
A Real-World Segmentation Model for Melanocytic and Nonmelanocytic
Dermoscopic Images
Eleonora Melissa
1 a
, Daria Riabitch
1 b
, Linda Lazzeri
2
, Federica La Rosa
1 c
, Chiara Benvenuti
1
,
Mario D’Acunto
3 d
, Giovanni Bagnoni
2
, Daniela Massi
4 e
and Marco Laurino
1 f
1
Institute of Clinical Physiology, National Research Council, Pisa, Italy
2
Uniti of Dermatologia, Specialist Surgery Area, Department of General Surgery, Livorno Hospital, Azienda Usl Toscana
Nord Ovest, Italy
3
Institute of Biophysics, National Research Council, Pisa, Italy
4
Department of Health Sciences, Section of Pathological Anatomy, University of Florence, Florence, Italy
Keywords:
Segmentation, Skin Lesions, Deep Learning, Border Features.
Abstract:
Segmentation is a critical step in computer-aided diagnosis (CAD) systems for skin lesion classification. In
this study, we applied the Deeplabv3+ network to segment real dermoscopic images. The model was trained on
public datasets and tested both on public and on a disjoint set of images from the TELEMO project, covering
six clinically significant skin lesion types: basal cell carcinoma, squamous cell carcinoma, melanoma, benign
nevi, actinic keratosis and seborrheic keratosis. Our model achieved a testing global accuracy of 97.88% on
public dataset and of 92.62% on TELEMO dataset, outperforming literature models. Although some misclassi-
fications occurred, largely due to class imbalance, the model demonstrated strong generalization to real-world
clinical images, a critical achievement for deep learning in medical imaging. To evaluate the clinical relevance
of our segmentation, we extracted ten key features related to lesion border and diameter. Notably, the ”Diame-
ters Mean” and ”Area to Perimeter Product” features revealed significant differences between melanoma-nevi
and basal cell carcinoma-nevi classes, with strong effect sizes. These findings suggest that border features
are crucial for distinguishing between multiple skin lesion types, highlighting the model’s potential for aiding
dermatological diagnoses.
1 INTRODUCTION
Diagnosis of skin lesions has become increasingly im-
portant in clinical practice, especially when the early
diagnosis of skin tumors can increase patient survival
rates (Rojas et al., 2022). Melanoma poses significant
challenges in clinical dermatology, as it is the deadli-
est form of skin cancer with a steadily increasing inci-
dence over recent decades. It results from the malig-
nant transformation of melanocytes, which can arise
from healthy skin or develop from congenital nevi.
Despite its aggressiveness, melanoma has favorable
survival rates when detected early. However, its iden-
tification can be challenging, as it may be confused
a
https://orcid.org/0009-0005-8892-0470
b
https://orcid.org/0009-0007-7502-695X
c
https://orcid.org/0000-0003-1671-6057
d
https://orcid.org/0000-0003-4233-1943
e
https://orcid.org/0000-0002-5688-5923
f
https://orcid.org/0000-0003-4798-5196
with other nonmelanocytic skin cancers that present
similar dermoscopic features.
Nonmelanoma skin cancers (NMSCs) are more
prevalent, with a global incidence of 7.7 million cases
(Society, 2019). This category includes basal cell
carcinoma (BCC), squamous cell carcinoma (SCC),
and actinic keratosis (AK). BCC is a high survival
rate skin tumor (100% over 5 years (Society, 2019))
and is recognized as the most common malignant ep-
ithelial cancer. SCC is the second most widespread
nonmelanoma skin cancer behind BCC and it is of-
ten seen in elderly populations (et al., 2021; et al.,
2012). AK is a sun-induced precancerous lesion and
it may progress to SCC; its dermoscopic characteris-
tics present diagnostic challenges that require careful
evaluation (et al., 2021). Seborrheic keratosis (SK),
although it is a benign lesion, can mimic melanoma
and AK and complicates the differential diagnosis
with its varied color and unique structures (et al.,
2019; Hafner and Vogt, 2008).
