de-
veloping reliable CAD systems. fMRI data from
539 individuals with ASD and 573 matched controls
were compiled by the Autism Brain Imaging Data
Exchange (ABIDE) (Di Martino et al., 2014) con-
sortium. The data was sourced from 17 sites, cre-
ating an unparalleled opportunity for extensive ASD
research. In our work, we used the ABIDE dataset
along with the fMRI dataset provided by the Healthy
Brain Network of the Child Mind Institute (CMI-
HBN) (Alexander et al., 2017). CMI-HBN includes
publicly shared identified data on various behavioral,
psychiatric, cognitive and lifestyle factors (such as fit-
ness and diet), along with multimodal brain imaging
(MRI), electroencephalography (EEG), digital video
and voice recordings, and genetic information.
Among many researchers, convolutional neural
networks (CNN) were commonly employed to design
CAD systems based on fMRI data for differentiating
people with ASD from total control (TC), using the
ABIDE database (Husna et al., 2021). They extracted
key features to analyze and differentiate patients with
ASD from total control (TC) (Manaswi et al., 2018);
(Sherkatghanad et al., 2020). CNNs have been the
cornerstone of many deep learning applications, par-
ticularly in computer vision. However, they have sev-
eral limitations; for example, they rely on convolu-
tional layers that process local regions of an image
through small receptive fields. While it enables CNNs
to identify spatial hierarchies, their capability to de-
tect long-range relationships or global context in an
image is restricted. Moreover, CNNs have a strong in-
ductive bias due to their architecture, which assumes
that local spatial relationships are the most important.
This bias limits their ability to learn more complex
or abstract features. Furthermore, these approaches
struggled because they were data-driven and relied
on the availability of large image data to train their
model. Moreover, training deep learning models from
scratch is known to be computationally demanding,
time-consuming, and often requires powerful hard-
ware. These above-outlined issues are addressed in
our approach through two key strategies:
1. We employed cross-domain transfer learning
along with knowledge distillation (KD) loss to
solve the need for a large fMRI dataset to train
a deep neural network. We used the transfer
learning technique, where a model that has been
developed for one task is used as the starting
point for a different yet related task (Pan and
Yang, 2009). By using the model which has been
pre-trained on large-scale datasets like ImageNet
(Deng et al., 2009), as a starting point. Those
features that were already learned by the model
during the pre-training task are crucial when re-
fining the model. The already trained model can
subsequently be fine-tuned on the small-scale tar-
get datasets, enhancing performance without the
need for a large amount of labeled data (Yosin-
ski et al., 2014). Since we used transfer learn-
ing along with KD loss to fine-tune a pre-trained
model rather than training one from scratch, the
time required and computational resources needed
were significantly reduced. This efficiency en-
ables faster model deployment and experimenta-
tion, making the approach accessible to those with
limited resources. Moreover, fine-tuning the pre-
trained model on a domain-specific dataset allows
the model to learn relevant features specific to
the new domain while benefiting from the gen-
eral knowledge acquired during the pre-training.
This adaptability is essential in fields like health-
care care, where direct data transfer is often chal-
lenging due to the unavailability of large-scale
databases.
2. Further, we used the TinyViT (Wu et al., 2022)
models which are a new family of compact and ef-
ficient vision transformers derived from the orig-
inal vision transformers (ViT) (Dosovitskiy et al.,
2020) which has surfaced as a powerful alterna-
tive and overcomes the limitations of CNNs and
traditional ViT models. Unlike CNNs, TinyViT
processes images as a sequence of patches, using
window-attention mechanisms that allow them
to consider relationships between all patches si-
multaneously, regardless of their spatial distance.
This enables TinyViTs to capture global context
and long-range dependencies more effectively,
improving performance in tasks where global in-
formation is critical. Also, it has a less pro-
nounced inductive bias compared to CNNs. They
do not assume that local spatial relationships al-
low them to learn more complex and abstract
features from the data. In addition, recent ViT
models have a large number of parameters, mak-
ing them less suitable for devices with limited
resources. To address this, TinyViT was pre-
trained on the ImageNet dataset using a fast dis-
tillation framework. In this process, knowledge
was transferred from larger pre-trained models
to smaller ones, enabling the smaller models to
benefit from huge pretraining data. Specifically,
HEALTHINF 2025 - 18th International Conference on Health Informatics
54

knowledge transfer is achieved through distilla-
tion during the pre-training phase. To minimize
memory and computational demands, the logits
from large teacher models were sparsified and
saved beforehand. The tiny student transform-
ers were then scaled down automatically from a
large pre-trained model, taking into account com-
putation and parameter constraints. This makes
TinyViTs more versatile than traditional ViTs and
adaptable across different tasks with small-scale
datasets, often leading to better generalization.
Our research focuses on designing a computer-
aided diagnosis method using models that are al-
ready trained on large-scale image data repositories
and employing transfer learning methods for cross-
domain adaptation to diagnose autism and attention
maps were visualized to verify that the model is learn-
ing relevant features.
The structure of this paper is as follows: the next
section presents a review of related works in this
field. Section (III) discusses the datasets utilized in
our study. The detailed methodology is covered in
section (IV), while section (V) outlines the experi-
mental settings and implementation details. In sec-
tion (VI), we present the results obtained and discuss
our key findings in section (VII). Section (VIII) offers
the conclusions drawn from our work. Lastly, section
(IX) addresses future directions, highlighting poten-
tial areas for further research and development.
2 RELATED WORKS
Significant interest has been drawn to recent advances
in detecting Autism Spectrum Disorder (ASD) utiliz-
ing fMRI data. This is largely because the ABIDE ini-
tiative has made functional and structural brain imag-
ing datasets from multiple global imaging venues ac-
cessible (Craddock et al., 2013). ABIDE was used as
the primary dataset in the development of many re-
search works (Heinsfeld et al., 2018); (Iidaka, 2015);
(Chen et al., 2016). Some researchers have selected
specific demographic subsets from this dataset to test
their proposed methods. The controlled demographic
variability helps in understanding how ASD manifests
across different groups. For instance, (Iidaka, 2015)
used a probabilistic neural network to classify rest-
ing state (rs-fMRI) data of 312 subjects with ASD
and 328 healthy controls (all under 20 years of age),
achieving an accuracy of approximately 90%. (Plitt
et al., 2015) used two sub sets of rs-fMRI data: one
consisting of 118 male subjects (59 ASD and 59 typi-
cally developing (TD)) and another with 178 individ-
uals matched by age and IQ (89 ASD and 89 TD),
achieving a classification accuracy of 76.67%.
The primary challenge in diagnosing autism us-
ing fMRI images is identifying the key features from
complex fMRI images. One effective method for
identifying ASD involves the use of a machine learn-
ing technique called Support Vector Machine (SVM).
