
3D View Reconstruction from Endoscopic Videos for Gastrointestinal
Tract Surgery Planning
Xiaohong W. Gao
1a
, Annisa Rahmanti
1b
and Barbara Braden
2c
1
Department of Computer Science, Middlesex University, London, U.K.
2
Medical Department B, University of Münster, Germany
Keywords: Deep Learning, 3D Reconstruction, Nerfs, Gastrointestinal Tract, SfM, 2D Endoscopic Video, Endoscopic
Artefacts.
Abstract: This paper investigates the application of neural radiance field (NeRF) to reconstruct a 3D model from 2D
endoscopic videos for surgical planning and removal of gastrointestinal lesions. It comprises three stages. The
first one is video preprocess to remove frames with artefact of colour misalignment based on a deep learning
network. Then the remaining frames are converted into NeRF compatible format. This stage includes
extraction of camera information regarding intrinsic, extrinsic and ray pathway parameters as well as
conversion to NeRF format based on COLMAP library, a pipeline built upon structure-from-motion (SfM)
with multi-view stereo (MVS). Finally the training takes place for establishment of NeRF model implemented
upon Nerfstudio library. Initial results illustrate that this end-to-end, i.e. from 2D video input to 3D model
output deep learning architecture presents great potentials for reconstruction of gastrointestinal tract. Base on
the two sets of data containing 2600 images, the similarity measures of SSIM, PSNR and LPIPS between
original (ground truth) and rendered images are 19.46 ± 2.56, 0.70 ± 0.054, and 0.49 ± 0.05 respectively.
Future work includes enlarging dataset and removal of ghostly artefact from rendered images.
1 INTRODUCTION
Gastrointestinal tract (GI) cancers (oesophagus,
stomach, bowel), were responsible for 26.3% of
cancer cases and 35.4% of deaths worldwide in 2018
(Lu 2021). As the 2
nd
largest death caused by cancer
in the world (after lung cancer), GI cancers have very
low 5-year survival rate (<20%). At present, the only
curative and most effective treatment for early GI
cancer or lesion is the removal of concerned lesion
endoscopically, especially, for a lesion confined to the
mucosal layer, the surface columnar epithelium and
the first of four layers on the GI wall. In this
procedure, a substance is injected first under the target
to act as a cushion. Then a surgical plan by marking
cutting lines is conducted. Finally, the dissection takes
place at submucosal layer under the concerned lesion
following the planning boundary. The key to success
of this surgery is that the endoscopists have a clear
view of the lesion, the planned lines and surroundings
a
https://orcid.org/0000-0002-8103-6624
b
https://orcid.org/0000-0001-9478-6267
c
https://orcid.org/0000-0002-8534-6873
from varying view angles throughout in order to
perform precise dissection.
The challenges here are that all the views are
confined into a narrow (~2cm in diameter) cylindrical
food path or tube where the endoscopic camera travels
in one direction, resulting in some concerned tissues,
anatomy and planned boundary being invisible. In
addition, in this complex surgical scene, the
endoscopist/surgeon/clinician has to compete with
various motions coming from respiration, heartbeat,
camera as well as muscles. Because of this, at present,
all these operations can only be conducted by expert
endoscopists, which put significant amount of pressure
in health care systems. Figure 1 demonstrates the
process of resection of a polyp in the stomach
endoscopically. While the lesion in Figure 1 is benign,
if left untreated, it could progress into cancer. Hence
resection is in need. Once the target is confirmed
(Figure 1 (a)), an injection of a dedicated substance is
carried out to highlight and alleviate the lesion whereby
Gao, X. W., Rahmanti, A. and Braden, B.
3D View Reconstruction from Endoscopic Videos for Gastrointestinal Tract Surgery Planning.