316
Melissa, E., Riabitch, D., Lazzeri, L., La Rosa, F., Benvenuti, C., D’Acunto, M., Bagnoni, G., Massi, D. and Laurino, M.
A Real-World Segmentation Model for Melanocytic and Nonmelanocytic Dermoscopic Images.
DOI: 10.5220/0013129400003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 316-323
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

Recent advancements in artificial intelligence (AI)
have revolutionized the diagnostic landscape for skin
lesions. Automated tools can facilitate their identifi-
cation, particularly skin tumors, making the diagnos-
tic process standardized and less operator dependent.
This advancement enhances the screening and early
detection of skin cancers, potentially improving pa-
tient outcomes and treatment success.
One of the fundamental steps of automatic skin
lesion classification is segmentation, which poses an
important challenge in the field of computer-aided di-
agnosis (CAD) systems due to the low contrast and
different quality of dermoscopic images. As a result,
many segmentation methods have been investigated
and developed over the years (Oliveira et al., 2016).
Thresholding-based methods, such as Otsu’s method
(Otsu, 1979) and Xu method (Xu et al., 1999), sepa-
rate lesion and background pixels choosing a thresh-
old based on the histogram intensity levels of the der-
moscopic image. In Canny’s edge detector (Canny,
1986) variations of the gradient magnitude are used to
identify the borders of the lesion. Chen-Vese method
(Chan and Vese, 2001) is based on active curves
which move toward the boundaries respecting an en-
ergy minimization principle. Region-based methods
(Oliveira et al., 2016) rely on identifying similar prop-
erties among neighbouring pixels to segment the input
image into different regions with a growing criterion.
Reddy et al (Reddy et al., 2022) employed such an ap-
proach in their work, leveraging pixel color and con-
trast similarity as the underlying principle for region
homogeneity.
Nevertheless, in recent years deep-learning ap-
proaches have shown better performances in terms of
skin lesion segmentation and border detection. The
earliest attempts to use deep networks for skin lesion
segmentation focused on the application of Convo-
lutional Neural Network (CNN) (Ronneberger et al.,
2015) and Fully Convolutional Network (FCN) (Bi
et al., 2018; Yuan et al., 2017), but then the U-Net
emerged as the leading deep network for biomedi-
cal image segmentations (Seeja and Suresh, 2019).
Many extensions of the U-Net, such as ResUNet, Re-
sUNet++, Fuzzy U-Net and attention U-Net (Ashraf
et al., 2022; Bindhu and Thanammal, 2023; Tong
et al., 2021), were eventually developed to obtain bet-
ter results, but also new nets were employed for this
task. Deeplabv3 (Wang et al., 2018) is a deep network
specifically designed for semantic segmentation and
its advanced version Deeplabv3+ has been used for
skin lesions segmentation combined with other archi-
tectures such as VGG19-Unet (Ali et al., 2019) and
MobileNetv2 (Zafar et al., 2023) with good results.
However, as far as we know, none of these studies has
tried to apply their models to real-world clinically ac-
quired images: training and testing are performed on
the same dataset usually split into two parts. Some-
times (Yuan et al., 2017), when a public dataset is
employed, the training and testing images are divided
by the database itself. Despite this, the quality of im-
ages collected from public databases differs signifi-
cantly from that of images acquired during actual der-
matological inspections, which can affect the perfor-
mances of the methods.
In this context, we developed a Deeplabv3+ net-
work for the segmentation of real dermatoscopic im-
ages collected as part of the TELEMO project. We
trained the model using public datasets and tested it
both on a split portion of the public datasets and on
a completely disjoint dataset. Furthermore, to evalu-
ate the efficacy of our model, we extracted ten of the
most relevant features related to border and diameter,
which are closely associated with segmentation qual-
ity. We then assessed whether these features could
effectively distinguish among six clinically relevant
types of skin lesions: basal cell carcinoma, squamous
cell carcinoma, melanoma, benign nevi, actinic ker-
atosis and seborrheic keratosis.
