
An Interpretable Machine Learning Model for Meningioma Grade
Prediction
Ali Golbaf
1a
, Damjan Veljanoski
2b
, Prutha Chawda
2 c
, Swen Gaudl
1d
,
C. Oliver Hanemann
2e
and Emmanuel Ifeachor
1f
1
School of Engineering, Computing and Mathematics, University of Plymouth, Plymouth, U.K.
2
Peninsula Schools of Medicine, University of Plymouth University, Plymouth, U.K.
Keywords: Meningiomas, Grading, Radiomics, MRI, Interpretable Techniques.
Abstract: Accurate preoperative prediction of meningioma grade is crucial for enhancing the clinical management of
these tumours. In this study, we developed a non-invasive machine learning (ML) model to predict
meningioma grade using clinical features and radiomics features from preoperative MRI scans, focusing on
interpretability to improve clinical adoption of such models. A dataset of 94 patients from The Cancer Imaging
Archive (TCIA) was analysed. Clinical features and radiomics features from T1-weighted contrast-enhanced
(T1C) and T2-weighted Fluid Attenuated Inversion Recovery (T2 FLAIR) scans were utilised. Two feature
subsets were constructed: one using radiomics features alone and the other combining clinical and radiomics
features. Feature selection was performed using a modified Least Absolute Shrinkage and Selection Operator
(LASSO) technique. Four ML models: Logistic Regression (LR), Support Vector Machine (SVM), Random
Forest (RF), and Gradient Boosting (GB), were developed. SHapley Additive exPlanations (SHAP) was
employed to address the blackbox nature of ML models by providing radiomics overall feature importance
scores and model interpretation. Results using the clinical-radiomics subset showed that the SVM
outperformed others (test AUC: 0.83), indicating its reliability for predicting meningioma grade. SHAP
highlights discriminative radiomics features and their interaction with clinical features, thereby enhancing the
clinical adoption of such models.
1 INTRODUCTION
Meningiomas, the most common primary brain
tumours, are among the most understudied tumours
within the central nervous system (Low et al. 2022).
However, a significant proportion of meningiomas
(20% - 30%), show aggressive behaviour, and high
recurrence rate (Zhang et al. 2019). These tumours
are categorised into three grades, according to 2021
World Health Organisation (WHO) guidelines (Louis
et al. 2021). High-grade meningiomas (Grades II and
III) show more aggressive behaviour than low-grade
cases (Grade I), leading to a 5-year progression free
survival probability (Wang, Nassiri, et al. 2023).
a
https://orcid.org/0000-0002-8104-5600
b
https://orcid.org/0000-0002-4951-8586
c
https://orcid.org/0009-0004-0345-9017
d
https://orcid.org/0000-0003-3116-3761
e
https://orcid.org/0000-0002-1951-1025
f
https://orcid.org/0000-0001-8362-6292
They are also challenging to be completely resected
using invasive treatment strategies and often require
adjunctive radiotherapy (Fountain, Young, and
Santarius 2020). Thus, accurate grading of these
tumours is important in enhancing the clinical
management of meningiomas.
The gold standard for grading of meningiomas
still relies on invasive methods such as
histopathology and biopsy (Herrgott et al. 2023).
However , invasive methods may not be applicable to
tumours that are surgically inaccessible and patients
with multiple diseases. Moreover, biopsies may not
accurately reflect the heterogeneity of meningiomas
due to limited sampling (Islim et al. 2020; Tagle et al.
Golbaf, A., Veljanoski, D., Chawda, P., Gaudl, S., Hanemann, C. O. and Ifeachor, E.
An Interpretable Machine Learning Model for Meningioma Grade Prediction.
DOI: 10.5220/0013114000003911
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 65-74
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
65

2002). Consequently, there is a growing need for the
development of non-invasive models that accurately
predict the grade of meningiomas.
Currently, MRI serves as the primary non-
invasive method in the clinical management of
meningiomas (Zhang et al. 2020). However, some
conventional MRI features of different meningioma
grades overlap, which can potentially lead to
misdiagnosis (Spille et al. 2019). In this context,
radiomic, which is a quantitative approach for
medical image analysis, has emerged as a novel way
to extract imaging features that carry valuable
biological information about tumours which are not
accessible by conventional image analysis (Lambin et
al. 2012). Machine learning has also demonstrated
potential in developing non-invasive predictive
models by capturing complex patterns within these
features (Langs et al. 2018). Such models have been
developed for the diagnosis, prognosis, and treatment
of meningiomas, and have particularly shown
promise in meningioma grading (Patel et al. 2023).
However, radiomics and machine learning have
not yet been adopted in the clinical management of
meningiomas. The blackbox nature of machine
learning models make their outputs difficult to
interpret (Patel et al. 2023). The application of
interpretability techniques may mitigate the inherent
blackbox nature of machine learning models (Reyes
et al. 2020). However, only a few studies have
focused on improving the interpretability of machine
learning models in the clinical management of
meningiomas. SHAP, which is used to assess the
contribution of each radiomic feature to model
performance, has been used to interpret a machine
learning model for evaluating the post-surgical
recurrence of high-grade meningiomas (Park, Choi, et
al. 2022). Relevance-weighted Class Activation
Mapping, an explanation method for visualising class
relevance, has been employed to explain a machine
learning model for meningioma segmentation (Jun et
al. 2023). Additionally, Local Interpretable Model-
Agnostic Explanations (LIME), an estimator
technique, which approximates models locally for
interpretability, has been applied to interpret machine
learning models, for predicting glioma grades but
not for the prediction of meningioma grade (Wang et
al. 2019).
In this paper, we developed an interpretable
machine learning model for predicting meningioma
grade using both clinical and radiomics features. The
aim is to enhance the adoption of radiomics and
machine learning in the clinical management of
meningiomas by establishing links between
meningioma grade, radiomics features, and their
interactions with clinical features.
