
Privacy-Preserving Mortality Prediction in ICUs Using Federated
Learning
Pedro Vieira
a
, Eva Maia
b
and Isabel Prac¸a
c
Instituto Superior de Engenharia do Porto, Polit
´
ecnico do Porto, Porto, Portugal
{pmrrv, egm, icp}@isep.ipp.pt
Keywords:
Federated Learning, Machine Learning, Artificial Intelligence, Mortality Prediction.
Abstract:
Managing multiple patients in an Intensive Care Unit (ICU) can be extremely challenging. By predicting
patient mortality, healthcare professionals can provide more efficient treatment and manage resources more
effectively. This allows for more precise and useful interventions, potentially preventing fatalities. Although
artificial intelligence (AI) is making significant advancements in this field, traditional Machine Learning (ML)
continues to be the most widely used AI method, though it raises concerns about data security in collaborative
environments. Since ensuring the safe handling of patients’ private data is crucial, Federated Learning (FL)
has emerged as a viable alternative. Its intrinsic characteristics offer a valuable solution for training predictive
models securely, as raw data does not need to be shared between participants. In this study, FL was used
to develop models capable of predicting ICU patient mortality while protecting data privacy. Using data
from the MIMIC-IV dataset, the most accurate model achieved an accuracy of 0.886, a recall of 0.817, and
a specificity of 0.965, surpassing all the analyzed studies. A comparison between FL and traditional ML
approaches revealed similar performance results. Moreover, three FL aggregation algorithms were evaluated,
a less common focus in this area of research. Federated Averaging performed best with some classifiers,
while delivering results comparable to FederAdagrad and FedAdam with others. In conclusion, the findings
demonstrate that FL can be as effective as traditional ML for mortality prediction, with the added benefit of
enhanced data privacy.
1 INTRODUCTION
Predicting mortality in Intensive Care Units (ICUs)
can offer various advantages to healthcare profession-
als and facilities. Firstly, it enables the hospitals and
medical professionals to allocate resources more ef-
fectively by identifying patients at higher risk of de-
terioration. This allows for prioritization in monitor-
ing, treatment, and intervention strategies, potentially
reducing preventable deaths. Furthermore, mortality
prediction tools, often powered by Machine Learn-
ing (ML), can enhance decision-making regarding
treatment plans and end-of-life care, providing fam-
ilies and medical teams with data-driven insights to
support difficult choices. Additionally, by predict-
ing mortality, it is possible to do a more efficient re-
sources allocation. In this context, Artificial Intelli-
gence (AI) shows itself as a weapon to forecast and
fight deadly outcomes (Holmstr
¨
om et al., 2023).
a
https://orcid.org/0009-0003-9103-896X
b
https://orcid.org/0000-0002-8075-531X
c
https://orcid.org/0000-0002-2519-9859
However, a critical issue that must always be
addressed when applying AI in healthcare is pri-
vacy (Khalid et al., 2023). While the vast amount
of healthcare data offers immense potential for ad-
vancing medical research and innovations, it also re-
quires measures to protect the security and privacy
of individuals (Rieke et al., 2020). Ensuring the
confidentiality, integrity, and secure management of
patient data is essential, especially considering the
highly sensitive nature of health information (Stan-
fill and Marc, 2019). To address this challenge,
Federated Learning (FL) has emerged as an innova-
tive ML approach designed to balance data privacy
with data storage needs (Mammen, 2021). In FL,
multiple clients collaborate with one or more central
servers in decentralized ML setups. This approach
enables models to learn from distributed data sources
without exposing sensitive information, ensuring pri-
vacy while promoting collaborative insights (Mam-
men, 2021).
Even if FL brings several advantages, mainly con-
sidering the privacy of the data, it also presents some
Vieira, P., Maia, E. and Praça, I.
Privacy-Preserving Mortality Prediction in ICUs Using Federated Learning.
DOI: 10.5220/0013120600003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 87-95
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
87

issues. Integrating FL seamlessly into the healthcare
domain while navigating the details of varied data
sources, guaranteeing model accuracy, and address-
ing regulatory conformity requires innovative strate-
gies and robust frameworks. The decentralization of
the data encompasses different types of data partition-
ing, and selecting the appropriate partitioning method
is critical to the success of the model. The most
common approaches include: Horizontal Federated
Learning (HFL), where different clients have data
on the same features but for different individuals or
cases (Yang et al., 2019); Vertical Federated Learning
(VFL), where multiple sources each provide differ-
ent features for the same group of individuals or cases
(Zhuang et al., 2016); and Federated Transfer Learn-
ing (FTL), which applies transfer learning techniques
to build a new model using a pre-trained one (Yang
et al., 2023). HFL is the most common approach in
the scope of healthcare in the literature (Sharma and
Guleria, 2024). VFL is pertinent in scenarios where
different domains collaborate to train a global model
using shared data that are not linked (Zhuang et al.,
2016). This methodology allows the collaboration
and utilization of data across unrelated domains while
preserving the confidentiality of sensitive information
unique to each domain. In other words, in HFL the
features are the same in every client, while in VFL
the clients have different features. FTL employs the
conventional ML-based transfer learning technique
to train a new requirement on a pre-trained frame-
work that has already undergone training on a simi-
lar dataset. This way it is possible to address an en-
tirely distinct problem. The fundamental concept be-
hind FTL revolves around the diversity in characteris-
tics among different participants. It addresses issues
related to limited or inadequate data by effectively
leveraging knowledge transfer while simultaneously
ensuring the security (Yang et al., 2023).
Beyond the choice of partitioning strategy, the ag-
gregation of model updates from decentralized data
sources presents additional challenges. To address
this, various aggregation techniques are employed,
each designed to cope with the issues of non-IID
(non-independent and identically distributed) data
across clients (Lazzarini et al., 2023). Federated Av-
eraging (FedAvg) is one of the most common meth-
ods, where client updates are averaged to create the
global model, but it may struggle with heterogeneous
data (Nilsson et al., 2018). More advanced optimizers
like FedAdam (C¸ elik and G
¨
ull
¨
u, 2023) and FedAda-
grad (C¸ elik and G
¨
ull
¨
u, 2023) apply adaptive learn-
ing rate strategies from classical optimization tech-
niques to better handle variations in data distribu-
tion. FedAdam builds on the Adam optimizer, ad-
justing the learning rate based on momentum, while
FedAdagrad adapts learning rates per parameter, im-
proving convergence for clients with differing data
scales. Selecting the right aggregation technique is
crucial for ensuring model performance, fairness, and
generalizability across diverse and partitioned health-
care datasets.