DOI: 10.5220/0013125000003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 221-228
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
221
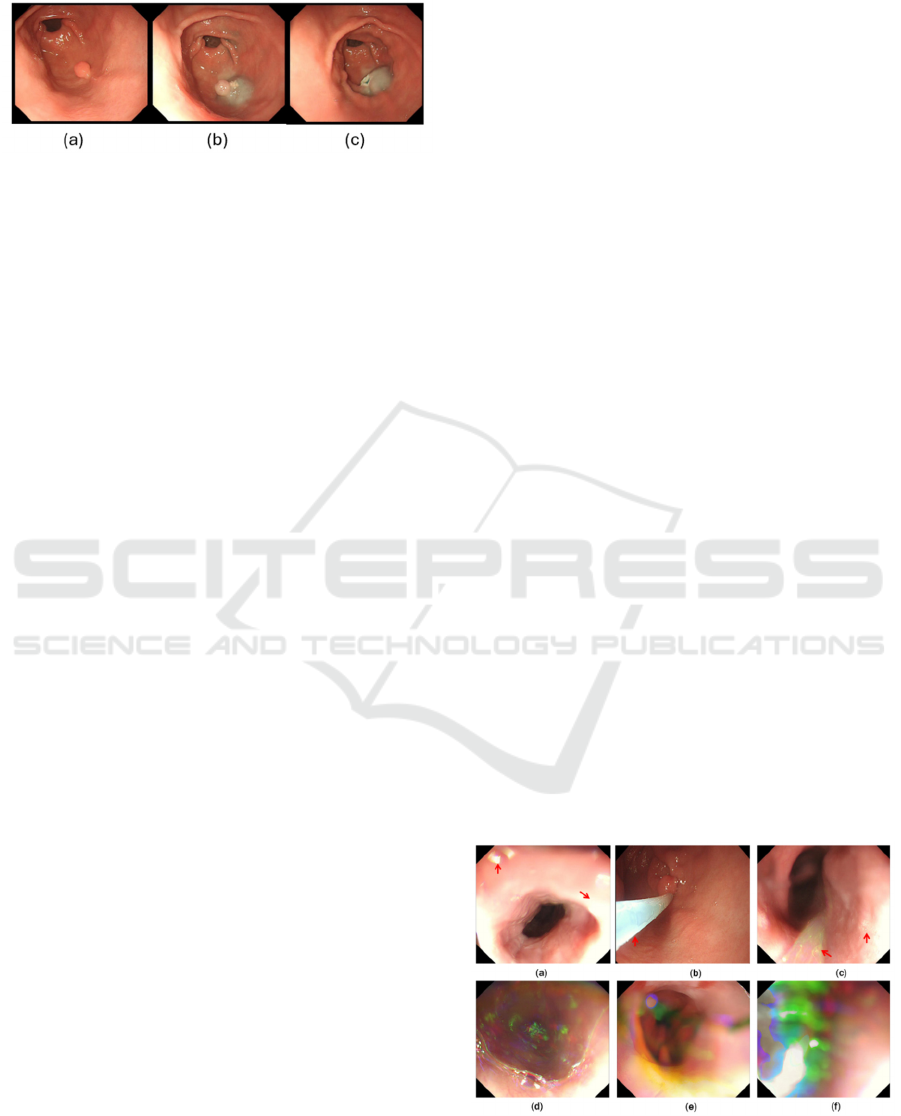
a surgical planning can be made (Figure 1(b)). Then the
lesion can be removed safely (Figure 1(c)).
Figure 1: Demonstration of lesion resection endoscopically.
(a) a lesion is detected; (b) cushion is injected; (c) successful
removal of the concerned lesion.
Hence reconstruction of 3D view of concerned
lesions plays an important role in endoscopic surgical
planning and lesion removal. 3D reconstruction for GI
from endoscopic videos has been studied by a number
of researchers. For example, Ali et al (Ali 2021a) has
established a physical model of oesophagus that is
applied to develop an AI system to quantify Barrett’s
oesophagus. The researchers as detailed in (Prinzen,
2015) has been established the shape of oesophagus to
allow visualization of additional contextual and
geometrical information of oesophagus, from
panorama image to 3D points cloud then to regular
triangulation mesh. Recently, 3D shape reconstruction
of whole stomach based on structure-from-motion
(SfM) (Widya 2019) is investigated by spreading
indigo carmine (IC) dye on the stomach surface to
present colour texture of the stomach. This is because
endoscopic videos present weak texture of GI surface.
On the other hand, the approach of SfM appears to
present robust results when it comes 3D
reconstruction from 2D videos. Further study for deep
multi-view stereo for dense 3D reconstruction (Bae
2020) is also conducted based on SfM and is consisted
of 3 steps, which are sparse reconstruction via SfM,
monocular depth estimation and embedding vector
generation via patch embedding network.