2 MATERIALS AND METHODS
2.1 Datasets
Our model was trained on different public datasets:
the ISIC 2017, ISIC 2019, ISIC 2020, PH2, 7-point
criteria evaluation database and a custom dataset by
Department of Dermatology of Hospital Italiano de
Buenos Aires (HIBA) (Lara et al., 2023), for a total
of 26597 images. A portion of the training dataset
(10% of the total images) was used to test the model
to enable comparison with literature models, which
utilized splitted datasets for training and testing. The
model was further tested on a separate test dataset col-
lected as part of the TELEMO project.
2.1.1 TELEMO Dataset
TELEMO (‘An Innovative TELEmedicine System for
the Early Screening of Melanoma in the General Pop-
ulation’) is a project whose primary objective is to de-
velop a teledermatology platform integrated with AI
tools for the analysis of skin lesions. A total of 479
dermoscopic images were obtained from patients with
an average age of 55 years (52 years for females and
57 years for males), using either the FotoFinder Medi-
acam 1000 (FotoFinder Systems GmbH, Germany) or
the Heine Delta 30 (HEINE Optotechnik, Germany).
A Real-World Segmentation Model for Melanocytic and Nonmelanocytic Dermoscopic Images
317

Figure 1: Comparison between public dataset images (first
row) used as training set and TELEMO images (second
row) used as testing set.
The image quality produced by these clinical in-
struments, as previously highlighted, is very different
to that of public datasets (Figure 1), making the seg-
mentation task on real-world images more challeng-
ing. The skin lesion types of the TELEMO dataset
were distributed as follows: 53 melanoma (MEL),
232 benign nevi (NV), 20 squamous cell carcinoma
(SCC), 101 basal cell carcinoma (BCC), 15 actinic
keratosis (AK), 23 seborrheic keratosis (SK).
2.2 Image Segmentation
Table 1: This table summarizes the training options used for
the deep learning model.
Options Value
Backbone ResNet50
Optimizer sgdm
Max Epochs 30
Mini-batch size 16
Initial Learning Rate 0.001
L2 regularization 0.0005
Momentum 0.9
DeepLabv3+ is a network that uses a ResNet
backbone and integrates Atrous Spatial Pyramid
Pooling (ASPP) to capture multi-scale context, along
with a decoder module to refine object boundaries
(Ali et al., 2019; Zafar et al., 2023). The net param-
eters used to train our Deeplabv3+ network in Mat-
lab®2023b are shown in Table 1. The ground-truth
segmentations for each image were already provided
online in the case of the ISIC 2017 and 7-point cri-
teria evaluation database. In the other cases, we first
attempted to obtain reference segmentation using the
Topological Process Imaging method (Vandaele et al.,
2020), used in the work of Brutti et al (et al., 2023).
When this method proved unsuccessful, we manually
derived the segmentations with guidance from expert
dermatologists. The net required the resizing of all
the images to a standard dimension of 224x224 pixels.
Data augmentation techniques such as translation, ro-
tation, scaling, saturation, hue and brightness jittering
were applied to enhance the training set. The follow-
ing metrics were assessed to evaluate the performance
of our model:
1. Mean Accuracy: percentage of the correctly clas-
sified pixels of all classes in all the images.
2. Global Accuracy: percentage of the correctly
classified pixels with respect to the total number
of pixels without the distinction of classes.
3. Mean IoU: also known as the Jaccard similarity
coefficient, is evaluated as (1):
IoU index =
T P
T P + FP + FN
(1)
where TP is the number of true positive pixels, FP
is the number of false positive pixels and FN is the
number of false negative pixels.
4. Wheighted IoU: value of IoU for each class
weighted by the number of that class’ pixels.