The aim of this work is to develop a FL approach
that allows to predict the mortality of ICU patients
while keeping in mind the privacy of health data.
For that, an HFL solution was developed to analyse
the performance of different algorithms, combined
with different FL aggregation algorithms, using a sub-
set of Medical Information Mart for Intensive Care
(MIMIC) IV dataset (Johnson et al., 2023). Logis-
tic Regression, Decision Tree, Random Forest, Sup-
port Vector Classifier (SVC), and Multi-Layer Per-
ceptron (MLP) models were employed, alongside dif-
ferent aggregation methods, to assess and compare
the impact of FL on prediction accuracy. This way
we were able to perform a comparative analysis be-
tween different aggregation algorithms, an aspect of-
ten missing in most studies in this field, and between
traditional ML and FL approaches. Furthermore, un-
like most previous research, this work also includes a
time efficiency comparison, highlighting which clas-
sifiers and algorithms provide faster predictions of pa-
tient mortality. By optimizing time, we also reduce
energy and resource consumption, a crucial factor in
today’s context. This not only results in significant
cost savings (Kim et al., 2023) but also contributes to
environmental sustainability (Rosen, 2021), aligning
financial efficiency with ecological responsibility.
The paper is structured into different sections. The
first section introduced the topic. Section 2 aims to
present and discuss works related to the scope. Sec-
tion 3 focus on the methodology employed in the
study. Section 4 presents and discusses the results
obtained by the authors. Lastly, Section 5 displays
a conclusion, while showing options for future work.
2 RELATED WORK
FL has been used to predict patients mortality in the
recent years. Randl, et al.’s (Randl et al., 2023) work
concludes that FL can be efficiently employed to pre-
dict early mortality in ICU patients. This conclu-
sion is supported by the fact that the FL approach
utilized by the authors also presented comparable re-
sults to those presented by traditional ML. More-
over, the study adds that utilizing the F1-Score met-
ric can result in a more efficient model performance.
Therefore, it was possible for Randl et al. to ad-
HEALTHINF 2025 - 18th International Conference on Health Informatics
88

mit that FL can be a reliable solution to predict pa-
tients outcome, while securing their privacy. Further-
more, Georgoutsos’ work (Georgoutsos, 2023) esta-
bilished another comparison between FL and tradi-
tional ML. This time, the author concluded that, in
settings with non-IID datasets, FL models outperform
privacy-preserving Local Machine Learning (LML)
models in terms of AUROC, AUPRC, and F1-Score.
Nevertheless, he also concluded they generally per-
form slightly worse than Centralized Machine Learn-
ing (CML) models. Furthermore, his study stated that
the effectiveness of specific FL algorithms depends
on the characteristics of the FL elements, such as
data size and class representation. Mondrejevski et
al. (Mondrejevski et al., 2022) also delved into the
comparison between traditional ML and FL. The au-
thors came to the conclusion that CML and FL have
comparable performances when judging metrics such
as AUPRC and F1-Score, which goes against the find-
ings of the previous study. Just as Georgoutsos con-
cluded, they also stated that FL performs better than
LML, which seems to confirm this tendency. Despite
focusing on predicting patients mortality prediction,
none of the three studies seemed to focus on the dif-
ferent aggregation algorithms performance, which is
a pivot matter in this study.
Vieira et al. (Vieira et al., 2024) evaluated the
performance of several algorithms, with and without
FL techniques, to assess and compare the impact of
FL on predicting mortality in acute pancreatitis pa-
tients. Their results indicate that both traditional and
FL methods are highly effective in predicting mor-
tality in this patient population. The authors exam-
ined different aggregation algorithms, with FedAvg
emerging as the best choice, although the three ag-
gregation methods yielded similar results. However,
the study is limited to the specific medical condition
of acute pancreatitis, a very focused area. Expanding
the scope to encompass a broader range of diseases
would not only increase the dataset size but also al-
low for a deeper analysis of how FL models perform
with larger and more diverse datasets.
3 METHOD
The following subsections will explain the approach
used in creating both the FL and ML models,
designed for comparing and evaluating purposes.
Firstly, the dataset that was selected will be elaborated
on. Following that, the pre-processing steps will be
detailed, finalizing with the information regarding the
FL procedure.
3.1 Dataset
Several healthcare datasets have been employed in
previous studies, such as various versions of the
MIMIC dataset family and the eICU Collabora-
tive Research Database (eICU-CRD) (Pollard et al.,
2018). In this study, the MIMIC-IV dataset (John-
son et al., 2023) was used, which is one of the largest
publicly accessible healthcare datasets. This dataset
includes detailed records of patients admitted to In-
tensive Care Units (ICUs) at a major tertiary care hos-
pital. It offers a vast range of medical data, such as vi-
tal signs, medications, laboratory test results, provider
notes, fluid balance, procedure and diagnostic codes,
imaging reports, hospital stay durations, survival in-
formation, and more. It is important to highlight,
however, that access to this dataset is restricted and
requires prior approval. To gain access, individuals
must complete a credentialing process, undergo re-
quired training, and sign a data use agreement.
MIMIC-IV is organized into various tables, each
containing distinct types of data. For the purposes of
this study, only a subset of these tables, specifically
those containing data relevant to the research objec-
tives, were selected. The tables used in this research
include the following:
admissions. This table is responsible for having data
regarding the admission of patients to the hospi-
tal. Even though a patient can be admitted more
than once, each row represents a single admission
exclusively.
patients. The patients table contains demographic
information for each admitted patient, including
details such as gender and age.
chartevents. This table records measurements and
observations about patients during their hospital
stay. It is one of the most crucial tables, as it con-
tains a wide range of medical data, primarily vital
signs, along with laboratory results, intake/output
measurements, and other clinical observations.
d items. This table functions as a map between code
and name for items recorded in the chartevents.
Each ID from the previous table is mapped to its
corresponding meaning here.