The steps to construct a 3D model from 2D image
usually include image collection, feature or/and
keypoint detection and extraction, keypoint tracking
and matching, structure from motion to determine
camera intrinsic, extrinsic and orientation parameters,
and key point-cloud reconstruction, such as mesh
reconstruction, mesh refinement and mesh texturing.
With the current advances of state-of-the-art
(SOTA) deep learning (DL) techniques, many
innovative approaches have been developed towards
reconstructing 3D deformable objects.
In this study, 3D scene reconstruction based on 2D
endoscopic videos is conducted based on SOTA
neural radiance field (NeRF) so that a lesion can be
viewed from all viewing angles, allowing correct
recognition of concerned lesions, tissue types and
related anatomy, leading to an assistant system in an
operative room allowing multiple views while
performing lesion removal.
NeRF (Mildenhall 2020) addresses the long-
standing problem in computer vision field, which is to
reconstruct a 3D representation of a scene from sparse
2D images. NeRF method synthesizes a new view by
directly optimising parameters of a continuous 5D
scene representation to minimize the error of
rendering a set of captured images. NeRF represents
a scene using a fulling connected deep network with
an input of a single continuous 5D coordinate (i.e. 3D
spatial + 2D view direction angles). The output of this
network is the volume density and view-dependent
emitted radiance at that spatial location.
2 METHODOLOGY
2.1 Image Pre-Process to Remove
Artefact of Colour Misalignment
Due to the confinement of narrow space of GI, it is
quite common that as many as a quarter video frames
contain several types of artefacts, such as water
bubbles, instrument, and saturation. While these
artefacts are present most of the time, the concerned
image features are still visible. However, the artefact
of colour misalignment, where coloured frames of red,
green and blue are acquired at different locations
because of the combination of movements when an
endo camera travels, should be removed not only
because most of interested contents are not present but
also the presence of these artificial colours will affect
the detection of camera light of rays, hence affecting
the accuracy of training. Figure 2 illustrates a number
of artefacts present in a video clip, where the top row
Figure 2: Examples of frames with artefacts (arrow). (a)
saturation; (b) instrument; (c) bubbles; (d)(e)(f) colour
misalignment (all over images).
BIOIMAGING 2025 - 12th International Conference on Bioimaging
222

of artefacts (red arrows) still contains visible GI
features but bottom row of colour misalignment
(whole images) mis-represents GI contents.
To detect artefacts, many deep learning-based
systems are developed offering promising
performance (Ali 2020, Ali 2021b,
Bissoonauth-Daiboo
2023). In this study, the processing time also plays an
important role for the future development of real-time
3D systems. Hence the real-time system, real-time
instance segmentation system, YOLACT (Bolya
2019, Gao 2023) is enhanced and applied for artefact
classification. Figure 2 presents the architecture of the
network to classify frames with normal or artefact
features (instrument, bubbles, saturation or colour
misalignment (CMA)). In this pre-processing stage,
only frames with CMA will be discarded. This is
because the subsequent 3D modelling and
reconstruction of lesioned GI depends on the
information of colour and intensity attributes, i.e.
structure from motion and neutral radiance fields.
Figure 3: The architecture of YOLACT network for
detection and segmentation of artefact. The artefacts to
distinguish are instrument, bubbles, saturation, and colour
misalignment (CMA). The mask or ground truth of frames
with CMA is the whole frame.
For this end-to-end detection system of YOLACT
(Bolya 2019) (Figure 3), the basic underline model
employs ResNet101 to extract initial feature maps.
The object segmentation is accomplished through two
parallel subnets (ProtoNet and Prediction Head),
which generate a set of prototype masks and predict
per-object mask coefficients respectively.
More specifically, ProtoNet employs a fully
connected network (FCN) accommodating the largest
pyramid feature layer (𝑃3), to produce a set of image-
sized prototype masks. These 𝑘 mask prototypes (𝑘
32 in this study) are then applied to deliver predictions
for the entire image in relation to classification,
segmentation and detection (Gao 2023).