2 METHODS
2.1 Dataset
The dataset used in this study was obtained from
TCIA, a publicly available database (Clark et al.
2013). It comprises a cohort of 96 patients who were
diagnosed with meningioma between 2010 and 2019
(Vassantachart). Low-grade and high-grade
meningiomas were identified according to the 2016
WHO guidelines. Clinical features were also recorded
by two experienced neuropathologists and one
neuropathology fellow. All patients underwent pre-
operative T1C, and T2 FLAIR MRI scans. A detailed
description of the imaging protocol can be found in
(Vassantachart et al. 2022). In this study, cases with
inconsistent histopathological records and suboptimal
image qualities were excluded, yielding a final cohort
of 94 patients. The clinical features of the patients are
summarised in Table 1.
2.2 Model Development
Figure 1 shows the workflow for developing an
interpretable model. The TCIA dataset was
processed, with MRI data standardised to the Brain
Imaging Data Structure (BIDS) format for
consistency and reproducibility (Gorgolewski et al.
2016). Radiomics features were extracted, and the
most discriminative features selected. These features
trained various ML models to predict meningioma
grade. SHAP was then applied for determining
overall radiomics feature importance scores and
model interpretation. The final model can predict
meningioma grade in new cases.
2.2.1 Image Processing and Radiomics
Feature Extraction
The dataset had undergone a prior image processing
pipeline, ensuring data consistency and quality. As
detailed in (Vassantachart et al. 2022), anatomically
co-registered T1C and T2 FLAIR MRI scans were
obtained, with any misalignment corrected using the
automated rigid registration software, VelocityAI. T2
FLAIR scans were resampled into their
corresponding T1C scans, followed by isovoxel
resampling. In the present study, further image
processing techniques, including bias field correction
and normalisation, were applied based on the
HEALTHINF 2025 - 18th International Conference on Health Informatics
66
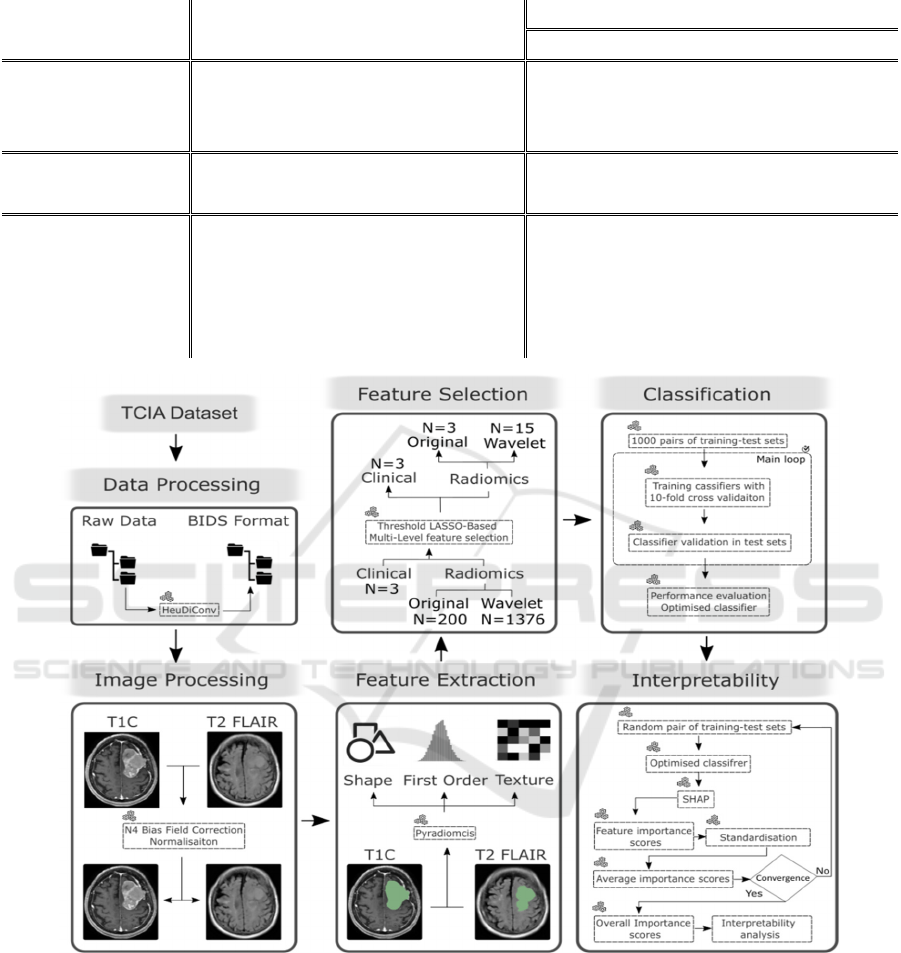
Table 1: Histopathological and demographic characteristics of the patients.
Features Groups
Total Female Male
94 67 (71.3%) 27 (28.7%)
Age
Min 25 25 29
Mean 55.39 54.53 57.51
Max 88 88 85
Grade
Low-grade 53 (56.4%) 46 (68.7%) 07 (26.0%)
High-grade 41 (43.6%) 21 (31.3%) 20 (74.0%)
Location
Anterior and middle cranial fossa 45 (47.9%) 33 (49.3%) 12 (44.5%)
Convexity 19 (20.2%) 09 (13.4%) 10 (37.0%)
Falx and parasagittal 16 (17.0%) 12 (17.9%) 04 (14.8%)
Posterior cranial fossa 12 (12.8%) 11 (16.4%) 01 (03.7%)
Lateral ventricle 02 (02.1%) 02 (03.0%) 00 (00.0%)
Figure 1: Study workflow.
radiomics standardisation protocol for brain MRI
scans outlined in (Carré et al. 2020), using the
SimpleITK N4BiasFieldCorrection and
NormaliseImage filters (Yaniv et al. 2018). Manually
delineated tumour lesions were also available within
T1C and T2 FLAIR MRI scans. These annotations
were created by a medical student and a radiation
oncology resident and then reviewed by a radiation
oncologist with over 5 years of experience
(Vassantachart et al. 2023). Radiomics features
including shape, first-order, and texture features were
subsequently extracted from these lesions.