5. Mean BF score: boundary F1 score, it indicates
how well the model predicted the boundaries of
the lesion for all the images.
2.3 Feature Extraction and Statistical
Analysis
The ABCD rule defines four primary feature classes
used to describe the appearance of skin lesions: asym-
metry, border, color, and diameter. These features
are regarded as the gold standard for distinguishing
melanoma from nonmelanoma skin lesions (Rao and
Ahn, 2012). In this study, we focused on the first three
feature classes, which can be evaluated solely based
on the segmentation of dermoscopic images. Fur-
thermore, we explored whether these features could
also be applied to differentiate other types of skin le-
sions beyond melanoma. Hence we first performed
the Levene’s test and the Shapiro-Wilk test to check if
the conditions of normality and homogeneity of vari-
ance were met for each feature. Since the results of
these tests showed a non-Gaussian distribution of the
data for every feature, we proceeded by performing
the Kruskall-Wallis test and the post-hoc Dunn test.
Additionally, for the significant differences, we cal-
culated the epsilon squared index (ε
2
), an effect size
measure that indicates whether the differences also
have real clinical significance.
2.3.1 Asymmetry
The asymmetry of the lesion is commonly measured
with the Normalized E-Factor (NEF), defined by
Sancen-Plaza et al (et al., 2018) as (2):
NEF =
p
4 ×
√
A
(2)
BIOIMAGING 2025 - 12th International Conference on Bioimaging
318

where p is the perimeter of the lesion and A is its area.
2.3.2 Border
Khan et al. (Khan et al., 2020) identified three pri-
mary descriptors of lesion border quality with respect
to its irregularities:
1. Area to Perimeter Ratio (B1): ratio between the
area and the perimeter of the segmented lesion (3)
A
p
. (3)
2. Smooth index (B2): ratio between the area and the
square of the perimeter of the segmented lesion
(4)
4 ×π ×A
p
2
. (4)
3. Product of Area and Perimeter (B3): Product be-
tween the area and the perimeter of the segmented
lesion (5)
A ×p. (5)
Moreover, we also considered the fractal dimension,
an index of the lesion’s complexity and morphology,
which is calculated using the bounding-box algorithm
(Messadi et al., 2021; Ali et al., 2020).
2.3.3 Diameter
The real diameter of the lesion can’t be derived from
the dermoscopic images unless the magnification fac-
tor of the original image is known. Since this parame-
ter in not supplied for all the images, we derived other
descriptors of the lesion diameter which are (Khan
et al., 2020; Cavalcanti and Scharcanski, 2013; et al.,
2017):
1. Mean diameter (D1): mean between the two ap-
parent diameters (Da and Db) of the segmented
lesion (6)
D
a
+ D
b
2
with D
a
=
r
4 ×A
π
, D
b
=
D + d
2
(6)
where D is the major axis and d is the minor axis
of the segmented lesion.
2. Equivalent diameter: measure of diameter equiv-
alent to the real one: (7)
3 ×A
D × pi
. (7)
3. Axis Ratio: Ratio between the major (D) and mi-
nor (d) axis of the segmented lesion (8)
D
d
. (8)
4. Circularity: Index of the level of circularity of the
lesion (9)
4 × pi ×A
D × pi
(9)
3 RESULTS
3.1 Image Segmentation
Figure 2 (b) shows the confusion matrix for the public
images test dataset, while Figure 2 (a) corresponds
to the model performance on the TELEMO dataset.
The percentages in the confusion matrix refer to the
number of pixels which were correctly classified as
lesion pixels or background pixels.
In Table 2 the metrics obtained by our net are com-
pared with that of previous works.
3.2 Statistical Analysis
Figure 3 shows the box plots for all 10 features ex-
tracted for each class. The plot also includes brack-
ets with asterisks that connect the classes of fea-
tures showing significant differences between multi-
ple pairs of skin lesions. The number of asterisks in-
dicates the level of significance; the more asterisks,
the stronger the significance.