On the other hand, Prediction Head contains three
branches, which are c class confidence (c=5 for
‘Instrument’, ‘Bubbles’, ‘Saturation’, ‘artefact-text’,
‘CMA’), 4 bounding box regressors (=[x
top-left-corner
,
y
top-left-corner,
width, height])), and a vector of mask
coefficients, one for each prototype to be processed in
parallel. Subsequently, the branch of
‘Crop+thresholding’ in Figure 2 delivers a vector size
of 4 𝑐 𝑘 for each anchor or region of interest
(RoI), As a result, for each instance, one or more
masks will stem from that instance by linearly
combining (plus or minus) the work from both
prototype and mask coefficient strands, leading to the
production of final masks ( 𝑀 ) by a sigmoid
nonlinearity as formulated in Eq. (1).
𝑀𝜎𝑃𝐶
(1)
where 𝑃 is an ℎ𝑤𝑘 matrix of prototype masks
and 𝐶 is a 𝑛 𝑘 matrix of mask coefficients for 𝑛
instances that have passed score thresholding and
initial 𝑁𝑀𝑆 , the maxima suppression technique
(Bolya 2019). NMS determines whether an instance
should be kept or discard. For example, duplicated
detections are suppressed not only for the same class
but also for cross-class boundary boxes depending on
the probability of boxes, i.e. the box with higher
probability suppresses the one with lower
probability. In Eq. (1), 𝐶
indicates the transpose of
𝐶 Matrix.
The calculation of the loss function is the same as
for YOLACT (Bolya 2019). Three loss functions are
utilised to train this end-to-end detection model as
formulated in Eq. (2), which are classification loss
(ℒ
, box regression loss ( ℒ
and mask loss
ℒ
where the weights of 1, 1.5, and 1.5 are
applied for them respectively to give more weight to
classification.
ℒℒ
1.5 ℒ
1.5 ℒ
(2)
In particular,
ℒ
𝐵𝐶𝐸𝑀,𝑀
(3)
where the binary cross entropy 𝐵𝐶𝐸 is formulated
using Eq. (4).
𝐵𝐶𝐸𝑝, 𝑦
∑
𝑦
log
𝑝
1
𝑦
log 1 𝑝
(4)
where 𝑦 represents the label and 𝑝 is the predicted
probability of the point being a label for all 𝑁 points.
𝑀 and 𝑀
are calculated in Eq. (1).
After removing the frames with CMA artefact, the
remaining are applied to train the 3D model takes
place based on NeRF.
3D View Reconstruction from Endoscopic Videos for Gastrointestinal Tract Surgery Planning
223

2.2 3D View Reconstruction for
Concerned Lesioned GI Based on
NeRF
For 3D scene modelling, neural radiance fields
(NeRFs) (Mildenhall 2020) takes 5-degree
coordinates as an input. The 5D refer to each 3D point
at (𝑥, 𝑦,𝑧) when viewing with a camera ray of light
emitting direction at (𝜃, 𝜙). Hence, NeRF enables
learning novel view synthesis, scene geometry and
reflectance properties by optimising a deep fully-
connected neural network as a multilayer perceptron
(MLP). As such, NeRF represents this 5D function by
regressing from a single 5D coordinate (𝑥,𝑦,𝑧,𝜃,𝜙
to a single volume density and view-dependent RGB
colour. As a result, to render NeRF from a specific
viewpoint, camera rays are marched through the
scene, and a neural network produces colours and
densities for 3D points, which are then accumulated
into a 2D image.
Figure 4 schematically illustrates the process of
representing scenes as neural radiance fields for view
synthesis.
Figure 4: The process flow of NeRF. (a) 5D input; (b) 4D
output; (c) volume rendering at a new viewing direction.
Firstly, a scene (Figure 4(a)) is represented using a
5D vector-valued function with an input of a 3D
location 𝒙 𝑥,𝑦,𝑧 and 2D viewing direction 𝒅
𝜃,𝜙. The output of this function is an emitted colour
𝒄 𝑟,𝑔,𝑏 and volume density
(Figure 4(b)) at
each ray. This 5D function is approximated applying
a multilayer perceptron (MLP) network 𝐹
:
𝒙, 𝒅
→
𝒄, 𝝈 to optimise its weights
in order to map each
input 5D coordinate to its corresponding volume
density and directional emitted colour. Finally, based
on the classical volume rendering approach (Kajiya
1984), the colour of any light ray passing through the
scene is rendered (Figure 4(c)) whereas the volume
density 𝝈𝒙 is interpreted as the differential
probability of a ray terminating at location 𝒙. Hence
the expected colour 𝐶𝒓 of camera ray light 𝒓
𝑡
𝒐𝑡𝒅 , starting at the original location 𝒐, with near
and far bounds 𝑡
and 𝑡
, is expressed in Eq. (5)
(Mildenhall 2020).