2.2.2 Clinical and Radiomics Feature
Selection
Feature selection is a key step in the development of
ML models, as radiomics features often show strong
An Interpretable Machine Learning Model for Meningioma Grade Prediction
67

correlations, potentially resulting in redundant
information, that can detrimentally affect model
interpretability and generalisability (Reyes et al.
2020). LASSO, a widely used feature selection
technique for analysing high-dimensional data,
improves model performance and interpretation,
although, highly correlated features may undermine
its efficiency (Zou and Hastie 2003). To tackle this
issue, we perform a multi-level feature selection
method based on LASSO coefficient thresholds
(Wang, An, et al. 2023). Clinical features were also
analysed using the t-test for age and the chi-square
test for gender and tumour location, with p-values
below 0.05 as statistically significant.
2.2.3 ML Model to Classify Meningioma
Grade
ML models for classifying low-grade and high-grade
meningiomas were developed using LR, SVM, RF,
and GB classifiers. To ensure robustness and
generalisability, we conducted 1000 random training-
test splits (1:4 ratio), generating training-test set pairs
with 70 training and 24 test cases (An et al. 2021).
Models were trained using 10-fold cross-validation
within each training set. Model performance was
evaluated by averaging AUC, Accuracy, Precision,
Recall, and F1-score. The best-performing model was
selected for interpretability analysis.
2.2.4 Model Interpretability
SHAP is a well-established technique for enhancing
ML model interpretability. However, random
perturbation-based sampling in SHAP implies that
with different random seeds, a high ranked feature in
one iteration may be considered as a low ranked
feature in the next iteration (Xiang et al. 2023). To
mitigate this issue, we determined overall radiomics
feature importance scores by generating multiple
training-test sets. The iteration process was
terminated when the change in average importance
scores for each feature was equal or less than 0.01
between two consecutive iterations. Overall
radiomics feature importance scores were then
considered as these averages. SHAP Kernel Explainer
was utilised to determine radiomics feature
importance scores in each iteration and scores were
normalised by the sum of all feature importances.
2.2.5 Implementation
In this study, Python 3.8 was used for data conversion
to BIDS format, image processing, radiomics feature
extraction, and feature selection, as well as for ML
development, and interpretability analyses. The
HeuDiConv tool (version 0.9.0,
https://github.com/nipy/heudiconv), facilitated the
conversion of DICOM files into BIDS format
(Halchenko, Goncalves, and Castello 2020). Image
processing was implemented using SimpleITK
package (version 2.3.0). The open-source package
Pyradiomics (version 3.1.0, https://github.com/AIM-
Harvard/pyradiomics) was used for feature extraction
(Van Griethuysen et al. 2017). Clinical categorical
variables were encoded numerically. The Scikit-learn
package (version 1.3.2) was used for radiomics
feature selection, ML model development, and
evaluation. Interpretability techniques was performed
using SHAP (version 0.43.0) package.
3 RESULTS
3.1 Radiomics Feature Extraction
A total of 1576 radiomics features were extracted,
including 14 shape features describing the size and
contours of the tumours, 18 first-order features
characterising the distribution of voxel intensities
within the lesions, and 68 texture features measuring
the variation of voxel intensities across T1C and T2
FLAIR MRI scans. Texture features were extracted
using 22 Grey Level Co-occurrence Matrix (GLCM),
16 Gray-Level Run-length Matrix (GLRLM), 16
Gray-Level Size Zone Matrix (GLSZM), and 14 Gray
Level Difference Matrix (GLDM). Subsequently, 688
wavelet radiomics features were evaluated by
applying wavelet decomposition on the original
images at both high and low frequencies.
3.2 Feature Selection and Model
Performance
A subset of 18 radiomics features was identified as the
most discriminative. Significant differences in age,
gender, and tumour location between low-grade and
high-grade meningiomas were observed, with p-values
lower than 0.05. These clinical features were added to
the radiomics subset to form a clinical-radiomics
subset. The specifics of the feature subsets are outlined
in Table 2, and the performance of ML models in
training and test sets are shown in Table 3. The
classifiers exhibited high accuracy and precision in
distinguishing between tumour grades. Among the
models, the SVM using the clinical-radiomics subset
achieved the highest performance with AUC (0.90 ±
0.12 and 0.83 ± 0.07), Accuracy (0.83 ± 0.13 and 0.84
± 0.06), Precision (0.84 ± 0.18 and 0.82 ± 0.10), Recall
HEALTHINF 2025 - 18th International Conference on Health Informatics
68

(0.80 ± 0.21 and 0.80 ± 0.11), and F1-score (0.80 ±
0.16 and 0.80 ± 0.08) in training and test sets,
respectively.
3.3 Model Interpretability
Figure 2 depicts overall radiomics feature importance
scores. Figure 3 presents the SHAP violin summary
plot, which illustrates the distribution and variability
of SHAP values for each feature in distinguishing
between low-grade and high-grade meningiomas.
Higher SHAP values indicate greater impact on the
model output, with wider violins showing higher
density and more frequent values. Figure 4 represents
feature SHAP dependence plots for the 3 top-ranked
radiomics features. Figure 5 shows the interaction
between the top-ranked radiomics feature with
clinical features. In these figures each dot represents
a prediction related to feature values, with the x-axis
showing actual values and the y-axis showing SHAP
values.
4 DISCUSSIONS
In this study, using clinical-radiomics feature
subset, SVM model was the best-performing model
with the highest average values of AUC (0.90 ± 0.12
and 0.83 ± 0.07), Accuracy (0.83 ± 0.13 and 0.84 ±
0.06), Precision (0.84 ± 0.18 and 0.82 ± 0.10),
Recall (0.80 ± 0.21 and 0.80 ± 0.11), and F1-score
(0.80 ± 0.16 and 0.80 ± 0.08) in the training and test
sets, respectively.