For example, (Abraham et al., 2017) trained a sup-
port vector classifier on rs-fMRI data from the ABIDE
dataset and achieved an accuracy of 67%. Similarly,
(Monté-Rubio et al., 2018) applied an SVM algorithm
on the ABIDE dataset and reached a 62% accuracy.
Recently, neural networks including deep neural
networks (DNNs), autoencoders, and long-short-term
memory networks (LSTM) have gained substantial
popularity for their applications in the diagnosis of
ASD(Guo et al., 2017); (Bi et al., 2018); (Brown
et al., 2018); (Dvornek et al., 2017a); (Khosla et al.,
2018). For instance, (Brown et al., 2018) introduced
an element-wise layer for deep neural networks that
integrated data-driven structural priors, achieving a
classification accuracy of 68.7% on a dataset of 1013
subjects consisting of 539 healthy controls and 474
individuals with ASD. (Sherkatghanad et al., 2020)
achieved an accuracy of 70.22% with a CNN model
applied to the ABIDE dataset. Similarly, (Dvornek
et al., 2018), and (Shahamat and Abadeh, 2020)
trained a CNN model on the ABIDE dataset for ASD
classification and also reported an accuracy of ap-
proximately 70%. Hand-crafted feature extractors
form the basis of these methods, which face limita-
tions in generalizing to new image samples. Data-
driven approaches encounter difficulties and chal-
lenges related to big data (Koirala et al., 2019).
In our approach, we eliminated the need for a large
fMRI dataset to build a reliable deep neural network
by utilizing cross-domain transfer learning along with
knowledge distillation loss. Models pre-trained on
large-scale natural image datasets like ImageNet were
used. To adapt these models for classifying patients
with autism (ASD) and total controls (TC), they were
initially fine-tuned on the ABIDE dataset. Subse-
quently, the teacher-student method, combined with
knowledge distillation loss, was applied to further re-
fine the models using the CMI-HBN dataset. This
approach enhances the model’s generalization capa-
bilities for diagnosing autism spectrum disorder. The
implemented method is explained in detail in the sub-
section (5.2).
3 DATASETS
Functional Magnetic Resonance Imaging (fMRI) is
a brain imaging method that enables researchers to
Cross-Domain Transfer Learning for Domain Adaptation in Autism Spectrum Disorder Diagnosis
55
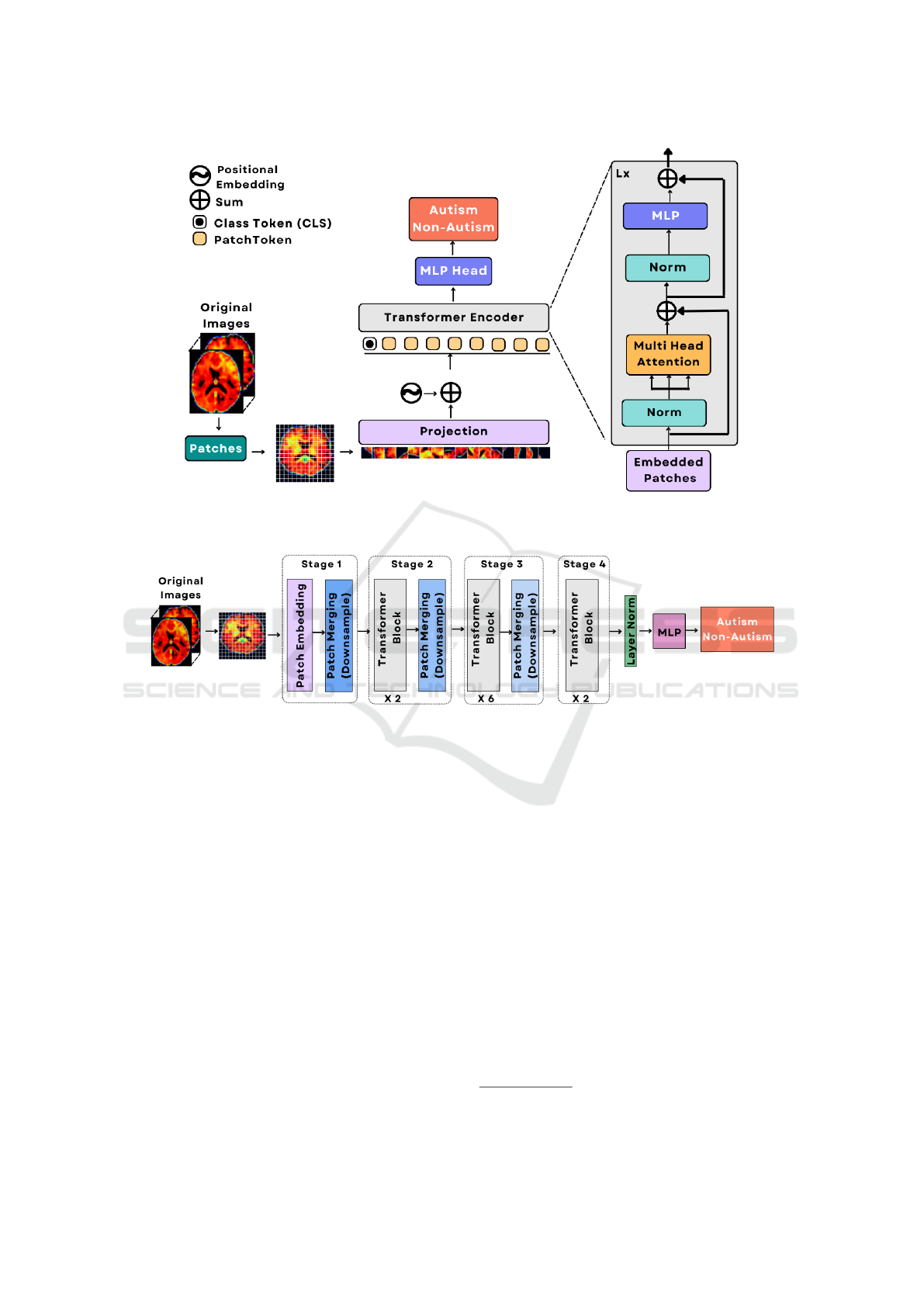
Figure 1: Outline of the components of traditional ViT model. The backbone is a vision transformer (ViT) encoder and an
optimized MLP.
.