𝐶
𝑟
𝑇
𝑡
𝜎𝒓
𝑡
𝑐
𝒓
𝑡
,𝒅
𝑑𝑡
(5)
where
𝑇
𝑡
exp
𝜎
𝒓
𝑠
𝑑𝑠 (6)
As pointed out by Mildenhall et al [6], operating a
network 𝐹
directly on 𝑥𝑦𝑧𝜃𝜙 can result in poorly
rendering performance when colour and geometry
have high-frequency variations. Hence, 𝐹
is
formulated as a composite of two functions, i.e., 𝐹
𝐹
∘𝛾
, where 𝐹
is learned applying a regular MLP
network, from which the estimated colour 𝐶
𝒓
can
be expressed in Eq. (7) as a weighted sum of all
sampled colours 𝑐
along the ray.
𝐶
𝒓
∑
𝑤
𝑐
(7)
where
𝑤
𝑇
1 exp
𝜎
𝛿
(8)
𝑇
exp
∑
𝜎
𝛿
(9)
and
𝛿
𝑡
𝑡
(10)
𝛿
in Eq (10) refers to the distance between adjacent
samples.
On the other hand, 𝛾
is not learnt but a mapping from
ℝ space into a higher dimensional space ℝ
as
computed in Eq. (11).
𝛾
𝑝
sin
2
𝜋𝑝
, cos
2
𝜋𝑝
, … , sin
2
𝜋𝑝
,cos 2
𝜋𝑝
(11)
Where 𝑝 𝑥,𝑦,𝑧,𝜃,𝜙 respectively. 𝐿 10 when
𝑝 𝑥,𝑦,𝑧,𝜃,𝜙 and 𝐿 4 when 𝑝 𝜃,𝜙 .
Hence the loss between ground true colour 𝐶𝒓)
and predicted pixel colours for both coarse 𝐶
𝒓
and
fine 𝐶
𝒓
renderings is calculated in Eq. (12).
ℒ
∑
𝐶
𝒓
𝐶𝒓
∈ℛ
𝐶
𝒓
𝐶𝒓
(12)
where ℛ is the set of rays in each batch.
2.3 Implementation
To model 3D view of endoscopic view based on
NeRFs, Nerfstudio (Tancik 2023) is implemented.
Nerfstudio tools are a repository collecting an family of
simple python-based extensive application programing
interface (API) functions to allow visualisation of
modelling of scenes based on neural radiance fields
BIOIMAGING 2025 - 12th International Conference on Bioimaging
224

(NeRFs). By providing a simplified end-to-end process
of creating, training and testing NeRFs, these APIs
allow viewing and interacting these processes through
an internet browser.
In this study, the input data are a clip of endoscopic
video containing RGB frames. After removal of
artefact of CMA as explained in Section A, these
frames/images are analysed to extract needed
information, including ground truth information such as
camera intrinsic and extrinsic data. To obtain the
endoscopic camera information, COLMAP system is
employed. COLMAP (
Schonberger 2016)
is the
structure-from-motion (SfM) package that can be
employed to estimate the ground truth information
regarding to camera poses, camera intrinsic parameters,
and scene boundaries. In addition, this pre-processing
stage converts those input images into a format that is
compatible with NerfStudio
1
, i.e. a JSON format.
Then training takes place based on the pre-
processed images to create a configuration file and a
model.
2.4 Similarity Measurements
Three common measures are employed to calculate
the similarity between original (ground truth) and
rendered images, which are structural similarity
(SSIM) (Wang 2004) (Eq. (13)), peak signal-to-noise
ratio (PSNR) (Eq. (14)), and more recently Learned
Perceptual Image Patch Similarity (LPIPS) (Eq.(15)).
𝑆𝑆𝐼𝑀
𝑥,𝑦
,
(13)
In Eq. (13) of SSIM, 𝜇
,𝜇
are the averages of 𝑥,
𝑦 , with 𝜎
,𝜎
being the variances of 𝑥
, 𝑦
respectively and 𝜎
,
the covariance of 𝑥
and 𝑦. The
variables of 𝑐
and 𝑐
are applied to stabilize the
division when a small denominator occurs and are set
to be 0.01𝐿
and 0.03𝐿
respectively, whereby L
stands for the dynamic intensity range of an image,
e.g. L=255 for an 8-bit image. 𝑥
,𝑦 refer to original
and rendered images respectively.