The admissions and patients tables were used
to identify the first admission for each adult patient.
Additionally, the patients table provided key demo-
graphic information such as age and gender, both
of which were utilized as categories in the model’s
training and testing phases. The necessary medi-
cal data for model creation was extracted from the
chartevent table, made possible through the d Items
Privacy-Preserving Mortality Prediction in ICUs Using Federated Learning
89
table, which provided the definitions for each code
found in chartevents.
3.2 Pre-Processing
The first step when dealing with the data provided by
the MIMIC-IV dataset was removing all the under-
age patients. This was needed due to the fact that mi-
nors may introduce variables that come with the risk
of impacting the prediction significantly (Bavdekar,
2013). Removing all the lines that included null val-
ues was also an employed technique, as we were
dealing with large portions of data and the removal
did not harm the quality and the size of the dataset.
Next, all the variables were converted into binary for-
mat. The majority of models benefit from binary data,
which allows them to have better results. Moreover,
the dataset also exhibits an imbalance in the “hospi-
tal expire flag” category, which serves as the depen-
dent variable indicating whether a patient survived or
died. This imbalance could potentially introduce bias
into the model’s predictions. Therefore, undersam-
pling and SMOTE techniques were both tested in or-
der to balance it. As undersampling, with 50/50 rep-
resentation, presented the best results, it was chosen
to fight the imbalance problem, resulting in the final
dataset utilized to train the models. Additionally, un-
like SMOTE, undersampling has the advantage of re-
lying solely on real data, without the need for synthet-
ically generated samples.
3.3 Federated Learning
FL enables distributed model training on multiple de-
vices or servers, each of which retains its local data,
eliminating the need for data sharing between loca-
tions. Instead of transferring raw data to a central
server, FL allows models to be trained locally, and
only the resulting model updates or gradients are sent
to a central server for aggregation. In this work, the
authors focus on a network of hospitals collaborating
to create a unified model for predicting mortality in
ICU patients. So, to coordinate the three hospitals,
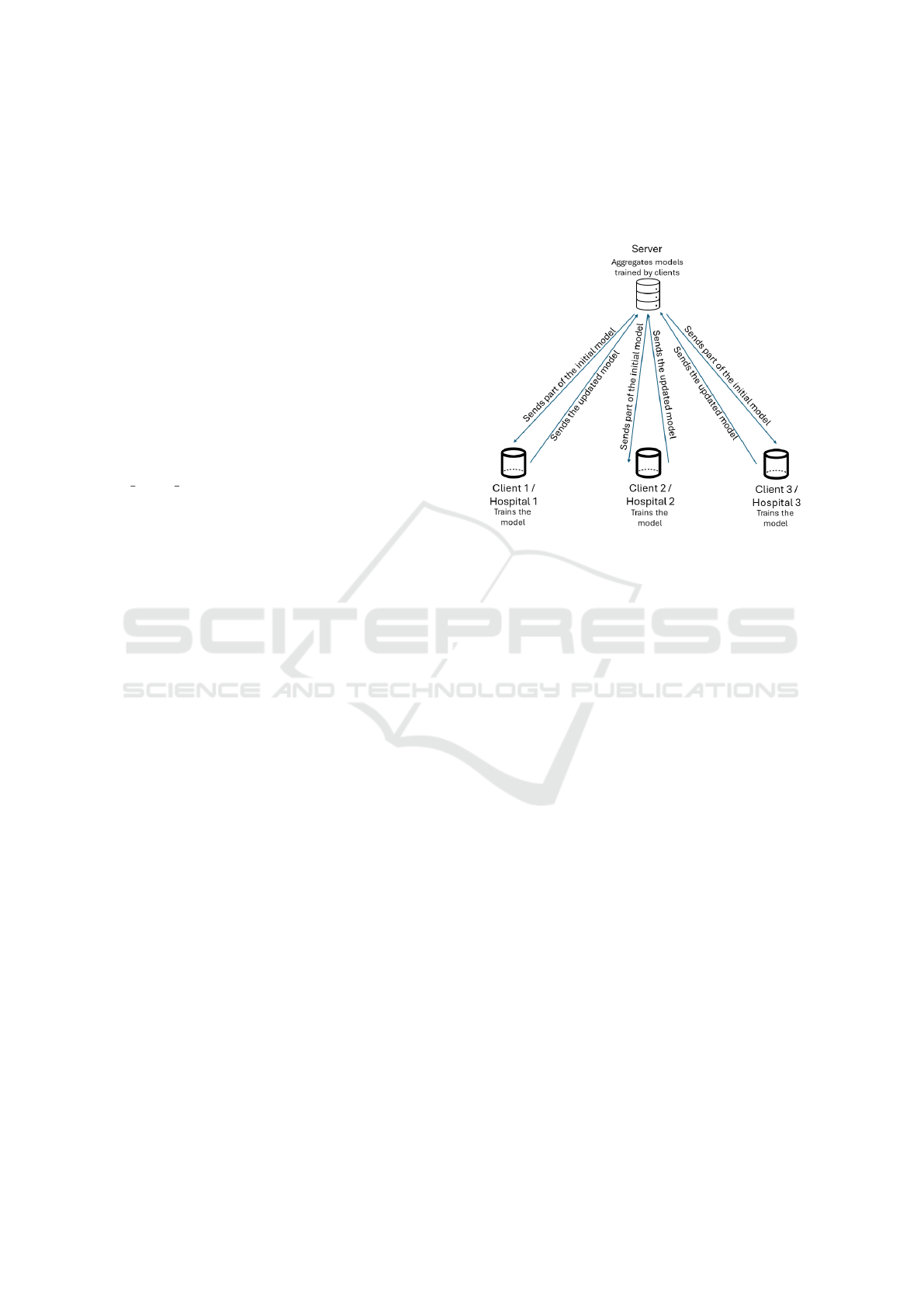
a server was linked to them, resulting in a healthcare
network, as illustrated in Figure 1. The server starts
by initializing a model and splitting it between the
three clients, which will locally train the model and
send it back to the server. The server then aggregates
the models received and once again splits them be-
tween the clients (Nilsson et al., 2018). This happens
for six rounds, which are iterations of the training pro-
cess, until a final model is obtained. Moreover, it is
important to clarify that each one of the clients also
had 70% of data distributed for training and 30% for
testing. FL is particularly suited for this scenario, as
it balances the need for data privacy with the collab-
orative nature of the hospitals’ efforts in building a
shared predictive model.