Figure 2: Illustration of the basic building blocks of the TinyViT model used for autism spectrum disorder (ASD) prediction.
examine brain activities (Lindquist, 2008). In fMRI
data, the brain is divided into numerous small cubic
units referred to as voxels, with each voxel contain-
ing a time series that records its activity over a spec-
ified period. Resting-state fMRI (rs-fMRI) is a type
of fMRI scan that is conducted while the subject is
resting, and it is a commonly employed method for
studying various brain disorders. In rs-fMRI scans
subjects were instructed to allow their minds to wan-
der (i.e., think freely) while focusing on a crosshair
or keeping their eyes closed. No specific motor, per-
ceptual, or cognitive tasks were required (Gonzalez-
Castillo et al., 2021). Our work used rs-fMRI data
from ABIDE and CMI-HBN.
3.1 ABIDE
The original fMRI and demographic data were ob-
tained from ABIDE, which permits unrestricted use
for noncommercial research purposes. Our study used
the pre-processed ABIDE dataset. It includes 1112
resting-state fMRI (rs-fMRI) scans from both ASD
and healthy individuals, gathered at 17 venues. Of
these, 505 were subjects with ASD, and 530 were
healthy controls. The ABIDE provides mean time
series data from 7 sets of regions of interest (ROIs),
based on distinct brain atlases. For our experiments,
the dataset pre-processed using the C-PAC pipeline
(Craddock et al., 2013) was used. Additionally, it
was suggested by studies such as (Power et al., 2014),
and (Power et al., 2012) that an FD value
2
exceed-
ing 0.2mm can corrupt fMRI data. As a result, fMRI
images with a mean FD greater than 0.2mm were ex-
cluded. After filtering, data from 424 patients with
ASD and 510 healthy controls were retained. Details
on the class membership for each site are provided in
Figure (3).
2
Framewise Displacement (FD) measures head move-
ment during an MRI scan
HEALTHINF 2025 - 18th International Conference on Health Informatics
56

Figure 3: The number of individuals for each class from
different ABIDE sites, following FD filtering.
.
3.2 CMI-HBN
The Healthy Brain Network aimed to create a large-
scale, transdiagnostic dataset that reflects the diverse
range of mental health and learning disability found
in mental disorders. The Child Mind Institute (CMI)
started an initiative to collect and administer a bio-
bank containing data from 10,000 youngsters falling
between the age range of 5 to 21 years in New York
City. The Healthy Brain Network (HBN) bio-bank is
a limited access database that includes data on psy-
chiatric, behavioral, cognitive, and lifestyle pheno-
types (such as fitness and diet), along with multimodal
brain imaging, electroencephalography (EEG), digital
voice and video recordings, genetic information, and
actigraphy. From this dataset (Alexander et al., 2017),
fMRI data from 359 ASD and 359 healthy subjects
across four locations (RU, CBIC, CUNY, SU)
3
were
used in our study.
The data pre-processing steps involved slice tim-
ing correction, motion correction, removal of nui-
sance signals, correction for low-frequency drifts, and
normalization of voxel intensities for both datasets.
It is important to note that each site employed dis-
tinct parameters and scanning protocols. Differences
between sites include factors such as repetition time
(TR), voxel count, echo time (TE), amount of vol-
umes, and whether participants had their eyes open
or closed during the scans.
4 METHODOLOGY
This section provides a detailed explanation of the
methodology. Since the available datasets were small,
the objective was to use a lightweight model that of-
3
SI: Staten Island, RUBIC: Rutgers University Brain
Imaging Center, CBIC: Citigroup Cornell Brain Imaging
Center, CUNY: City College of New York.
fers performance comparable to the vision transform-
ers and has a small number of parameters. TinyViT
showed reliable performance in this aspect for de-
tecting people with ASD with a small-scale dataset.
Figure (2) provides a summary of the basic building
blocks of the TinyViT model. The decoder used was
a lightweight optimized multilayer perceptron (MLP).
Furthermore, Figure (1) provides an overview of the
traditional ViT encoder used to establish a baseline in
our approach. The decoder for this ViT model was
also a lightweight multilayer perceptron (MLP) opti-
mized for the model.
4.1 Baseline: ViT Model Structure
Unlike CNNs, vision transformers (ViTs) process im-
age data differently. Instead of processing the whole
image at once, they divide it into smaller patches and
treat those patches as input tokens. Figure (4) demon-
strates an outline of the components of the ViT model.
Given an fMRI image X ∈ R
H×W×3
as an input, it
is divided into smaller patches, each of size 16 × 16.
These patches are flattened into 1D vectors and com-
bined with an additional class token representing the
entire image. The number of patches is calculated by:
P ∈ R
(
H×W
16×16
+1)×C
(1)
where
H×W
16×16
gives the number of patches extracted
from the image, and +1 accounts for the class token.
C denotes the channel dimension, which is the size
of the feature embedding for each patch. Each patch
(along with the class token) is treated as a token and
fed into a series of transformer blocks. Each trans-
former block has two key components, the multi-head
self-attention (MHA) layer the and multi-layer per-
ceptron (MLP) layer. In the first step of the i + 1-
th transformer block, the tokens from the previous
block P
i
are first normalized using Layer Normal-
ization (LN) and passed through the multi-head self-
attention layer. Each token interacts with every other
token in the self-attention mechanism, which helps in
capturing relationships across the entire image. The
output of this operation is added back to the original
input P
i
, forming a residual connection. This process
can be written as:
P
′
i+1
= MHA
i+1
((LN(P
i
))) + P
i
(2)
where, P
′
i+1
is the intermediate output of the
transformer block after the self-attention operation;
MHA
i+1
represents the MHA layer of the i + 1-th
transformer block; LN(P
i
) is the layer normalization
applied to P
i
, and P
i
is added back to the attention
output to preserve the original information (residual
connection).
Cross-Domain Transfer Learning for Domain Adaptation in Autism Spectrum Disorder Diagnosis
57
Later, the output P
′
i+1
from the self-attention step
is normalized again and passed through MLP. It helps
in further refining the learned representations. The re-
sult of this step is also added to P
′
i+1
through another
residual connection, ensuring stability and flow of in-
formation through the network. The final update for
the tokens after the i + 1-th transformer block can be
described as:
P
i+1
= MLP
i+1
(LN(P
′
i+1
)) + P
′
i+1
(3)
where, P
i+1
is the final output of the transformer
block; MLP
i+1
represents the MLP layer of the i + 1-
th transformer block, and LN(P
′
i+1
) is the Layer Nor-
malization applied to the intermediate output P
′
i+1
.
4.2 TinyViT Model Structure
The TinyViT model architecture resembles the hierar-
chical vision transformer architecture. More specifi-
cally, the base model is composed of 4 stages. Similar
to Swin transformer (Liu et al., 2021), there is a grad-
ual downsizing of the image size in each stage. The
patch embedding block is constructed with two con-
volutions, featuring a kernel size of 3, and a stride
of 2. Lightweight and efficient MBConvs (Howard
et al., 2019) and down-sampling blocks were applied
in Stage 1, as convolutions in initial layers are effec-
tive at capturing low-level representations because of
their high inductive biases. The remaining 3 stages
have transformer blocks, and window attention to re-
duce computational costs. Attention biases (Graham
et al., 2021) and a 3 × 3 depth-wise convolution be-
tween MLP and attention modules were put in place
to gather localized information (Codella et al., 2019).