Table 3 presents the epsilon squared values for
each significant difference (indicated by the brackets
connecting classes in the plots of Figure 3) for each
feature.
4 DISCUSSION AND
CONCLUSIONS
Our application of the Deeplabv3+ network for image
segmentation yielded highly promising results, partic-
ularly in challenging cases where traditional methods
often struggle due to low contrast between lesions and
surrounding skin or the complex structure of lesion
boundaries.
This is evident in Figure 4: in image (a), the refer-
ence and Deeplabv3+ segmentations are nearly iden-
tical; in images (b) and (c), where lesion borders are
less clear and more complex, our segmentations show
only slight deviations from the reference. Greater in-
accuracies are observed in image (d), the most diffi-
cult case with very unclear boundaries. Nonetheless,
our segmentation remains quite similar to the refer-
ence, whereas other methods have completely failed
A Real-World Segmentation Model for Melanocytic and Nonmelanocytic Dermoscopic Images
319

(a) (b)
Figure 2: Testing confusion matrix of Deeplabv3+ net for TELEMO dataset (a) and public dataset (b).
Table 2: Performance of our model (internal testing on public dataset and external testing on TELEMO dataset) and previous
models.
Reference Method MA GA MIoU WIoU BF
(Ali et al., 2019) VGG19-Unet + Deeplabv3+ - 0.935 0.763 - -
(Zafar et al., 2023) MobileNetv2 + Deeplabv3+ 0.9147 0.9864 0.8814 0.9839 0.7836
(Vandaele et al., 2020) TPI** - 0.809 - - -
(Bi et al., 2018) FCN** - - 0.778 - -
(Tong et al., 2021) ASCU-Net** - 0.926 0.847 - -
(Yuan et al., 2017) FCN** 0.9555 - 0.847 - -
Our model Deeplabv3+ (internal testing) 0.9721 0.9788 0.9434 0.9589 0.9254
Our model Deeplabv3+ (external testing) 0.8895 0.9262 0.7901 0.8697 0.7553
*MA = Mean accuracy; GA = Global accuracy; MIoU = Mean IoU; WIoU = Weighted IoU; BF = Mean BF score.
** TPI = Topological Process Imaging; FCN = FUlly Convolutional Network; ASCU-Net = Attention Gate, Spatial and
Channel Attention U-Net.
Table 3: Features Kruskall-Wallis p-values and relatives ef-
fect size indexes.
Feature p-value ε
2
Asymmetry index 6.0940 x10
-5
0.0505
Area to perimeter ratio 3.1431x10
-7
0.0771
Area to perimeter product 1.8324x10
-11
0.1250
Smoothness index 1.1095 x10
-6
0.0708
Fractal dimension 0.0011 0.0351
Diameters mean 9.3245 x10
-11
0.1171
Equivalent diameter 8.9616 x10
-9
0.0948
Axis ratio 0.0626 0.0127
Circularity 0.0033 0.0294
in these cases. One notable issue is the misclassifi-
cation of some background pixels as lesion pixels, as
our segmentations tend to be slightly larger than the
reference ones. This is also reflected in the confu-
sion matrices percentages (Figure 2) and can be at-
tributed to class imbalance, as the skin region domi-
nates most images with background pixels accounting
for an average of 76.20% and lesion pixels making
up 23.80%. This highlights the importance of using
more balanced datasets and paying close attention to
pixel-level class distributions in future refinements.
Despite this limitation, our model achieved a
mean IoU of 94.34% and a mean accuracy of 97.21%
when tested on a split portion of the training dataset,
outperforming all the methods listed in Table 2. The
only metrics that are lower are global accuracy and
weighted IoU, compared to the second model in Table
2, which, however, is a more complex and computa-
tionally demanding network.