Figure 1: FL Configuration.
For the technical implementation of this scenario,
Flower (Beutel et al., 2022), a multi-platform, open-
source framework designed for secure execution of
FL, was selected. The FL architecture built using
Flower consists of various components for both the
server and the clients (Figure 2). On the server
side, three aggregation algorithms are represented:
FedAvg, FedAdam, and FedAdagrad. These algo-
rithms, which are utilized one at a time, provide dif-
ferent methods for aggregating the model updates
from the clients. Pivotal to the server’s operations
is the FL Loop, which coordinates the iterative pro-
cess of distributing the global model to the clients
and aggregating their locally trained models, accord-
ing to the selected aggregation algorithm. Communi-
cation between the server and clients is facilitated by
the gRPC server, which manages the transmission of
global model parameters to the clients and the recep-
tion of updated parameters from them. On the client
side, each identical client comprises three main com-
ponents: the gRPC client, the Flower client, and the
local data. The gRPC client handles the communi-
cation with the server’s gRPC server, ensuring the
smooth exchange of model parameters. The Flower
client, implemented in Python, is responsible for in-
tegrating the local ML environment with the FL pro-
cess. It receives the global model parameters from
the server, applies them to the local model, trains the
model using the local data, and sends the updated
model parameters back to the server.
HEALTHINF 2025 - 18th International Conference on Health Informatics
90

Figure 2: Flower Architecture.
4 EXPERIMENTAL RESULTS
In addition to implementing FL models, the authors
opted to develop traditional ML models using the
same pre-processed dataset to ensure a fair compar-
ison between the two approaches. The models de-
veloped for both methods include Logistic Regres-
sion, Decision Tree Classifier, Random Forest Clas-
sifier, Support Vector Classifier (SVC), and Multi-
Layer Perceptron (MLP) Classifier. As previously
mentioned, for the FL approach, three aggregation
algorithms were utilized: FedAvg, FedAdam, and
FedAdagrad. It is important to note that due to the
lengthy training time required for the SVC model, the
scenario was divided into two approaches: in the first,
only 10% of the dataset was used to allow for SVC
training; in the second, the complete dataset was used,
excluding the SVC model from training. This two-
pronged strategy enabled the authors to obtain results
for the SVC model while still using the full dataset for
the other classifiers.
Table 1 presents the results obtained for ML and
FL approaches, with only 10% of the dataset. It fa-
cilitates a rightful comparison between several classi-
fiers and aggregation algorithms, while also compar-
ing ML and FL approaches. These results are capable
of providing different insights into the performance
of ML and FL algorithms in predicting mortality for
ICU patients.
Upon comparing all the trained models, it be-
comes evident that the Random Forest and Decision
Tree models, both trained using traditional ML, ex-
hibit the highest metric values. The Decision Tree
had the highest Precision and F1-Score, with Random
Forest having similar values. Additionally, the FL
MLP paired with FedAdam demonstrated the highest
specificity, and the FL SVC combined with FedAda-
grad achieved the best recall.
Nonetheless, other conclusions can be drawn. It
can be observed that, in general, the FedAvg mod-
els perform comparably to traditional ML models, as
supported by literature (Tu et al., 2022). However, the
MLP experienced a significant decline in metric val-
ues relative to other models. Furthermore, the MLP
also performed worse when using other aggregation
algorithms. It is also worth noting that three out of
five classifiers exhibited similar performance across
the three aggregation algorithms. However, two ex-
ceptions stand out: FedAdagrad produced notably
poor results when training both the SVC and MLP
compared to other algorithms, a trend supported by
other studies on other scopes (Vieira et al., 2024), in-
dicating this may be a recurring pattern with this algo-
rithm. Similarly, the MLP also underperformed with
FedAdam compared to FedAvg, as previously noted.
Another key goal of this experiment was to ana-
lyze the training times of the various algorithms. Tra-
ditional ML models were found to train faster than
FL models, which is expected since ML models do
not require aggregation of multiple models and do
not involve multiple training rounds, unlike FL. De-
spite this, the time required for the best-performing
models was not excessively long. The SVC is the
slowest classifier, making it an unsuitable choice if
time, energy, and resources are a priority. In con-
trast, Logistic Regression and Decision Tree were the
fastest, completing training in less than 0.02 min-
utes. Random Forest also trained quickly in the ML
setting, though it was slightly slower in FL, taking
around 0.30 minutes. Nevertheless, considering its
strong performance metrics, it remains a viable op-
tion. It’s important to note that only 10% of the
dataset was used in this analysis, meaning the models
would require more time to train on the complete pre-
processed dataset, as will be discussed next. Finally,
the results show that the three aggregation algorithms
Privacy-Preserving Mortality Prediction in ICUs Using Federated Learning
91
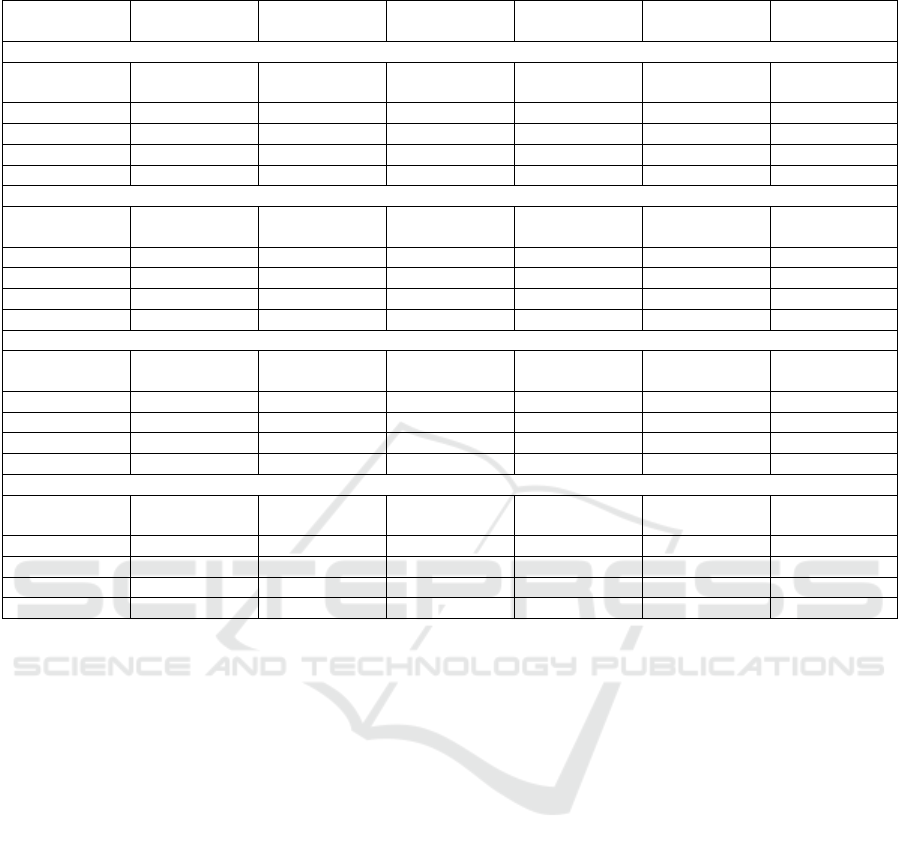
Table 1: ML and FL results to General Diseases Mortality Prediction – 10% of the dataset.