Residual connections (He et al., 2016) were utilized
in every block of stage 1, in the MLP blocks and
the attention modules. GELU (Hendrycks and Gim-
pel, 2016) was used for all activation functions. The
normalization layers for convolution and linear oper-
ations were BatchNorm (Ioffe, 2015) and LayerNorm
(Lei Ba et al., 2016), respectively.
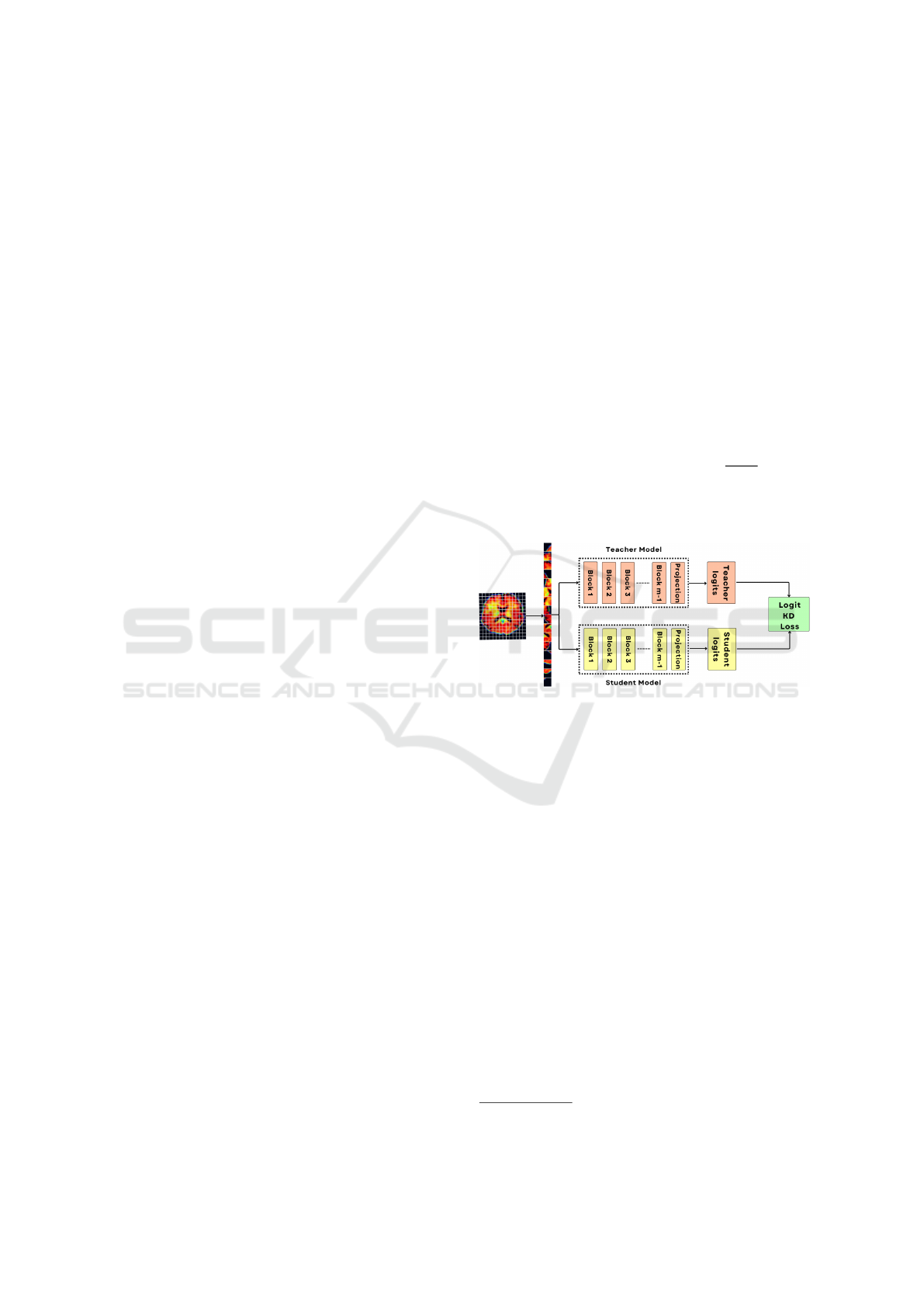
4.3 Model Transferability
The transfer of knowledge from the large teacher
model to the student model is done through distil-
lation within a teacher-student framework (Hinton,
2015).In this process, the teacher’s logits are utilized
to enhance the efficiency of the training process. In
our approach, the models used were trained on the Im-
ageNet21K and then fine-tuned on the ImageNet1K
dataset. We further fine-tuned a model on the ABIDE
dataset and used it as the teacher model. The teacher-
student framework used is demonstrated in Figure
(4). The student model was also pre-trained on Im-
ageNet21K. The distillation loss L
distill
was used to
improve the fine-tuning of the student model. In a nut-
shell, we fine-tuned the student model with the L
f inal
loss which is a regulated combination of L
model
and
L
distill
, refer to equation (4). The logit loss of the stu-
dent model is denoted by L
model
, and the Kullback-
Leibler (KL) divergence loss (Chien, 2018) between
the teacher and student logits is represented by L
distill
.
This way, the teacher helps the student model to learn
the domain knowledge faster. These losses are de-
fined as follows.
L
f inal
= L
model
∗ α + L
distill
∗ (1 − α) (4)
L
distill
= KL(M||N) =
∑
x
M(x)log(
M(x)
N(x)
) (5)
and α
4
was the hyper-parameter to offset the L
f inal
loss.
Figure 4: Pre-trained distillation method. The above branch
is for processing teacher logits and the bottom branch is for
processing student logits. The two branches are indepen-
dent.
5 EXPERIMENTS
5.1 Experimental Setup
Class weighing was used to balance the ASD and to-
tal control samples during training. The experiments
used a 12 GB NVIDIA GeForce GTX 1080 Ti GPU.
Data augmentation methods like center crop, sharp-
ening, RGB shift, and random contrast were used to
expand the amount of training data.
5.2 Implementation Details
To establish a baseline, the ViT model (ViT_B_16)
was used by both the teacher and the student. Initially,
we fine-tuned ViT_B_16 on ABIDE for 65 epochs.
4
α = 0.5 was used through out the experiments.