In the current study, the number of extracted
radiomics features from T1C, and T2 FLAIR MRI
scans was almost the same, highlighting the
importance of using multi-parametric MRI scans in
the relevant studies (Park, Shin, et al. 2022). Previous
studies used clinical features and radiomics features
from MRI scans to predict meningioma low-grade
and high-grade. Duan et al. developed a radiomics
nomogram with AUC of 0.95, using clinical and
LASSO-selected radiomics features from T1C MRI
scans (Duan, Zhou, et al. 2022). They also developed
seven ML models, with the SVM model achieving an
AUC of 0.88 (Duan, Li, et al. 2022). Similarly, Chu
et al. used LASSO-selected radiomics features from
T1, T1C, and T2 MRI scans to develop an LG model
with an AUC of 0.95 (Chu et al. 2021). While these
studies demonstrated strong predictive performance,
they lacked interpretability in their models. Although
complex machine learning models are effective in
capturing patterns in the data, they often result in
models that are difficult for clinicians to interpret,
where understanding the features influencing
predictions is crucial for clinical management.
One major hinderance to the adoption of
radiomics and ML models in the clinical management
of meningiomas is the blackbox nature of ML models.
To address this issue, SHAP was utilised to extract
the overall radiomics feature importance scores and
model interpretation. Here, GLRLM and GLSZM
radiomics features were identified as the most
discriminative features, showing high correlation
with meningioma grade prediction (Han et al. 2021).
GLSZM quantifies gray level zones within MRI
scans. A gray level zone is defined as the number of
connected voxels that share the same gray level
intensity. GLRLM features describes heterogeneity in
the distribution of run lengths (Traverso et al. 2020).
The majority of selected radiomics features (15 out of
18) were derived from the wavelet-filtered MRI
scans, which have been proved to be the most
discriminative features in meningioma grade
prediction (Han et al. 2021).
The violin plot depicted in Figure 3 indicates that
higher values of the first radiomics feature, wavelet-
HLH_glszm_ZoneEntropy_t1c, correspond to an
increased output probability of high-grade
meningiomas. A similar trend was observed for
the third radiomics feature, wavelet-
LHL_glszm_GrayLevelNonUniformityNormalized_
t1c. Conversely, lower values of the
second radiomics feature, wavelet-
LHH_glszm_GrayLevelNonUniformityNormalized_
t2f, were associated with an increased output
probability.
This study presented SHAP dependence plots to
illustrate the 3 top-ranked radiomics feature
interactions. The results presented in figure 4 a-c,
show that (i) higher values of the first radiomics
feature paired with lower values of the second
increase the probability of high-grade meningiomas,
while lower first feature values diminish this effect;
(ii) lower values of the first feature combined with
higher third feature values decrease probability, and
(iii) the second feature values consistently impact
probability regardless of the third feature values.
Interestingly, in this study, age emerged as a
statistically significant feature in predicting
meningioma grade based on t-test analysis. However,
as depicted in the Figure 5.a, age does not exhibit a
specific distribution that increases the output
probability, aligning with (Hu et al. 2020; Duan, Li,
et al. 2022).
An Interpretable Machine Learning Model for Meningioma Grade Prediction
69

Table 2: Total and statistically significant features.
Subset
Total features Statisticall
y
si
g
nificant features
Clinical
Radiomics
Clinical
Radiomics
Ori
g
inal Wavelet Ori
g
inal Wavelet
T1C
T2
FLAIR
T1C
T2
FLAIR
T1C
T2
FLAIR
T1C
T2
FLAIR
Radiomics - 100 100 688 688 - 2 1 8 7
Clinical-
Radiomics
3 100 100 688 688 3 2 1 8 7
Table 3: Prediction performance of ML models.
Classifie
r
Subset Set AUC Accurac
y
Precision Recall F1-Score
LR
radiomics
Trainin
g
0.83 ± 0.15 0.75 ± 0.14 0.73 ± 0.20 0.75 ± 0.23 0.72 ± 0.18
Test 0.76 ± 0.08 0.76 ± 0.08 0.71 ± 0.11 0.77 ± 0.14 0.73 ± 0.09
Clinical-
radiomics
Training 0.88 ± 0.13 0.78 ± 0.14 0.78 ± 0.20 0.78 ± 0.22 0.75 ± 0.17
Test 0.80 ± 0.08 0.80 ± 0.08 0.76 ± 0.11 0.79 ± 0.13 0.76 ± 0.09
SVM
radiomics
Trainin
g
0.87 ± 0.14 0.80 ± 0.20 0.81 ± 0.20 0.75 ± 0.24 0.75 ± 0.18
Test 0.80 ± 0.07 0.80 ± 0.07 0.79 ± 0.11 0.75 ± 0.14 0.76 ± 0.09
Clinical-
radiomics
Trainin
g
0.90 ± 0.12 0.83 ± 0.13 0.84 ± 0.18 0.80 ± 0.21 0.80 ± 0.16
Test 0.83 ± 0.07 0.84 ± 0.06 0.82 ± 0.10 0.80 ± 0.11 0.80 ± 0.08
RF
radiomics
Trainin
g
0.83 ± 0.15 0.76 ± 0.14 0.77 ± 0.22 0.72 ± 0.24 0.73 ± 0.20
Test 0.76 ± 0.08 0.77 ± 0.08 0.73 ± 0.11 0.73 ± 0.14 0.72 ± 0.10
Clinical-
radiomics
Trainin
g
0.86 ± 0.14 0.78 ± 0.14 0.80 ± 0.21 0.72 ± 0.24 0.73 ± 0.19
Test 0.77 ± 0.08 0.77 ± 0.08 0.75 ± 0.12 0.72 ± 0.14 0.72 ± 0.10
GB
radiomics
Trainin
g
0.79 ± 0.17 0.72 ± 0.15 0.71 ± 0.23 0.69 ± 0.24 0.67 ± 0.20
Test 0.73 ± 0.09 0.73 ± 0.09 0.67 ± 0.10 0.71 ± 0.14 0.68 ± 0.10
Clinical-
radiomics
Trainin
g
0.79 ± 0.17 0.72 ± 0.15 0.70 ± 0.23 0.68 ± 0.25 0.67 ± 0.20
Test 0.72 ± 0.08 0.72 ± 0.08 0.67 ± 0.11 0.69 ± 0.15 0.67 ± 0.10
Figure 2: The overall radiomics feature importance scores of features extracted by SHAP. t1c: contrast-enhanced T1-
weighted; t2f: T2-weighted fluid attenuated inversion recovery.