When tested on a disjoint real-world image
dataset, our model’s performance consistently de-
creased, although it still outperformed or remained
comparable to most of the models in Table 2. These
results underscore an important challenge in dermo-
scopic image segmentation: the need to generalize
models to real-world images. The quality of images
used for training, which are from public datasets, and
those captured in real clinical settings can differ sig-
nificantly. This discrepancy can lead to a drastic drop
in model performances when applied to images that
deviate from the ones in public datasets. One possible
solution would be to use real dermoscopic images for
training; however, this could compromise the model’s
robustness due to the limited number of available im-
ages. Therefore, it is essential to use public datasets,
BIOIMAGING 2025 - 12th International Conference on Bioimaging
320
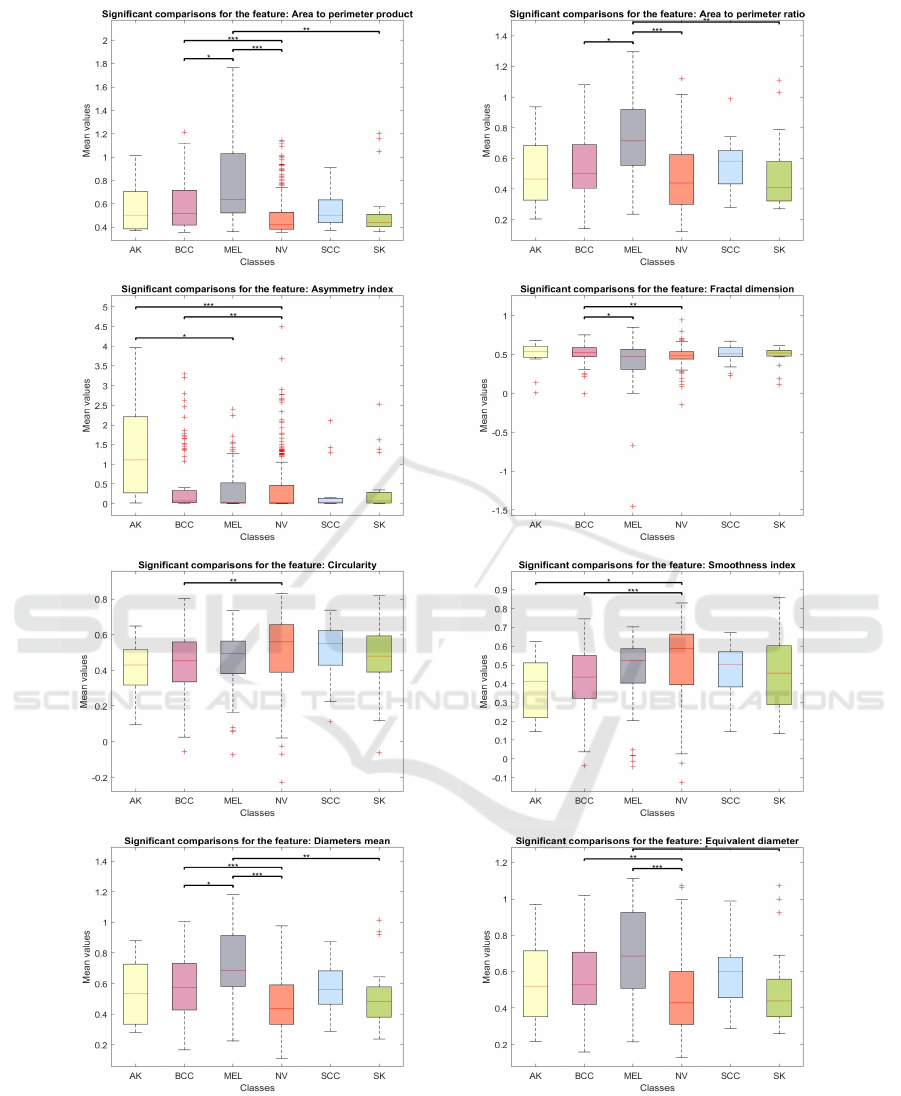
Figure 3: Features box plots. * stands for 0.01 < p −value < 0.05; ** stands for 0.001 < p −value < 0.01; *** stands for
p −value < 0.001.