HEALTHINF 2025 - 18th International Conference on Health Informatics
58

This acted as the teacher later, the student model was
fine-tuned using the approach discussed in the sub-
section (4.3) on the CMI-HBN dataset for 40 epochs
using the L
f inal
loss. The set of parameters
5
used
for fine-tuning both models were AdamW optimizer
with a learning rate (lr) of 3.6e − 05, a weight decay
(wd) of 1e − 4, and a multistep LR scheduler where
the lr was reduced by a factor of 0.1 every 10 epochs.
Further, we used two versions of TinyViT models
TinyViT_5m_224 and TinyViT_21m_224 which had
5 million and 21 million parameters respectively.
Both models had an input size of 224 X 224. The
TinyViT_5m_224 model was fine-tuned on ABIDE
for 100 epochs which acted as a teacher, and the stu-
dent model was tuned on the CMI-HBN dataset for
40 epochs. Again, the TinyViT_21m_224 model was
fine-tuned on ABIDE for 50 epochs which acted as
a teacher, and the student model was tuned on the
CMI-HBN dataset for 40 epochs. The set of param-
eters
5
used to fine-tune the above two sets of mod-
els were: Adam optimizer that has a learning rate
(lr) of 9.56e − 4, a weight decay of 1e − 4, and Re-
duceLRonPlateau scheduler that monitors the valida-
tion loss. If no improvement is seen for 3 epochs, the
learning rate is reduced by a factor of 0.5. Moreover,
the MLPs for the ViT_B_16, TinyViT_5m_224, and
TinyViT_21m_224 were optimized
5
.
Furthermore, we used four well-established CNN
models specifically, VGG16 (Simonyan and Zisser-
man, 2014), Alexnet (Krizhevsky et al., 2012), Re-
sent101 (He et al., 2016), and Mobilenet (Howard,
2017) to compare their performance with TinyVit and
ViT models. These models were fine-tuned using
the same teacher-student approach as described in the
sub-section (4.3). The teacher models were fine-tuned
for 60 epochs, and the student models were fine-tuned
for 40 epochs. The parameters used were an Adam
optimizer with a lr of 1e-3, a wd of 1e-4, and a multi-
step LR scheduler where the lr was reduced by a fac-
tor of 0.1 every 10 epochs.
6 RESULTS
In this section, the classification performance of var-
ious models with different settings introduced earlier
in the sub-section (5.2), is reported and analyzed. Ta-
ble (2) presents the results for each model setting. Ta-
ble (1) presents a performance comparison between
our approach and previous studies on diagnosing
ASD, highlighting that our method significantly out-
performed earlier techniques. The methods presented
5
Optimized using optuna (Akiba et al., 2019) .
Figure 5: Attention map visualization performed using the
Tiny_ViT_21M model. (a) fMRI of an individual with
autism along with the corresponding attention maps. (b)
shows the fMRI of a subject from the TC group and the as-
sociated attention maps.
Table 1: Performance comparison: our method with previ-
ous studies using the ABIDE dataset.
Studies Accuracy(%)
(Heinsfeld et al., 2018) 70
(Plitt et al., 2015) 69.7
(Dvornek et al., 2017b) 68.5
(Sherkatghanad et al., 2020) 70.22
(Nielsen et al., 2013) 60
Our approach 76.62
in Table (1) relied on traditional training approaches,
where models were trained entirely from scratch. As
a result, these methods encountered difficulties in
achieving satisfactory performance due to the lim-
ited size of the datasets. In our approach, we uti-
lized cross-domain transfer learning combined with
knowledge distillation loss, which can effectively uti-
lize pre-trained models to address data scarcity is-
sues and enhance performance in scenarios involv-
ing small datasets. The TinyViT model with 21M
parameters surpassed the performance of ViT_B_16
and ViT_B_32 refer Table (2), despite having approx-
imately one-fourth model size. As the TinyViT model
architecture was adapted from a hierarchical vision
transformer framework, it was able to capture features
at multiple scales, which the traditional Vision Trans-
former (ViT) architecture could not achieve.
These findings suggest that effective adaptation of
knowledge learned from natural images to fMRI data
is achieved through the recommended cross-domain
transfer learning approach. The student model ben-
efits from the feature learning enhancement provided
by the teacher model. The outcomes of this work may
suggest a promising approach, to use cross-domain
Cross-Domain Transfer Learning for Domain Adaptation in Autism Spectrum Disorder Diagnosis
59

Table 2: The classification performance of different transformer-based models.
Models Accuracy (%) Precision(%)
Recall/
TPR(%)
TNR/
Specificity(%)
FPR(%)
F1
Score(%)
Model
Size (Million)
Embedding
dim
VIT_B_16 72.53 77.35 63.72 81.33 18.67 69.88 86 768
VIT_B_32 73.8 78.3 65.4 82.6 17.4 71.18 88.22 768
TinyViT_5m_224 70.9 72.25 67.87 73.93 26.07 69.9 5 320
TinyViT_21m_224 76.62 72.23 86.48 66.75 33.25 78.72 21 576
transfer learning methods in data-intensive fields with
limited data samples. Furthermore, the transformer-
based models, including both ViT and TinyViT with
varying sizes, outperform CNN-based architectures,
highlighting the superiority of transformer models.
From Table (3), it can be noted that the performance
of the CNN-based models was not as expected. This
may be attributed to the inability of CNN-based mod-
els to directly capture long-range dependencies from
the image features, which hinders the rapid adaptation
of specific features learned from the ImageNet dataset
images to brain imaging data.
Additionally, even though the TinyViT_5M model
contains only 5M parameters, its performance was
comparable to ViT_B_16, which has 86M parame-
ters. Moreover, the ViT_B_32 did not show sig-
nificant performance improvement over ViT_B_16,
likely because the datasets were small, and most fea-
tures were already learned, leaving few new features
for the model to capture. In Figure (5), the atten-
tion maps for each of the 10 attention heads from the
TinyViT model are illustrated. It can be observed that
the model distributes its focus across the entire fMRI
scan, as indicated by the color scale. The yellowish-
red hues correspond to regions with positive attention
weights, while the bluish tones represent areas with
negative attention weights. This distribution of at-
tention suggests that the model is comprehensively
analyzing the fMRI data to capture relevant features
across different brain regions. Lastly, the attention
maps were utilized to evaluate whether the model was
focusing on meaningful brain areas, rather than learn-
ing irrelevant features. This step is crucial to build
confidence in the model’s predictions.
7 DISCUSSION
The proposed method employed a cross-domain
transfer learning approach. The pre-trained TinyViT
and ViT models were fine-tuned using a teacher-
student framework and knowledge distillation (KD)
loss. In contrast, the methods outlined in Table (1) uti-
lized traditional machine learning approaches, where
the models were trained from scratch. Those methods
did not achieve satisfactory results due to the limited
size of the dataset and the model’s inability to capture
critical features effectively. To address these limita-
tions, pre-trained TinyViT models were fine-tuned to
adapt effectively to the target dataset.