Algorithm Accuracy Precision Recall F1-score Specificity Training Time
(Minutes)
Machine Learning
Logistic Regres-
sion
0.839 0.832 0.790 0.810 0.863 0.00
Decision Tree 0.856 0.892 0.794 0.842 0.918 0.00
Random Forest 0.857 0.883 0.802 0.841 0.909 0.03
SVC 0.843 0.852 0.798 0.824 0.881 0.89
MLP 0.843 0.865 0.784 0.822 0.895 0.55
Federated Learning - FedAvg
Logistic Regres-
sion
0.833 0.822 0.806 0.814 0.855 0.02
Decision Tree 0.860 0.883 0.796 0.838 0.913 0.02
Random Forest 0.859 0.869 0.812 0.840 0.898 0.30
SVC 0.841 0.836 0.811 0.823 0.868 66.89
MLP 0.747 0.844 0.544 0.662 0.917 3.32
Federated Learning - FedAdam
Logistic Regres-
sion
0.827 0.820 0.798 0.809 0.805 0.02
Decision Tree 0.860 0.884 0.797 0.839 0.911 0.02
Random Forest 0.858 0.868 0.812 0.839 0.897 0.29
SVC 0.841 0.832 0.814 0.823 0.863 65.10
MLP 0.660 0.816 0.324 0.468 0.939 3.30
Federated Learning - FedAdagrad
Logistic Regres-
sion
0.818 0.819 0.795 0.807 0.845 0.02
Decision Tree 0.860 0.883 0.798 0.838 0.912 0.02
Random Forest 0.857 0.866 0.811 0.838 0.896 0.30
SVC 0.457 0.455 0.996 0.625 0.451 68.83
MLP 0.552 0.890 0.014 0.028 0.936 3.25
required a similar amount of time to finish training.
Therefore, when it comes to time, the choice between
them becomes almost negligible.
The results presented in Table 2 facilitate a sim-
ilar comparison to those in Table 1, but utilizing the
complete dataset. By comparing the trained models,
it becomes evident that the Random Forest trained
with traditional ML outperforms the others, achiev-
ing the highest Accuracy, Precision, and F1-Score.
The best Recall was found in both the FedAvg and
FedAdagrad Decision Trees, while FedAvg’s Random
Forest showed the highest Specificity. Once again,
the MLP underperformed in the FL setting, particu-
larly when trained with FedAdam and FedAdagrad, as
compared to traditional ML. However, all other clas-
sifiers demonstrated comparable performance to tra-
ditional ML models and among themselves. Despite
this, FedAvg showed a slight advantage, although the
difference was minimal across three out of four mod-
els. It is also clear that FL requires significantly more
time to train than traditional ML, due to its iterative
and aggregative processes, a tendency also shown in
the previous case. In terms of training time, Logistic
Regression and Decision Tree were the fastest, while
MLP was the slowest, a trend already observed in the
analysis of 10% of the dataset. Random Forest, on
the other hand, struck a good balance between perfor-
mance and training time.
As this topic has been explored in previous ML
studies, a comparison was made between the best ML
model trained in this work and traditional ML models
from other studies. Table 3 presents this comparison,
featuring some of the most notable works focused
on training ML models for ICU mortality prediction.
The ML Random Forest from this work provided the
best performance metrics in three out of the five an-
alyzed metrics. In other words, it showed the best
Accuracy, Precision and F1-Score, despite Iwase, et
al.’s Random Forest presenting the best Recall value.
It also presented the second-best Specificity, trailing
behind Alghatani et al.’s Random Forest.