For PSNR, Eq. (14) is calculated.
𝑃𝑆𝑁𝑅 20log
𝑀𝐴𝑋
10log
𝑀𝑆𝐸 (14)
where 𝑀𝐴𝑋
refers to the maximum possible value of
the image (e.g. 255 for 8-bit) and 𝑀𝑆𝐸 the mean
squared error between two concerned images.
In addition, LPIPS (Zhang 2018) metric refers to
Learned Perceptual Image Patch Similarity and is
formulated in Eq. (15). LPIPS is calculated by
comparing the activations of two image patches using
pre-defined neural network features. Specifically, it
1
https://docs.nerf.studio.
computes the distance between the feature
representations of the patches. The lower the LPIPS
score is, the more perceptually similar the patches are.
The distance between reference and rendered
patches 𝑥, 𝑥
with network ℱ is calculated in Eq. (15)
where 𝐻,𝑊,𝐶 refer to image patch high, width, and
channel.
𝑑
𝑥,𝑥
∑
∑
𝑤
⊙𝑦
𝑦
,
(15)
The feature stacks are extracted from layer 𝐿
where unit-normalization ( 𝑦
, 𝑦
corresponding to
𝑥,𝑥
respectively) in the channel dimension takes
place, and for layer 𝑙 , 𝑦
, 𝑦
∈ℝ
. The
weight 𝑤
is performing element-wise multiplication
(⨀, which is equivalent to computing cosine distance
(Zhang 2018).
3 RESULTS
Figure 5 demonstrates the training processing
implemented via Nerfstudio. The training processing
is visualised live on a browser, which allows users to
select any camera ray direction as exemplified at
Figure 5 (a)(b) to view detailed training (5(a)) as well
as each individual training image (5(b)) with a specific
camera location (Figure 5(c)).
Figure 5: The illustration of training process. (a)(b) training
takes place at varying camera rays. (c) An individual
training sample selected from (b) with a specific camera ray
direction.
3D View Reconstruction from Endoscopic Videos for Gastrointestinal Tract Surgery Planning
225
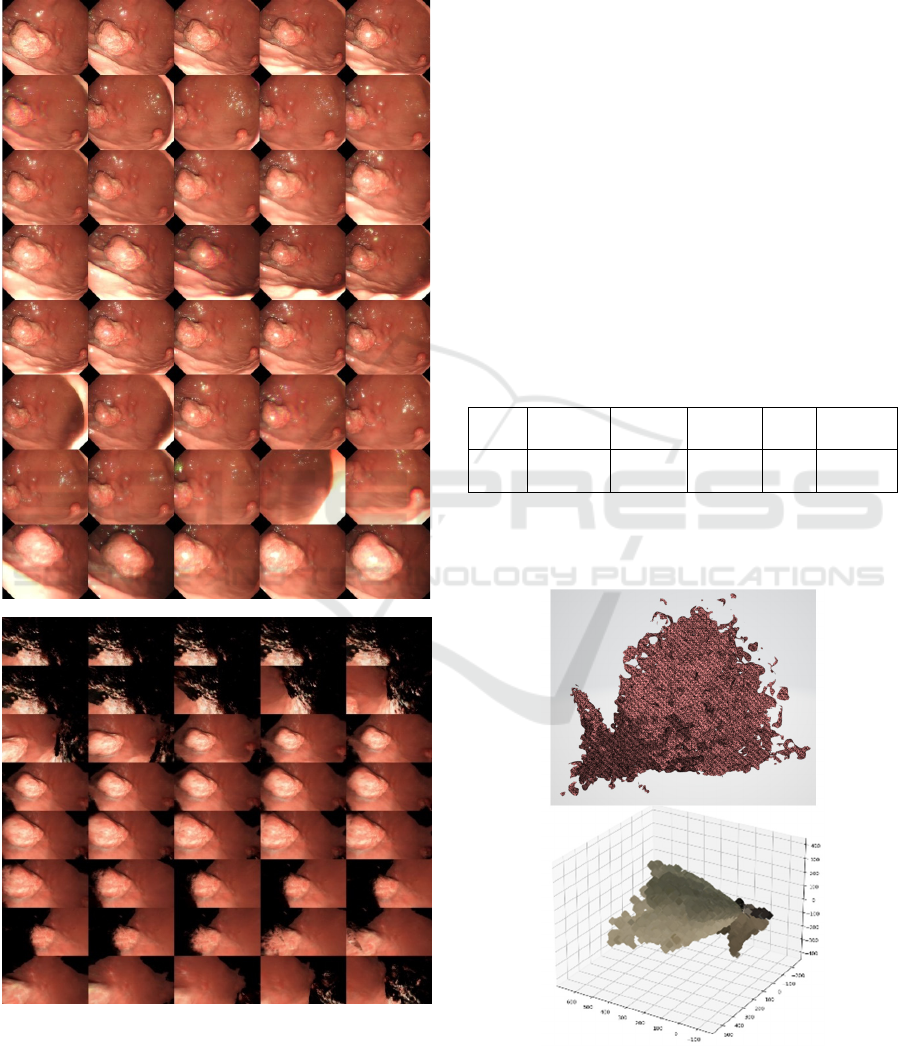
Figure 6 demonstrates the montage of training data
set (a) and the new view angle (b) rendered using the
trained model. The needed view path can be selected,
defined or requested through the visualisation tool
presented in Figure 5.