The advantages of using pre-trained TinyViT
models are multifaceted. Firstly, these models, having
been pre-trained and fine-tuned on large natural image
datasets, facilitate an efficient transfer of knowledge
from the natural image domain to the brain imag-
ing domain via cross-domain transfer learning and
KD loss. Consequently, the models learn critical fea-
tures more effectively and efficiently. Secondly, the
TinyViT architecture, being based on a hierarchical
transformer structure, processes images as sequences
of patches using window-attention mechanisms. This
design enables the models to consider relationships
between patches, irrespective of their spatial distance,
allowing them to capture global context and long-
range dependencies more effectively.
Additionally, the TinyViT models possess a less
pronounced inductive bias compared to convolutional
neural networks (CNNs). Unlike CNNs, TinyViTs do
not assume local spatial relationships, enabling them
to learn more complex and abstract features from the
data. Furthermore, the smaller number of parame-
ters in TinyViT models compared to other hierarchi-
cal transformers and traditional ViTs makes them par-
ticularly well-suited for scenarios involving smaller
datasets, ensuring both versatility and efficiency. This
combination of features positions TinyViT as an opti-
mal choice for the proposed approach.
The implementation of the proposed approach
in clinical settings would reduce clinicians’ reliance
on traditional methods, such as the use of ADOS
scores, and minimize the need for frequent follow-
ups with each patient to ensure reliable diagnostic
results. Computer-aided diagnostic (CAD) systems
developed based on our model would provide clini-
cians with tools to deliver more accurate and timely
diagnoses while supporting them in making well-
informed decisions. Moreover, the high-attention ar-
eas identified in the attention maps generated by the
model can be correlated with known brain regions and
their associated functions. This association provides
an opportunity to link machine learning predictions
with clinical knowledge, enhancing the interpretabil-
ity and reliability of the model’s outputs. Further-
more, the integration of such systems could stream-
HEALTHINF 2025 - 18th International Conference on Health Informatics
60

Table 3: The classification performance of different CNN-based models.
Models Accuracy(%) Precision(%)
Recall/
TPR(%)
TNR/
Specificity(%)
FPR(%)
F1
Score(%)
VGG16 64.3 67.2 59.3 38.5 61.05 58.12
Alexnet 60.6 62.8 57.2 40.2 58.6 59.86
Resnet101 67.3 70.2 60.6 64.4 39.8 65.06
MobileNet 66.8 69.4 59.2 60.3 42.6 63.89
line the diagnostic workflow, allowing clinicians to
focus more on personalized treatment planning and
intervention and a deeper understanding of the neuro-
logical underpinnings of autism.
8 CONCLUSION
To address issues related to limited available data in
the medical domain that deals with the brain imag-
ing domain, a cross-domain transfer learning method
was introduced in this work. The TinyViT and ViT
models, pre-trained and fine-tuned on the ImageNet-
21K and ImageNet-1K datasets, respectively, were
employed. The teacher-student fine-tuning approach,
along with knowledge distillation loss, was then ap-
plied to fine-tune these models on the ABIDE and
CMI-HBN datasets. Sequential fine-tuning on two
datasets using a teacher-student framework further
improves the models’ robustness, generalizability,
and diversity. The results suggest that effective trans-
fer of knowledge from the natural image domain
to the brain imaging domain can be achieved using
cross-domain transfer learning along with KD loss.
The final fine-tuned models were evaluated and
compared to previous studies. Our approach demon-
strated superior performance and achieved an accu-
racy of 76.62% and 78.72% F-1 score. Furthermore,
attention maps were visualized to understand how the
model processes and focuses on different parts of the
fMRI image. Also, using the attention maps we ver-
ified that the model is not learning any insignificant
features. Computer-aided diagnostic (CAD) systems
developed using this approach will enable clinicians
to make more accurate and timely diagnoses, and also
assist them in making informed decisions.
9 FUTURE WORK
Due to the small size of the datasets, a bottleneck
was encountered during the fine-tuning process, as the
models had already learned most of the features. As
a result, it was difficult to achieve an accuracy be-
yond 80%. However, we believe that the results of
our proposed approach—cross-domain transfer learn-
ing with knowledge distillation (KD) loss—can be
significantly enhanced by utilizing a comparatively
larger dataset containing more than 5K images. Fur-
thermore, data augmentation techniques, such as syn-
thetic data generation using GANs (Generative Ad-
versarial Networks) can also be explored to increase
the size and diversity of the datasets. The diversity in
the datasets would allow the model to become more
robust and generalizable.
Additionally, incorporating a multi-modal model
that fuses different modalities, such as Electroen-
cephalography (EEG), and fMRI data, along with
skeletal data would provide a more comprehensive
analysis of the subject’s condition. This multi-modal
integration would provide a more robust framework
for diagnosing autistic individuals. Additionally, at-
tention maps generated by the model can be corre-
lated with known brain regions and their associated
functions, improving the interpretability and reliabil-
ity of the model.
ACKNOWLEDGEMENTS
We want to thank EPSRC DTP HMT for funding this
project. Also, this manuscript was prepared using a
limited-access dataset obtained from the Child Mind
Institute Biobank, HBN dataset. This manuscript re-
flects the views of the authors and does not necessar-
ily reflect the opinions or views of the Child Mind
Institute.
REFERENCES
Abraham, A., Milham, M. P., Di Martino, A., Craddock,
R. C., Samaras, D., Thirion, B., and Varoquaux,
G. (2017). Deriving reproducible biomarkers from
multi-site resting-state data: An autism-based exam-
ple. NeuroImage, 147:736–745.
Akiba, T., Sano, S., Yanase, T., Ohta, T., and Koyama, M.
(2019). Optuna: A next-generation hyperparameter
optimization framework. In Proceedings of the 25th
ACM SIGKDD International Conference on Knowl-
edge Discovery and Data Mining.
Alexander, L. M., Escalera, J., Ai, L., Andreotti, C., Febre,
K., Mangone, A., Vega-Potler, N., Langer, N., Alexan-
der, A., Kovacs, M., Litke, S., O’Hagan, B., Ander-
Cross-Domain Transfer Learning for Domain Adaptation in Autism Spectrum Disorder Diagnosis
61

sen, J., Bronstein, B., Bui, A., Bushey, M., Butler,
H., Castagna, V., Camacho, N., Chan, E., Citera, D.,
Clucas, J., Cohen, S., Dufek, S., Eaves, M., Fradera,
B., Gardner, J., Grant-Villegas, N., Green, G., Gre-
gory, C., Hart, E., Harris, S., Horton, M., Kahn,
D., Kabotyanski, K., Karmel, B., Kelly, S. P., Klein-
man, K., Koo, B., Kramer, E., Lennon, E., Lord, C.,
Mantello, G., Margolis, A., Merikangas, K. R., Mil-
ham, J., Minniti, G., Neuhaus, R., Levine, A., Os-
man, Y., Parra, L. C., Pugh, K. R., Racanello, A.,
Restrepo, A., Saltzman, T., Septimus, B., Tobe, R.,
Waltz, R., Williams, A., Yeo, A., Castellanos, F. X.,
Klein, A., Paus, T., Leventhal, B. L., Craddock, R. C.,
Koplewicz, H. S., and Milham, M. P. (2017). An
open resource for transdiagnostic research in pediatric
mental health and learning disorders. Scientific Data,
4(1):170181.