0 0.2 0.4 0.6 0.8 1
wavelet-HLH_glszm_ZoneEntropy_t1c
wavelet-…
wavelet-…
wavelet-HLL_glrlm_RunVariance_t2f
wavelet-…
wavelet-…
wavelet-…
original_firstorder_Skewness_t1c
wavelet-…
original_glrlm_HighGrayLevelRunEmphasis_t1c
wavelet-HHH_firstorder_Skewness_t1c
wavelet-HLH_firstorder_Mean_t1c
wavelet-LHH_glszm_SizeZoneNonUniformityNormalized_t2f
wavelet-LHH_glszm_SmallAreaHighGrayLevelEmphasis_t2f
wavelet-HLH_glszm_SizeZoneNonUniformityNormalized_t2f
wavelet-HHL_glszm_SmallAreaLowGrayLevelEmphasis_t2f
original_glszm_HighGrayLevelZoneEmphasis_t2f
wavelet-HHL_glszm_HighGrayLevelZoneEmphasis_t1c
SHAP Overall Importance Scores
HEALTHINF 2025 - 18th International Conference on Health Informatics
70
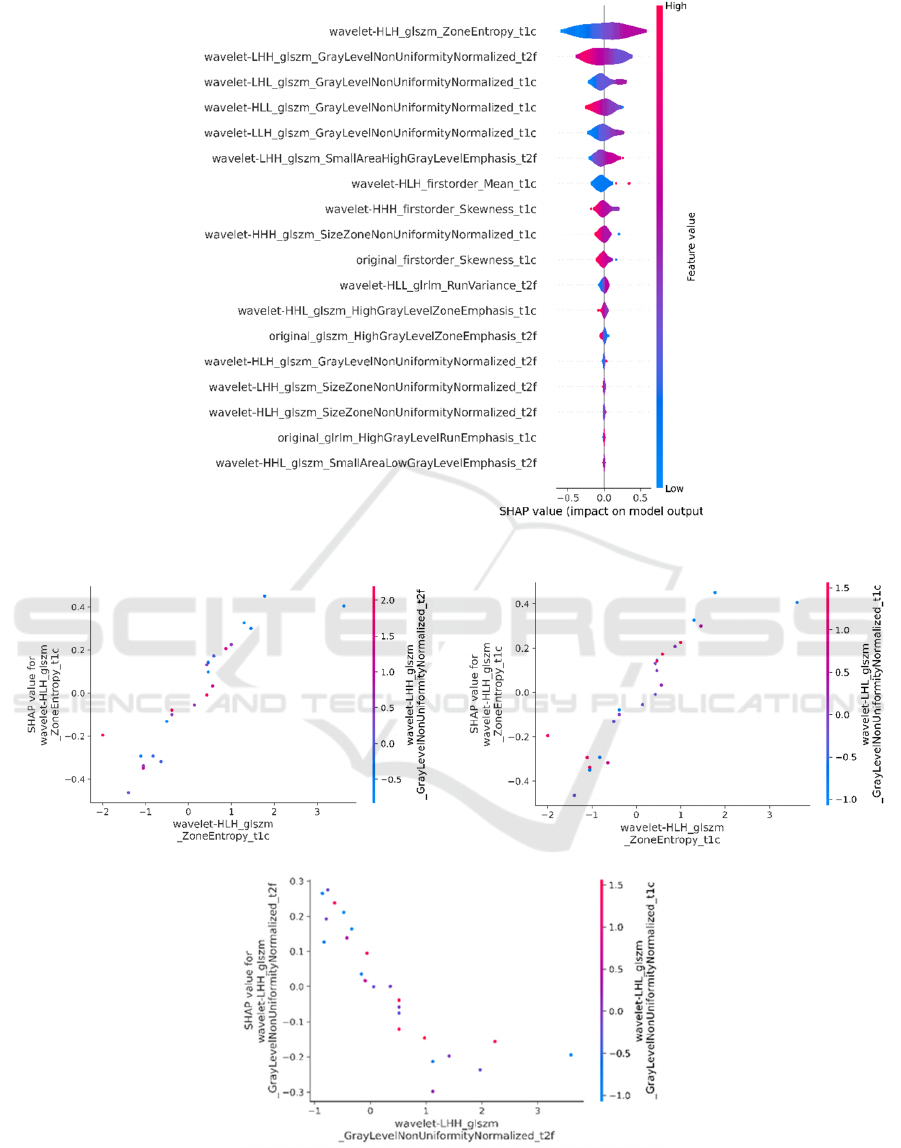
Figure 3: SHAP violin summary plot.
(a)
(b)
(
c
)
Figure 4: SHAP dependence plots for the three top-ranked radiomics features, illustrating the interactions between: (a) the
first and second features, (b) the first and third features, and (c) the second and third features.