In a similar way, a comparison was conducted be-
tween the best FL model trained in this study (the
Random Forest aggregated with FedAvg) and FL
models from other research, as shown in Table 4. This
table includes all the relevant works that employed
at least one of the metrics used in this work. How-
ever, since none of them applied all the same met-
HEALTHINF 2025 - 18th International Conference on Health Informatics
92
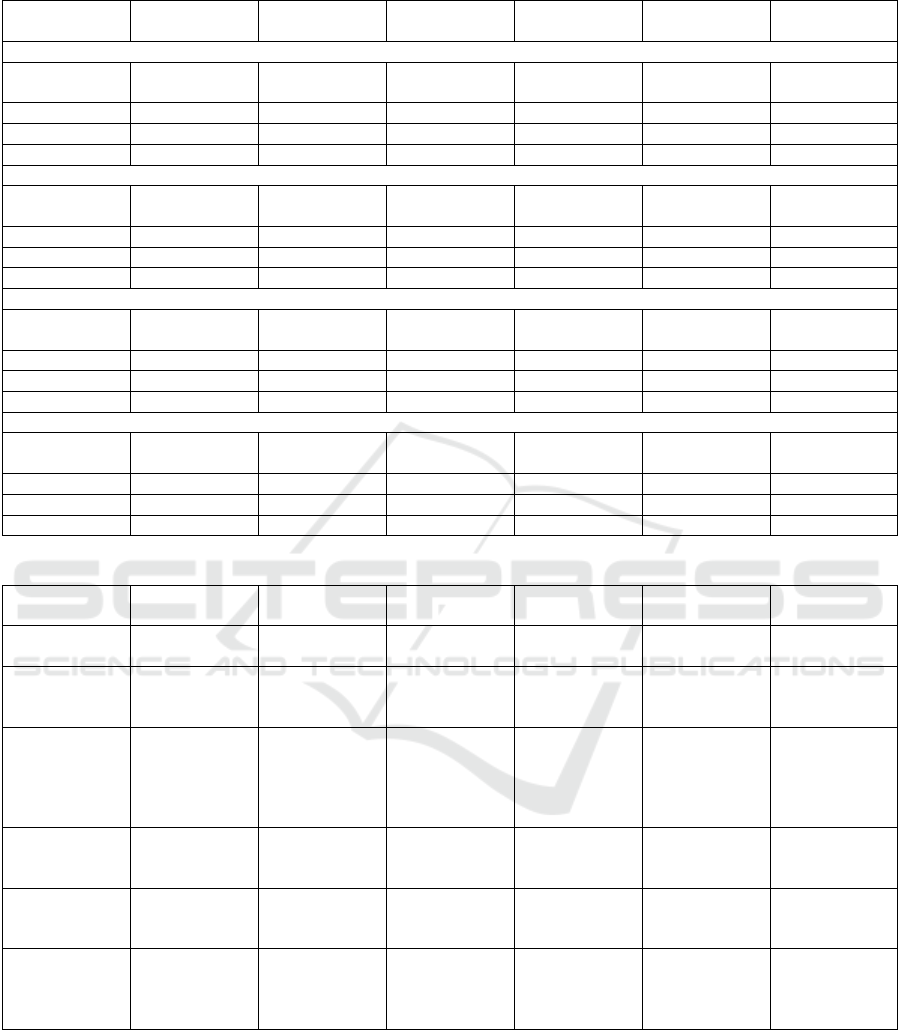
Table 2: ML and FL results to General Diseases Mortality Prediction – complete dataset.
Algorithm Accuracy Precision Recall F1-score Specificity Training Time
(Minutes)
Machine Learning
Logistic Regres-
sion
0.843 0.823 0.794 0.808 0.875 0.03
Decision Tree 0.892 0.933 0.820 0.873 0.951 0.01
Random Forest 0.893 0.936 0.821 0.875 0.953 0.34
MLP 0.843 0.865 0.784 0.822 0.895 4.575
Federated Learning - FedAvg
Logistic Regres-
sion
0.840 0.824 0.796 0.810 0.868 0.08
Decision Tree 0.892 0.933 0.822 0.874 0.948 0,17
Random Forest 0.886 0.935 0.817 0.870 0.965 3,04
MLP 0.756 0.720 0.749 0.734 0.818 36,78
Federated Learning - FedAdam
Logistic Regres-
sion
0.830 0.821 0.799 0.810 0.845 0.08
Decision Tree 0.890 0.930 0.821 0.871 0.951 0,02
Random Forest 0.884 0.928 0.808 0.864 0.938 3,02
MLP 0.546 0.900 0.454 0.603 0.832 35,88
Federated Learning - FedAdagrad
Logistic Regres-
sion
0.829 0.815 0.795 0.808 0.834 0.08
Decision Tree 0.889 0.928 0.822 0.856 0.950 0.19
Random Forest 0.885 0.927 0.820 0.875 0.942 3,12
MLP 0.546 0.645 0.102 0.176 0.435 36,56
Table 3: General Diseases Mortality Prediction – State-of-the-art ML results comparison.
Classifier Accuracy Precision Recall F1-Score Specificity Training Time -
Minutes
This Work’s Ran-
dom Forest
0.893 0.936 0.821 0.875 0.953 0.34
Iwase, et al.’s Ran-
domForest (Iwase
et al., 2022)
Unknown Unknown 0.865 Unknown 0.875 Unknown
Nistal-Nu
˜
no’s
Extreme Gradient
Boosting (XGB)
(Nistal-Nu
˜
no,
2022)
0.855 0.528 0.831 0.645 0.860 Unknown
Pang et al.’s XG-
Boost(Pang et al.,
2022)
0.834 0.842 0.822 0.831 0.846 Unknown
Chia, et al.’s best
XGB (Chia et al.,
2021)
0.819 0.420 0.615 0.499 0.689 Unknownn
Alghatani et al.’s
Random Forest
(Alghatani et al.,
2021)
0.885 0.840 0.095 0.171 0.997 Unknown
rics, it ended up being a challenge in making a direct
and complete comparison. Furthermore, even though
some studies used certain metrics to evaluate their
models, a comparison with their findings was not pos-
sible, as those particular metrics were not used in this
work. First, Randl et al. (Randl et al., 2023) pro-
vided the most comprehensive set of metrics, allow-
ing for a fairer comparison with their work compared
to other authors. Although they presented multiple re-
sults due to the variety of models used, only their best
FL model was selected for this comparison. It be-
comes evident that their results are generally inferior
to most of the models trained in this study, both in
ML and FL. Unfortunately, Georgoutsos (Georgout-
Privacy-Preserving Mortality Prediction in ICUs Using Federated Learning
93
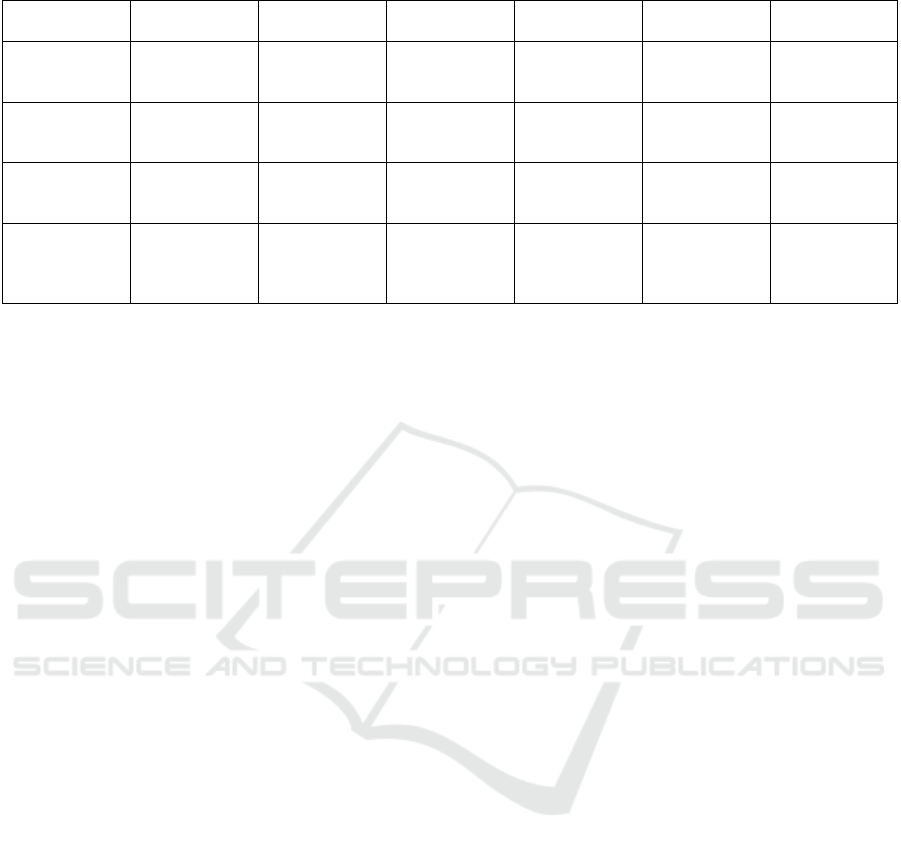
Table 4: General Diseases Mortality Prediction – State-of-the-art FL results comparison.