Ali, N. (2020). Autism spectrum disorder classification on
electroencephalogram signal using deep learning al-
gorithm. IAES International Journal of Artificial In-
telligence (IJ-AI), 9:91.
Bi, X.-A., Liu, Y., Jiang, Q., Shu, Q., Sun, Q., and Dai,
J. (2018). The diagnosis of autism spectrum disorder
based on the random neural network cluster. Frontiers
in human neuroscience, 12:257.
Brown, C. J., Kawahara, J., and Hamarneh, G. (2018).
Connectome priors in deep neural networks to predict
autism. In 2018 IEEE 15th international symposium
on biomedical imaging (ISBI 2018), pages 110–113.
IEEE.
Cao, X. and Cao, J. (2023). Commentary: Machine learning
for autism spectrum disorder diagnosis–challenges
and opportunities–a commentary on schulte-rüther et
al.(2022). Journal of Child Psychology and Psychia-
try, 64(6):966–967.
Chen, H., Duan, X., Liu, F., Lu, F., Ma, X., Zhang,
Y., Uddin, L. Q., and Chen, H. (2016). Mul-
tivariate classification of autism spectrum disor-
der using frequency-specific resting-state functional
connectivity—a multi-center study. Progress in
Neuro-Psychopharmacology and Biological Psychia-
try, 64:1–9.
Chien, J.-T. (2018). Source separation and machine learn-
ing. Academic Press.
Codella, N., Rotemberg, V., Tschandl, P., Celebi, M. E.,
Dusza, S., Gutman, D., Helba, B., Kalloo, A., Liopy-
ris, K., Marchetti, M., et al. (2019). Skin lesion anal-
ysis toward melanoma detection 2018: A challenge
hosted by the international skin imaging collaboration
(isic). arXiv preprint arXiv:1902.03368.
Craddock, C., Benhajali, Y., Chu, C., Chouinard, F., Evans,
A., Jakab, A., Khundrakpam, B. S., Lewis, J. D., Li,
Q., Milham, M., et al. (2013). The neuro bureau pre-
processing initiative: open sharing of preprocessed
neuroimaging data and derivatives. Frontiers in Neu-
roinformatics, 7(27):5.
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., and Fei-
Fei, L. (2009). Imagenet: A large-scale hierarchical
image database. In 2009 IEEE conference on com-
puter vision and pattern recognition, pages 248–255.
Ieee.
Di Martino, A., Yan, C.-G., Li, Q., Denio, E., Castel-
lanos, F. X., Alaerts, K., Anderson, J. S., Assaf, M.,
Bookheimer, S. Y., Dapretto, M., Deen, B., Delmonte,
S., Dinstein, I., Ertl-Wagner, B., Fair, D. A., Gal-
lagher, L., Kennedy, D. P., Keown, C. L., Keysers,
C., Lainhart, J. E., Lord, C., Luna, B., Menon, V.,
Minshew, N. J., Monk, C. S., Mueller, S., Müller, R.-
A., Nebel, M. B., Nigg, J. T., O’Hearn, K., Pelphrey,
K. A., Peltier, S. J., Rudie, J. D., Sunaert, S., Thioux,
M., Tyszka, J. M., Uddin, L. Q., Verhoeven, J. S.,
Wenderoth, N., Wiggins, J. L., Mostofsky, S. H., and
Milham, M. P. (2014). The autism brain imaging data
exchange: towards a large-scale evaluation of the in-
trinsic brain architecture in autism. Mol. Psychiatry,
19(6):659–667.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. arXiv preprint arXiv:2010.11929.
Durstewitz, D., Koppe, G., and Meyer-Lindenberg, A.
(2019). Deep neural networks in psychiatry. Mol. Psy-
chiatry, 24(11):1583–1598.
Dvornek, N. C., Ventola, P., and Duncan, J. S. (2018).
Combining phenotypic and resting-state fmri data
for autism classification with recurrent neural net-
works. In 2018 IEEE 15th International Symposium
on Biomedical Imaging (ISBI 2018), pages 725–728.
IEEE.
Dvornek, N. C., Ventola, P., Pelphrey, K. A., and Dun-
can, J. S. (2017a). Identifying autism from resting-
state fmri using long short-term memory networks.
In Machine Learning in Medical Imaging: 8th Inter-
national Workshop, MLMI 2017, Held in Conjunc-
tion with MICCAI 2017, Quebec City, QC, Canada,
September 10, 2017, Proceedings 8, pages 362–370.
Springer.
Dvornek, N. C., Ventola, P., Pelphrey, K. A., and Duncan,
J. S. (2017b). Identifying autism from resting-state
fMRI using long short-term memory networks. In Ma-
chine Learning in Medical Imaging, Lecture notes in
computer science, pages 362–370. Springer Interna-
tional Publishing, Cham.
Gonzalez-Castillo, J., Kam, J. W. Y., Hoy, C. W., and
Bandettini, P. A. (2021). How to interpret resting-
state fMRI: Ask your participants. J. Neurosci.,
41(6):1130–1141.
Graham, B., El-Nouby, A., Touvron, H., Stock, P., Joulin,
A., Jégou, H., and Douze, M. (2021). Levit: a vision
transformer in convnet’s clothing for faster inference.
In Proceedings of the IEEE/CVF international confer-
ence on computer vision, pages 12259–12269.
Guo, X., Dominick, K. C., Minai, A. A., Li, H., Erickson,
C. A., and Lu, L. J. (2017). Diagnosing autism spec-
trum disorder from brain resting-state functional con-
nectivity patterns using a deep neural network with a
novel feature selection method. Frontiers in neuro-
science, 11:460.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
HEALTHINF 2025 - 18th International Conference on Health Informatics
62

the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Heinsfeld, A. S., Franco, A. R., Craddock, R. C., Buch-
weitz, A., and Meneguzzi, F. (2018). Identification of
autism spectrum disorder using deep learning and the
ABIDE dataset. NeuroImage: Clinical, 17:16–23.