(a)
(b)
Figure 6: Demonstration of training data (a) and rendered
data (b) at a specific view direction for a polyps in the
stomach.
While the rendered data (Figure 6(b)) have some
background information missing due to the lack the
sufficient data, the concerned lesion can be rendered
and viewed at any needed viewing angle, which is
important in a clinical setting as an endoscopic camera
can only travel in one direction within the narrow food
path whereas clinicians need to know all the
surrounding information for a surgical planning.
The accuracy of rendered images using the
aforementioned three metric measures is provided in
Table 1, which is based on two sets of data. The initial
video frames with 10 minutes each contains over
30,000 frames each. After pre-process for detection of
colour misalignment artefact, 6000 frames are kept.
When extracting camera ray information using
COLMAN approach, only 2600 images are found
related due to poor image quality, e.g. blurry with
floating objects or noise/artefact.
Table 1: The measurement of similarity between ground
truth (original) and rendered images. for psnr and ssim,
higher value implies more similar whereas lower lpips
referring more similar between the two.
Data
set
Image
number
PSNR SSIM
LPIPS
2 2600 19.46 ±
2.56
0.70 ±
0.054
0.49 ±
0.05
Figure 7 presents a mesh and point cloud for the
concerned lesion shown in Figure 5, which can be
views at any viewing angle or camera light ray.
Figure 7: Demonstration of mesh (top) and point cloud
(bottom) of the concerned lesion shown in Figure 5.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
226

4 CONCLUSION
This study investigates the feasibility of
reconstruction of 3D scene of GI from 2D endoscopic
videos as an end-to-end process, i.e. from an input 2D
video to an output 3D model, without the prior
knowledge of camera information of location,
intrinsic and extrinsic data. SOTA NeRF approach is
applied. Because of the challenges facing acquisition
of endoscopic video with less texture information on
the GI surface, the camera positional information
extracted from videos requires images with varying
view angles, which in our case, is limited. Hence the
ground truth images after pre-processing only contain
10% of the original input. However, even with only
1000 images for each lesion as one training dataset,
the 3D model is able to render high quality images
with various viewing angles. For the two training data
sets, the averaged measures of SSIM, PSNR and
LPIPS between original (ground truth) and rendered
images are 19.46 ± 2.56, 0.70 ± 0.054, and 0.49 ± 0.05
respectively. In comparison with the work of NeRF
where 31.01, 0.947 and 0.081 are obtained for natural
images [6], our results appear to be less performed.
However, in [6], around 100 views are acquired for
each filmed object with known camera information. In
our study, this information has to be extracted from
endoscopic videos themselves with much less viewing
angles due to the constraints of viewing space in the
food passage, leading to less image frames are
employed. In addition, because of the combination of
movements while performing endoscopic filming,
including heartbeat, respiration, and camera, many
images appear blurry to a certain extent. These blurry
images are usually ignored when applying COLMAP
library to track camera locations. This is because the
tracking of motion based on optical flow, i.e. the same
spot would appear similar intensity level in the
subsequent images, which is not the case for blurry
images.