Classifier Accuracy Precision Recall F1-Score Specificity Training Time -
Minutes
This Work’s Ran-
dom Forest - Fe-
dAvg
0.886 0.935 0.817 0.870 0.965 3.04
Randl, et al. best
FL model (Randl
et al., 2023)
Unknown 0.520 0.460 0.480 Unknown Unknown
Georgoutsos best
FL model (Geor-
goutsos, 2023)
Unknown Unknown Unknown 0.512 Unknown Unknown
Mondrejevski et
al. best FL model
(Mondrejevski
et al., 2022)
Unknown Unknown Unknown 0.830 Unknown Unknown
sos, 2023) and Mondrejevski et al. (Mondrejevski
et al., 2022) only reported the F1-Score, out of all
the metrics used in this work. Georgoutsos showed
a low F1-Score, which underperformed compared to
most of the models discussed here (with the excep-
tion of the MLP model using the FedAdagrad algo-
rithm). Lastly, Mondrejevski et al. [90] reported
the highest F1-Score among the three state-of-the-art
works. While this result surpasses some of the models
presented, it does not outperform the top-performing
models from Table 2, specifically the Random Forest
and Decision Tree trained with any of the aggregation
algorithms.
After carefully analysing the results for both ap-
proaches (10% of the dataset, and the total dataset), it
was possible to confirm some results previously pre-
sented. The MLP tends to be less accurate in a FL set
and the SVC performance with FedAdagrad was once
more much worse than in any other approach (both in
FL and traditional ML). Moreover, as expected, the
SVC is the slowest classifier, while Logistic Regres-
sion is the fastest. No significant difference between
the time needed. In terms of performance, most of
the aggregation algorithms’ models showed compa-
rable performances between each other and with the
ML models. However, traditional ML models still had
the best metrics, even though the difference was not
significant. In terms of FL only, FedAvg was slightly
better than the other two aggregation algorithms, even
though the performances were practically equivalent.
5 CONCLUSIONS AND FUTURE
WORK
In conclusion, this study aimed to predict ICU pa-
tient mortality using a privacy-preserving approach
through FL, comparing different aggregation algo-
rithms and traditional ML methods. The MIMIC-
IV dataset was used, and a network of hospitals was
simulated, where multiple clients collaborated to train
models using Horizontal Federated Learning.
The experimental results indicated that FL mod-
els demonstrated comparable performance in several
cases, particularly when utilizing the FedAvg aggre-
gation algorithm, consistent with findings in the ex-
isting literature. The study also emphasized the trade-
offs between model performance and training time,
noting that FL models generally require more time
due to the aggregation and iterative processes in-
volved. Nevertheless, since the differences in time
and performance are not substantial, FL remains a
viable option for scenarios where privacy is a sig-
nificant concern, which applies to most cases within
the scope of this research. Additionally, it was ob-
served that FedAvg was the most reliable aggregation
method for producing stable results across various
classifiers. Conversely, the MLP algorithm exhibited
underperformance when using FL, particularly with
the FedAdam and FedAdagrad algorithms.
Although this study provides important insights
into the use of FL for predicting ICU mortality, fu-
ture research could focus on optimizing FL models
and incorporating data from multiple datasets to en-
hance model performance further. The model shar-
ing between server and clients could also benefit from
cryptography, as it would lead into an even safer and
more privacy-friendly approach.
ACKNOWLEDGEMENTS
This research was funded by the Celtic-Next Project
iCare4NextG (C2020/1-8) and Project iCare4NextG
(POCI-01-0247-FEDER-072265) co-funded by Por-
tugal 2020. Furthermore, this work also received
funding from the project UIDB/00760/2020.
HEALTHINF 2025 - 18th International Conference on Health Informatics
94

REFERENCES
Alghatani, K., Ammar, N., Rezgui, A., and Shaban-Nejad,
A. (2021). Predicting intensive care unit length of stay
and mortality using patient vital signs: Machine learn-
ing model development and validation. JMIR Medical
Informatics, 9(5):e21347.
Bavdekar, S. B. (2013). Pediatric clinical trials. Perspec-
tives in clinical research, 4(1):89–99.
Beutel, D. J., Topal, T., Mathur, A., Qiu, X., Fernandez-
Marques, J., Gao, Y., Sani, L., Li, K. H., Parcollet, T.,
de Gusm
˜
ao, P. P. B., et al. (2022). Flower: A friendly
federated learning framework.