An Interpretable Machine Learning Model for Meningioma Grade Prediction
71
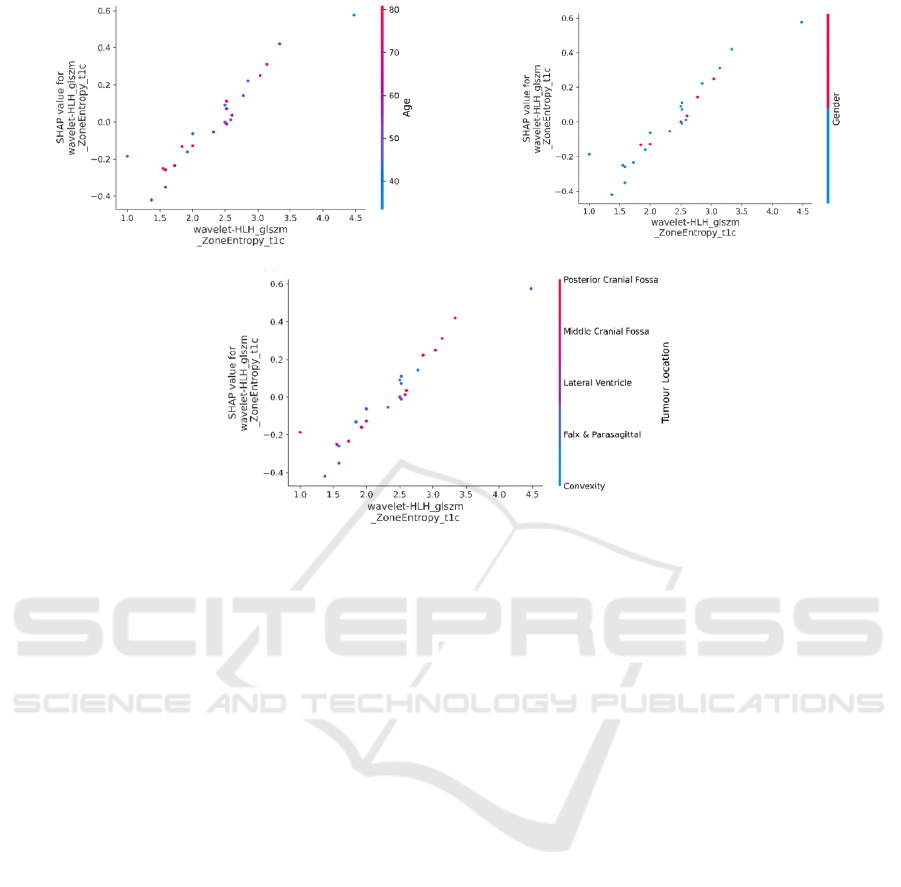
(a)
(b)
(
c
)
Figure 5: SHAP dependence plots for clinical and the top-ranked radiomics features, illustrating the interactions between the
top-ranked radiomics features and: (a) age, (b) gender and (c) tumour location.
Conversely, concerning gender (as depicted in figure
5.b), males (red dots) show a tendency to decrease the
output probability. Moreover, it is apparent that low
values of the first radiomics feature in males, tend to
decrease the output probability. This indicates that
relying solely on statistical tests is not sufficiently
reliable for predicting the effects of features on the
model predictions. It was also shown that for females
(blue dots), it generally increases the output
probability while lower values of the first radiomics
features tend to decrease the output probability.
Figure 5.c indicates that irrelevant to the tumour
location, lower values of the first radiomics feature
decrease the output probability while the combination
of the posterior cranial fossa and middle cranial fossa
locations and higher values of the first radiomics
feature tends to increase the output probability.
When a new case is presented, our model helps
clinicians to make better informed decisions by
providing insights into the factors influencing
predictions. Considering the interactions among
radiomics features themselves and their interaction
with clinical features may enable clinicians to
consider additional nuances in their clinical
judgments. Clinicians can also see which factors the
model considers most critical, helping them
understand the basis of the prediction. This also
enables comparison with previous cases. Clinicians
can compare the new case with similar past cases
where the model made predictions, seeing how the
new case aligns or differs, thereby validating the
model's prediction. Additionally, the model provides
detailed explanations for each prediction, breaking
down the contribution of each feature and offering a
clear rationale that clinicians can review. It also helps
identify anomalies. If a new case presents unusual
patterns or outliers in the data, interpretable models
can flag these anomalies, prompting further
investigation by clinicians to ensure the prediction is
accurate and relevant.
The current study has several limitations. TCIA,
the publicly available dataset used here was
retrospective, relatively small, and derived from a
single institution. Grade III meningiomas were also
excluded from the dataset due to their rare occurrence.
However, leveraging public datasets provides
researchers access to a diverse and extensive pool of
medical imaging data, enabling robust analysis and
enhancing the generalisability of findings across
various patient populations and clinical strategies.
Surprisingly, the utilisation of TCIA dataset accounts
for only 4% in the meningiomas-relevant studies (Patel
et al. 2023). This study only used two types of MRI
scans while other MRI scans such as ADC mapping
HEALTHINF 2025 - 18th International Conference on Health Informatics
72

were not considered. However, enhancing the clinical
management of meningiomas by constructing an
interpretable machine learning model that predicts
meningioma grade was the main objective of this
study.
5 CONCLUSIONS
Utilising clinical and radiomics features, the SVM
ML model, offers a reliable approach for preoperative
prediction of meningioma grade. By identifying
discriminative radiomic features and their
interactions with clinical features, SHAP supports the
potential for the enhanced clinical adoption of such
models. Future research should explore larger
datasets and diverse patients to validate and refine
these findings, further enhancing clinical adoption.
REFERENCES
An, Chansik, Yae Won Park, Sung Soo Ahn, Kyunghwa
Han, Hwiyoung Kim, and Seung-Koo Lee. 2021.
'Radiomics machine learning study with a small sample
size: Single random training-test set split may lead to
unreliable results', PloS one, 16: e0256152.