A Real-World Segmentation Model for Melanocytic and Nonmelanocytic Dermoscopic Images
321
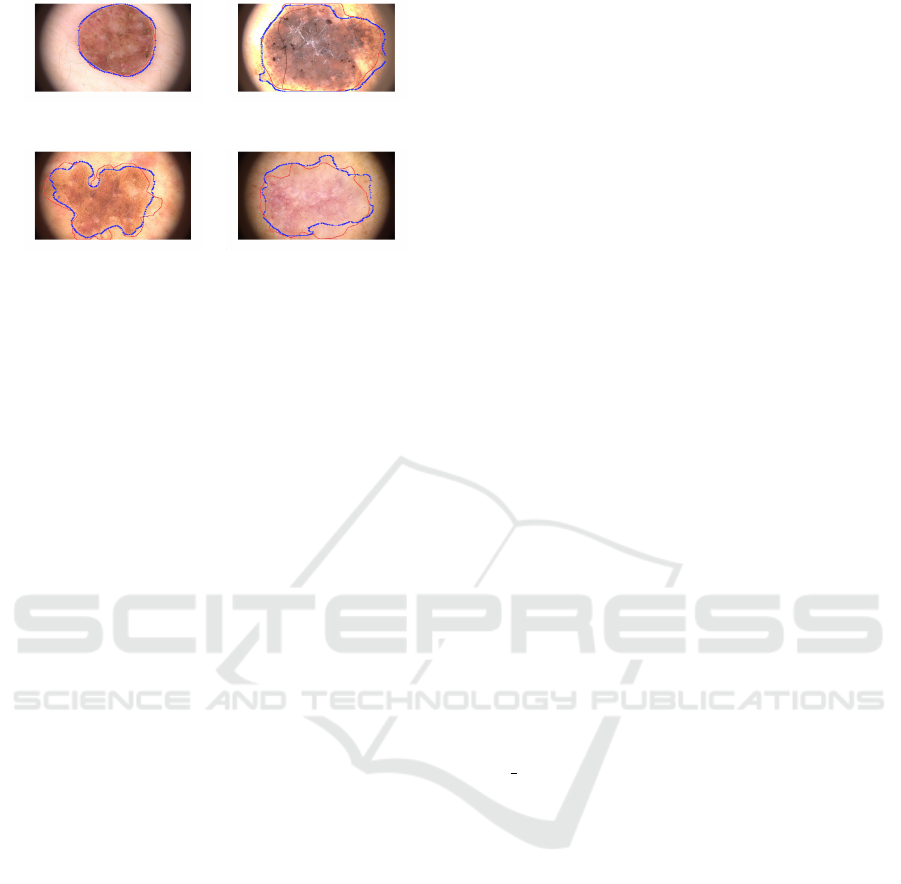
(a) (b)
(c) (d)
Figure 4: Examples of some Deeplabv3+ segmentation
(blue line) overlapped to the reference segmentation (red
line) of TELEMO images.
which contain tens of thousands of images, during the
training phase, but the model must also be specifically
designed to adapt to varying image qualities, as has
been done in this work.
Given the accurate recognition of lesion borders
of our image segmentations, we would have expected
that at least a few border-related features would show
significant differences between classes. As a matter
of fact, previous studies have shown that border and
color features are particularly useful for distinguish-
ing between skin lesions, especially melanoma and
common nevi (et al., 2017). This is confirmed in our
findings, as only one feature, the axis ratio, has shown
a non-significant p-value with also a small associated
effect size (Table3). From the boxplots of Figure 3,
the ”Diameters Mean” and ”Area to Perimeter Prod-
uct” features display two brackets with three asterisks,
indicating highly significant differences between the
MEL-NV and BCC-NV classes. These two features
also have the highest epsilon squared index values,
suggesting a strong effect size and practical impor-
tance of the results (Table 3). Our statistical analysis
therefore suggests that features from the ABCD rule
are not only relevant in the identification of melanoma
but also in distinguishing other skin tumors, such as
basal cell carcinoma.