Chia, A. H. T., Khoo, M. S., Lim, A. Z., Ong, K. E., Sun, Y.,
Nguyen, B. P., Chua, M. C. H., and Pang, J. (2021).
Explainable machine learning prediction of icu mor-
tality. Informatics in Medicine Unlocked, 25:100674.
Georgoutsos, A. (2023). Analysis of deep federated learn-
ing on early prediction of icu mortality risk.
Holmstr
¨
om, L., Zhang, F. Z., Ouyang, D., Dey, D., Slomka,
P. J., and Chugh, S. S. (2023). Artificial intelligence in
ventricular arrhythmias and sudden death. Arrhythmia
& Electrophysiology Review, 12.
Iwase, S., Nakada, T.-a., Shimada, T., Oami, T., Shi-
mazui, T., Takahashi, N., Yamabe, J., Yamao, Y., and
Kawakami, E. (2022). Prediction algorithm for icu
mortality and length of stay using machine learning.
Scientific Reports, 12(1).
Johnson, A. E., Bulgarelli, L., Shen, L., Gayles, A., Sham-
mout, A., Horng, S., Pollard, T. J., Hao, S., Moody,
B., Gow, B., et al. (2023). Mimic-iv, a freely acces-
sible electronic health record dataset. Scientific data,
10(1):1.
Khalid, N., Qayyum, A., Bilal, M., Al-Fuqaha, A., and
Qadir, J. (2023). Privacy-preserving artificial intel-
ligence in healthcare: Techniques and applications.
Computers in Biology and Medicine, page 106848.
Kim, Y., Torroba Hennigen, L., and Wang, P. (2023). New
technique enables faster training of machine-learning
models. MIT News. Accessed: 2024-09-26.
Lazzarini, R., Tianfield, H., and Charissis, V. (2023). Fed-
erated learning for iot intrusion detection. AI.
Mammen, P. M. (2021). Federated learning: Opportunities
and challenges. arXiv preprint arXiv:2101.05428.
Mondrejevski, L., Miliou, I., Montanino, A., Pitts, D.,
Hollm
´
en, J., and Papapetrou, P. (2022). Flicu: a
federated learning workflow for intensive care unit
mortality prediction. In 2022 IEEE 35th Interna-
tional Symposium on Computer-Based Medical Sys-
tems (CBMS), pages 32–37. IEEE.
Nilsson, A., Smith, S., Ulm, G., Gustavsson, E., and
Jirstrand, M. (2018). A performance evaluation of fed-
erated learning algorithms. In Proceedings of the sec-
ond workshop on distributed infrastructures for deep
learning, pages 1–8.
Nistal-Nu
˜
no, B. (2022). Developing machine learning mod-
els for prediction of mortality in the medical inten-
sive care unit. Computer Methods and Programs in
Biomedicine, 216:106663.
Pang, K., Li, L., Ouyang, W., Liu, X., and Tang, Y. (2022).
Establishment of icu mortality risk prediction mod-
els with machine learning algorithm using mimic-iv
database. Diagnostics, 12(5):1068.
Pollard, T. J., Johnson, A. E. W., Raffa, J. D., Celi, L. A.,
Mark, R. G., and Badawi, O. (2018). The eicu col-
laborative research database, a freely available multi-
center database for critical care research. Scientific
Data, 5(1).
Randl, K., Armengol, N. L., Mondrejevski, L., and Miliou,
I. (2023). Early prediction of the risk of icu mortality
with deep federated learning. In 2023 IEEE 36th In-
ternational Symposium on Computer-Based Medical
Systems (CBMS), pages 706–711. IEEE.
Rieke, N., Hancox, J., Li, W., Milletari, F., Roth, H. R.,
Albarqouni, S., Bakas, S., Galtier, M. N., Landman,
B. A., Maier-Hein, K., et al. (2020). The future of
digital health with federated learning. NPJ digital
medicine, 3(1):1–7.
Rosen, M. A. (2021). Energy sustainability with a focus on
environmental perspectives. Earth Systems and Envi-
ronment, 5(2):217–230.
Sharma, S. and Guleria, K. (2024). A comprehensive review
on federated learning based models for healthcare ap-
plications. Artif. Intell. Med., 146(C).
Stanfill, M. H. and Marc, D. T. (2019). Health information
management: implications of artificial intelligence on
healthcare data and information management. Year-
book of medical informatics, 28(01):056–064.
Tu, K., Zheng, S., Wang, X., and Hu, X. (2022). Adap-
tive federated learning via mean field approach. In
2022 IEEE International Conferences on Internet of
Things (iThings) and IEEE Green Computing & Com-
munications (GreenCom) and IEEE Cyber, Physical
& Social Computing (CPSCom) and IEEE Smart Data
(SmartData) and IEEE Congress on Cybermatics (Cy-
bermatics), pages 168–175. IEEE.
Vieira, P., Maia, E., and Prac¸a, I. (2024). Acute pancre-
atitis mortality prediction with federated learning. In
Progress in Artificial Intelligence - 23rd EPIA Con-
ference on Artificial Intelligence, EPIA 2024, Viana
do Castelo, Portugal, September 3-6, 2024, Proceed-
ings, volume II of Lecture Notes in Computer Science,
pages 3–15. Springer.
Yang, A., Ma, Z., Zhang, C., Han, Y., Hu, Z., Zhang, W.,
Huang, X., and Wu, Y. (2023). Review on applica-
tion progress of federated learning model and security
hazard protection. Digital Communications and Net-
works, 9(1):146–158.
Yang, Q., Liu, Y., Chen, T., and Tong, Y. (2019). Federated
machine learning: Concept and applications. ACM
Transactions on Intelligent Systems and Technology
(TIST), 10(2):1–19.
Zhuang, Y., Li, G., and Feng, J. (2016). A survey on entity
alignment of knowledge base. Journal of Computer
Research and Development, 53(1):165–192.
C¸ elik, E. and G
¨
ull
¨
u, M. K. (2023). Comparison of federated
learning strategies on ecg classification. In 2023 Inno-
vations in Intelligent Systems and Applications Con-
ference (ASYU), pages 1–4.
Privacy-Preserving Mortality Prediction in ICUs Using Federated Learning
95