Carré, Alexandre, Guillaume Klausner, Myriam Edjlali,
Marvin Lerousseau, Jade Briend-Diop, Roger Sun,
Samy Ammari, Sylvain Reuzé, Emilie Alvarez Andres,
and Théo Estienne. 2020. 'Standardization of brain MR
images across machines and protocols: bridging the gap
for MRI-based radiomics', Scientific reports, 10:
12340.
Chu, Hairui, Xiaoqi Lin, Jian He, Peipei Pang, Bing Fan,
Pinggui Lei, Dongchuang Guo, and Chenglong Ye.
2021. 'Value of MRI radiomics based on enhanced
T1WI images in prediction of meningiomas grade',
Academic radiology, 28: 687-93.
Clark, K., B. Vendt, K. Smith, J. Freymann, J. Kirby, P.
Koppel, S. Moore, S. Phillips, D. Maffitt, M. Pringle,
L. Tarbox, and F. Prior. 2013. 'The Cancer Imaging
Archive (TCIA): maintaining and operating a public
information repository', J Digit Imaging, 26: 1045-57.
Duan, CF, N Li, Y Li, F Liu, JC Wang, XJ Liu, and WJ Xu.
2022. 'Comparison of different radiomic models based
on enhanced T1-weighted images to predict the
meningioma grade', Clinical Radiology, 77: e302-e07.
Duan, Chongfeng, Xiaoming Zhou, Jiachen Wang, Nan Li,
Fang Liu, Song Gao, Xuejun Liu, and Wenjian Xu.
2022. 'A radiomics nomogram for predicting the
meningioma grade based on enhanced T 1WI images',
The British Journal of Radiology, 95: 20220141.
Fountain, Daniel M., Adam M. H. Young, and Thomas
Santarius. 2020. 'Chapter 24 - Malignant meningiomas.'
in Michael W. McDermott (ed.), Handbook of Clinical
Neurology (Elsevier).
Gorgolewski, Krzysztof J, Tibor Auer, Vince D Calhoun, R
Cameron Craddock, Samir Das, Eugene P Duff,
Guillaume Flandin, Satrajit S Ghosh, Tristan Glatard,
and Yaroslav O Halchenko. 2016. 'The brain imaging
data structure, a format for organizing and describing
outputs of neuroimaging experiments', Scientific data,
3: 1-9.
Halchenko, Y, M Goncalves, and MVDO Castello. 2020.
'nipy/heudiconv v0. 9.0', Published online December, 23.
Han, Yuxuan, Tianzuo Wang, Peng Wu, Hao Zhang,
Honghai Chen, and Chao Yang. 2021. 'Meningiomas:
Preoperative predictive histopathological grading based
on radiomics of MRI', Magnetic Resonance Imaging,
77: 36-43.
Herrgott, Grayson A, James M Snyder, Ruicong She,
Tathiane M Malta, Thais S Sabedot, Ian Y Lee, Jacob
Pawloski, Guilherme G Podolsky-Gondim, Karam P
Asmaro, and Jiaqi Zhang. 2023. 'Detection of
diagnostic and prognostic methylation-based signatures
in liquid biopsy specimens from patients with
meningiomas', Nature Communications, 14: 5669.
Hu, Jianping, Yijing Zhao, Mengcheng Li, Jianyi Liu, Feng
Wang, Qiang Weng, Xingfu Wang, and Dairong Cao.
2020. 'Machine learning-based radiomics analysis in
predicting the meningioma grade using multiparametric
MRI', European journal of radiology, 131: 109251.
Islim, Abdurrahman I, Midhun Mohan, Richard DC Moon,
Nitika Rathi, Ruwanthi Kolamunnage-Dona, Anna
Crofton, Brian J Haylock, Samantha J Mills, Andrew R
Brodbelt, and Michael D Jenkinson. 2020. 'Treatment
outcomes of incidental intracranial meningiomas:
results from the IMPACT cohort',
World Neurosurgery,
138: e725-e35.
Jun, Yohan, Yae Won Park, Hyungseob Shin, Yejee Shin,
Jeong Ryong Lee, Kyunghwa Han, Sung Soo Ahn,
Soo Mee Lim, Dosik Hwang, and Seung-Koo Lee.
2023. 'Intelligent noninvasive meningioma grading
with a fully automatic segmentation using
interpretable multiparametric deep learning',
European radiology: 1-10.
Lambin, Philippe, Emmanuel Rios-Velazquez, Ralph
Leijenaar, Sara Carvalho, Ruud GPM Van Stiphout,
Patrick Granton, Catharina ML Zegers, Robert Gillies,
Ronald Boellard, and André Dekker. 2012. 'Radiomics:
extracting more information from medical images using
advanced feature analysis', European journal of cancer,
48: 441-46.
Langs, G, S Röhrich, J Hofmanninger, F Prayer, J Pan, C
Herold, and H Prosch. 2018. 'Machine learning: from
radiomics to discovery and routine', Der Radiologe, 58: 1.
Louis, David N, Arie Perry, Pieter Wesseling, Daniel J Brat,
Ian A Cree, Dominique Figarella-Branger, Cynthia
Hawkins, HK Ng, Stefan M Pfister, and Guido
Reifenberger. 2021. 'The 2021 WHO classification of
tumors of the central nervous system: a summary',
Neuro-oncology, 23: 1231-51.
Low, Justin T, Quinn T Ostrom, Gino Cioffi, Corey Neff,
Kristin A Waite, Carol Kruchko, and Jill S Barnholtz-
Sloan. 2022. 'Primary brain and other central nervous
system tumors in the United States (2014-2018): A
An Interpretable Machine Learning Model for Meningioma Grade Prediction
73

summary of the CBTRUS statistical report for
clinicians', Neuro-oncology practice, 9: 165-82.
Park, Chae Jung, Seo Hee Choi, Jihwan Eom, Hwa Kyung
Byun, Sung Soo Ahn, Jong Hee Chang, Se Hoon Kim,
Seung-Koo Lee, Yae Won Park, and Hong In Yoon.