Hendrycks, D. and Gimpel, K. (2016). Gaussian error linear
units (gelus). arXiv preprint arXiv:1606.08415.
Hinton, G. (2015). Distilling the knowledge in a neural net-
work. arXiv preprint arXiv:1503.02531.
Howard, A., Sandler, M., Chu, G., Chen, L.-C., Chen, B.,
Tan, M., Wang, W., Zhu, Y., Pang, R., Vasudevan, V.,
et al. (2019). Searching for mobilenetv3. In Pro-
ceedings of the IEEE/CVF international conference
on computer vision, pages 1314–1324.
Howard, A. G. (2017). Mobilenets: Efficient convolutional
neural networks for mobile vision applications. arXiv
preprint arXiv:1704.04861.
Husna, R. N. S., Syafeeza, A., Hamid, N. A., Wong, Y., and
Raihan, R. A. (2021). Functional magnetic resonance
imaging for autism spectrum disorder detection using
deep learning. Jurnal Teknologi, 83(3):45–52.
Iidaka, T. (2015). Resting state functional magnetic reso-
nance imaging and neural network classified autism
and control. Cortex, 63:55–67.
Ioffe, S. (2015). Batch normalization: Accelerating deep
network training by reducing internal covariate shift.
arXiv preprint arXiv:1502.03167.
Jennings Dunlap, J. (2019). Autism spectrum disorder
screening and early action. The Journal for Nurse
Practitioners, 15(7):496–501. SI: SCOPE OF PRAC-
TICE.
Khosla, M., Jamison, K., Kuceyeski, A., and Sabuncu,
M. R. (2018). 3D convolutional neural networks for
classification of functional connectomes. In Interna-
tional Workshop on Deep Learning in Medical Image
Analysis, pages 137–145. Springer.
Klin, A. (2018). Biomarkers in autism spectrum disorder:
Challenges, advances, and the need for biomarkers
of relevance to public health. Focus (Am. Psychiatr.
Publ.), 16(2):135–142.
Koirala, A., Walsh, K. B., Wang, Z., and McCarthy, C.
(2019). Deep learning–method overview and review
of use for fruit detection and yield estimation. Com-
puters and electronics in agriculture, 162:219–234.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
agenet classification with deep convolutional neural
networks. Advances in neural information processing
systems, 25.
Lahiri, U., Welch, K., Warren, Z., and Sarkar, N. (2011).
Understanding psychophysiological response to a vir-
tual reality-based social communication system for
children with asd. 2011 International Conference on
Virtual Rehabilitation, ICVR 2011.
Lei Ba, J., Kiros, J. R., and Hinton, G. E. (2016). Layer
normalization. ArXiv e-prints, pages arXiv–1607.
Lindquist, M. A. (2008). The statistical analysis of fMRI
data.
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin,
S., and Guo, B. (2021). Swin transformer: Hierar-
chical vision transformer using shifted windows. In
Proceedings of the IEEE/CVF international confer-
ence on computer vision, pages 10012–10022.
Lord, C., Risi, S., Lambrecht, L., Cook, E. H., Leven-
thal, B. L., DiLavore, P. C., Pickles, A., and Rutter,
M. (2000). The autism diagnostic observation sched-
ule—generic: A standard measure of social and com-
munication deficits associated with the spectrum of
autism. Journal of autism and developmental disor-
ders, 30:205–223.
Manaswi, N. K., Manaswi, N. K., and John, S. (2018). Deep
learning with applications using python. Springer.
Monté-Rubio, G. C., Falcón, C., Pomarol-Clotet, E., and
Ashburner, J. (2018). A comparison of various MRI
feature types for characterizing whole brain anatomi-
cal differences using linear pattern recognition meth-
ods. Neuroimage, 178:753–768.
Nielsen, J. A., Zielinski, B. A., Fletcher, P. T., Alexander,
A. L., Lange, N., Bigler, E. D., Lainhart, J. E., and An-
derson, J. S. (2013). Multisite functional connectivity
mri classification of autism: Abide results. Frontiers
in human neuroscience, 7:599.
Pan, S. J. and Yang, Q. (2009). A survey on transfer learn-
ing. IEEE Transactions on knowledge and data engi-
neering, 22(10):1345–1359.
Plitt, M., Barnes, K. A., and Martin, A. (2015). Functional
connectivity classification of autism identifies highly
predictive brain features but falls short of biomarker
standards. NeuroImage: Clinical, 7:359–366.
Power, J. D., Barnes, K. A., Snyder, A. Z., Schlaggar, B.,
and Petersen, S. (2012). Spurious but systematic con-
ditions in functional connectivity MRI networks arise
from subject motion. Neuroimage, 59:2141–2154.
Power, J. D., Mitra, A., Laumann, T. O., Snyder, A. Z.,
Schlaggar, B. L., and Petersen, S. E. (2014). Methods
to detect, characterize, and remove motion artifact in
resting state fMRI. NeuroImage, 84:320–341.
Shahamat, H. and Abadeh, M. S. (2020). Brain mri analy-
sis using a deep learning based evolutionary approach.
Neural Networks, 126:218–234.
Sharda, M., Foster, N. E. V., Tryfon, A., Doyle-Thomas,
K. A. R., Ouimet, T., Anagnostou, E., Evans, A. C.,
Zwaigenbaum, L., Lerch, J. P., Lewis, J. D., and Hyde,
K. L. (2016). Language ability predicts cortical struc-
ture and covariance in boys with autism spectrum dis-
order. Cereb. Cortex, page bhw024.
Sherkatghanad, Z., Akhondzadeh, M., Salari, S., Zomorodi-
Moghadam, M., Abdar, M., Acharya, U. R., Khos-
rowabadi, R., and Salari, V. (2020). Automated detec-
tion of autism spectrum disorder using a convolutional
neural network. Frontiers in neuroscience, 13:1325.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Traut, N., Heuer, K., Lemaître, G., Beggiato, A., Ger-
manaud, D., Elmaleh, M., Bethegnies, A., Bonnasse-
Gahot, L., Cai, W., Chambon, S., et al. (2022). In-
sights from an autism imaging biomarker challenge:
Cross-Domain Transfer Learning for Domain Adaptation in Autism Spectrum Disorder Diagnosis
63

promises and threats to biomarker discovery. Neu-
roImage, 255:119171.
Wu, K., Zhang, J., Peng, H., Liu, M., Xiao, B., Fu, J., and
Yuan, L. (2022). Tinyvit: Fast pretraining distillation
for small vision transformers. In European conference
on computer vision, pages 68–85. Springer.
Yosinski, J., Clune, J., Bengio, Y., and Lipson, H. (2014).
How transferable are features in deep neural net-
works? Advances in neural information processing
systems, 27.
HEALTHINF 2025 - 18th International Conference on Health Informatics
64