In the future, more datasets will be evaluated. In
addition, post processing will be conducted including
to remove noises or ghostly artefact as recommended
more recently by Warburg et al (Warburg 2023).
Specifically, to make use as many video frames as
possible, especially for medical applications with
limited dataset, a new algorithm will be developed to
establish camera information based on the existing
available but less clear images through the application
of human vision models. While many frames are burry
with regard to motion tracking, human vision can still
perceive these motions easily and clearly. In this way,
the developed system will also become more
transparent.
ACKNOWLEDGEMENTS
This project is founded by the British Council as
Women in STEM Fellowship (2023-2024). Their
financial support is greatly acknowledged.
REFERENCES
Ali S, Zhou F, Braden B, et al (2020), “An objective
comparison of detection and segmentation algorithms
for artefacts in clinical endoscopy,” Nature Scientific
Reports, 10, Article number: 2748.
Ali S, Bailey A, Ash S, et al. (2021a), “A Pilot Study on
Automatic Three-Dimensional Quantification of
Barrett’s Esophagus for Risk Stratification and Therapy
Monitoring”, Gastroenterology, 161: 865-878.
Ali S, Dmitrieva M, Ghatwary N, et al (2021b), “Deep
learning for detection and segmentation of artefact and
disease instances in gastrointestinal endoscopy,”
Medical Image Analysis, 70:102002.
Bae G, Budvytis I, Yeung CK, Cipolla R (2020), “Deep
Multi-view Stereo for Dense 3D Reconstruction from
Monocular Endoscopic Video”, MICCAI 2020. Lecture
Notes in Computer Science, vol 12263. Springer.
Bissoonauth-Daiboo P, Khan MHM, Auzine MM, Baichoo
S, Gao XW, Heetun Z (2023), “Endoscopic Image
classification with Vision Transformers,” in ICAAI
2023, pp. 128-132.
Bolya D, Zhou C, Xiao F, Lee YJ (2019), YOLACT: real-
time Instance Segmentation, Proceedings of the ICCV
2019.
Gao XW, Taylor S, Pang W, Hui R , Lu X, Braden B (2023),
“Fusion of colour contrasted images for early detection
of oesophageal squamous cell dysplasia from
endoscopic videos in real time , Information Fusion,” 92:
64-79.
Kajiya JT, Herzen BPV (1984), “Ray tracing volume
densities,” Computer Graphics, SIGGRAPH.
Lu L, Mullins CS, Schafmayer C, Zeißig S, Linnebacher M
(2021), “A global assessment of recent trends in
gastrointestinal cancer and lifestyle-associated risk
factors,” Cancer Commun (Lond), 41(11): 1137-1151.
Mildenhall B, Srinivasan PP, Tancik M, et al. (2020), NeRF:
Representing Scenes as Neural Radiance Fields for
View Synthesis, in ECCV 2020.
Prinzen M, Trost J, Bergen T, Nowack S, Wittenberg T
(2015), “3D Shape Reconstruction of the Esophagus
from Gastroscopic Video,” In: H Handels, et al. (eds)
Image Processing for Medicine, pp. 173-178. Springer
Berlin, Heidelberg.
Schonberger JL, Frahm JM (2016), “Structure-from-motion
revisited,” In: CVPR’2016.
Tancik M, Weber E, Ng E, et al (2023), Nerfstudio: a
Modular Framework for Neural Radiance Field
Development, arXiv:2302.04264.
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004),
“Image quality assessment: From error visibility to
3D View Reconstruction from Endoscopic Videos for Gastrointestinal Tract Surgery Planning
227

structural similarity,” IEEE Transactions on Image
Processing, 13 (4) : 600-612.
Warburg F, Weber E, Tancik M, Hołyński A, Kanazawa A
(2023),” Nerfbusters: Removing Ghostly Artifacts from
Casually Captured NeRFs,” ICCV’2023.
Widya A, Monno Y, Imahori K, et al (2019), “3D
reconstructor of whole stomach from endoscope video
using structure-from-motion”, arXiv:1905,12988v1.
Zhang R, Isola P, Efros AA, Shechtman E, Wang O (2018),
“The unreasonable effectiveness of deep features as a
perceptual metric,” In: CVPR’2018.
BIOIMAGING 2025 - 12th International Conference on Bioimaging
228