2022. 'An interpretable radiomics model to select
patients for radiotherapy after surgery for WHO grade
2 meningiomas', Radiation Oncology, 17: 147.
Park, Yae Won, Seo Jeong Shin, Jihwan Eom, Heirim Lee,
Seng Chan You, Sung Soo Ahn, Soo Mee Lim, Rae
Woong Park, and Seung-Koo Lee. 2022. 'Cycle-
consistent adversarial networks improves
generalizability of radiomics model in grading
meningiomas on external validation', Scientific reports,
12: 7042.
Patel, Ruchit V, Shun Yao, Raymond Y Huang, and Wenya
Linda Bi. 2023. 'Application of radiomics to
meningiomas: a systematic review', Neuro-oncology,
25: 1166-76.
Reyes, Mauricio, Raphael Meier, Sérgio Pereira, Carlos A
Silva, Fried-Michael Dahlweid, Hendrik von Tengg-
Kobligk, Ronald M Summers, and Roland Wiest. 2020.
'On the interpretability of artificial intelligence in
radiology: challenges and opportunities', Radiology:
artificial intelligence, 2: e190043.
Spille, Dorothee Caecilia, Peter B Sporns, Katharina Hess,
Walter Stummer, and Benjamin Brokinkel. 2019.
'Prediction of high-grade histology and recurrence in
meningiomas using routine preoperative magnetic
resonance imaging: a systematic review', World
Neurosurgery, 128: 174-81.
Tagle, Patricio, Pablo Villanueva, Gonzalo Torrealba, and
Isidro Huete. 2002. 'Intracranial metastasis or
meningioma?: an uncommon clinical diagnostic
dilemma', Surgical neurology, 58: 241-45.
Traverso, Alberto, Michal Kazmierski, Ivan Zhovannik,
Mattea Welch, Leonard Wee, David Jaffray, Andre
Dekker, and Andrew Hope. 2020. 'Machine learning
helps identifying volume-confounding effects in
radiomics', Physica Medica, 71: 24-30.
Van Griethuysen, Joost JM, Andriy Fedorov, Chintan
Parmar, Ahmed Hosny, Nicole Aucoin, Vivek Narayan,
Regina GH Beets-Tan, Jean-Christophe Fillion-Robin,
Steve Pieper, and Hugo JWL Aerts. 2017.
'Computational radiomics system to decode the
radiographic phenotype', Cancer research, 77: e104-e07.
Vassantachart, A., Cao, Y., Shen, Z., Cheng, K., Gribble,
M., Ye, J. C., Zada, G., Hurth, K., Mathew.
"Segmentation and Classification of Grade I and II
Meningiomas from Magnetic Resonance Imaging: An
Open Annotated Dataset (Meningioma-SEG-CLASS)
(Version 1)." In, edited by The Cancer Imaging
Archive.
Vassantachart, April, Yufeng Cao, Michael Gribble,
Samuel Guzman, Jason C Ye, Kyle Hurth, Anna
Mathew, Gabriel Zada, Zhaoyang Fan, and Eric L
Chang. 2022. 'Automatic differentiation of Grade I and
II meningiomas on magnetic resonance image using an
asymmetric convolutional neural network', Scientific
reports, 12: 3806.
Vassantachart, April, Yufeng Cao, Zhilei Shen, Karen
Cheng, Michael Gribble, Jason C Ye, Gabriel Zada,
Kyle Hurth, Anna Mathew, and Samuel Guzman. 2023.
'A repository of grade 1 and 2 meningioma MRIs in a
public dataset for radiomics reproducibility tests',
Medical Physics.
Wang, Justin Z, Farshad Nassiri, Alexander P Landry,
Vikas Patil, Jeff Liu, Kenneth Aldape, Andrew Gao,
and Gelareh Zadeh. 2023. 'The multiomic landscape of
meningiomas: a review and update', Journal of Neuro-
oncology, 161: 405-14.
Wang, Ke, Ying An, Jiancun Zhou, Yuehong Long, and
Xianlai Chen. 2023. 'A novel Multi-Level feature
selection method for radiomics', Alexandria
Engineering Journal, 66: 993-99.
Wang, Xiuying, Dingqian Wang, Zhigang Yao, Bowen
Xin, Bao Wang, Chuanjin Lan, Yejun Qin, Shangchen
Xu, Dazhong He, and Yingchao Liu. 2019. 'Machine
learning models for multiparametric glioma grading
with quantitative result interpretations', Frontiers in
neuroscience, 12: 1046.
Xiang, Xu, Hong Yu, Ye Wang, and Guoyin Wang. 2023.
'Stable local interpretable model-agnostic explanations
based on a variational autoencoder', Applied
Intelligence: 1-15.
Yaniv, Ziv, Bradley C Lowekamp, Hans J Johnson, and
Richard Beare. 2018. 'SimpleITK image-analysis
notebooks: a collaborative environment for education
and reproducible research', Journal of digital imaging,
31: 290-303.
Zhang, Guobin, Yunsheng Zhang, Guijun Zhang, Da Li,
Zhen Wu, Yonggang Wang, and Junting Zhang. 2019.
'Outcome and prognostic factors for atypical
meningiomas after first recurrence', Journal of Clinical
Neuroscience, 63: 100-05.
Zhang, Jing, Kuan Yao, Panpan Liu, Zhenyu Liu, Tao Han,
Zhiyong Zhao, Yuntai Cao, Guojin Zhang, Junting
Zhang, and Jie Tian. 2020. 'A radiomics model for
preoperative prediction of brain invasion in
meningioma non-invasively based on MRI: A
multicentre study', EBioMedicine, 58: 102933.
Zou, Hui, and Trevor Hastie. 2003. 'Regression shrinkage
and selection via the elastic net, with applications to
microarrays', JR Stat Soc Ser B, 67: 301-20.
HEALTHINF 2025 - 18th International Conference on Health Informatics
74