For other features, even if the Kruskall-Wallis p-
values remain very low, the effect size is also moder-
ate or small. Additionally, all boxplots, except for
”Area to Perimeter Ratio”, show noticeable differ-
ences between the BCC and NV classes, suggesting
that border features are highly relevant in distinguish-
ing between these two classes. This warrants fur-
ther investigation with larger datasets, which could
also help determine whether the outlier values ob-
served, particularly in the ”Asymmetry Index”, ”Area
to Perimeter Product,” and ”Fractal Dimension”, are
due to the limited number of lesions analyzed in this
study or the high variability of these parameters in
dermoscopic images.
5 CONCLUSIONS
The Deeplabv3+ network developed in this work
for the segmentation of dermoscopic images demon-
strated high performance in handling challenging
cases where traditional methods often fail, particu-
larly when lesion boundaries are complex or indis-
tinct. This confers to our model a high generalization
ability to real-world clinical images, a crucial factor
in medical image analysis. Our accurate identifica-
tion of lesion borders also enhances the distinction
between certain lesion classes, underscoring the di-
agnostic value of these features. Future work should
focus on refining class balance and incorporating a
wider range of real clinical images to further vali-
date model performance. Furthermore, the potential
of border features for classifying various types of skin
lesions should be further explored by developing a
classification model based on the extraction of these
features from the segmentation technique developed
in this work.
ACKNOWLEDGEMENTS
We thank Tuscany Region that funded the TELEMO
project, the ’Covid-19 Research Call Tuscany’ as
well as the THE-Tuscany Health Ecosystem grant
ECS
00000017.
REFERENCES
Ali, A. R., Li, J., Yang, G., and O’Shea, S. J. (2020). A
machine learning approach to automatic detection of
irregularity in skin lesion border using dermoscopic
images. PeerJ Computer Science, 6:1–35.
Ali, R., Hardie, R. C., Narayanan Narayanan, B., and
De Silva, S. (2019). Deep learning ensemble meth-
ods for skin lesion analysis towards melanoma detec-
tion. In 2019 IEEE National Aerospace and Electron-
ics Conference (NAECON), pages 311–316.
Ashraf, H., Waris, A., Ghafoor, M. F., Gilani, S. O., and Ni-
azi, I. K. (2022). Melanoma segmentation using deep
learning with test-time augmentations and conditional
random fields. Scientific Reports, 12.
Bi, L., Feng, D., and Kim, J. (2018). Improving automatic
skin lesion segmentation using adversarial learning
based data augmentation. CoRR, abs/1807.08392.
Bindhu, A. and Thanammal, K. K. (2023). Segmentation of
BIOIMAGING 2025 - 12th International Conference on Bioimaging
322

skin cancer using fuzzy u-network via deep learning.
Measurement: Sensors, 26.
Canny, J. (1986). A computational approach to edge detec-
tion.
Cavalcanti, P. G. and Scharcanski, J. (2013). Macro-
scopic pigmented skin lesion segmentation and its in-
fluence on lesion classification and diagnosis, vol-
ume 6, pages 15–39. Springer Netherlands.
Chan, T. F. and Vese, L. A. (2001). Active contours without
edges.
et al., A. S.-P. (2018). Quantitative evaluation of binary dig-
ital region asymmetry with application to skin lesion
detection philip payne. BMC Medical Informatics and
Decision Making, 18.
et al., C. R. (2012). Dermoscopy of squamous cell carci-
noma and keratoacanthoma. Archives of Dermatology,
148:1386–1392.
et al., F. B. (2023). Artificial intelligence algorithms for
benign vs. malignant dermoscopic skin lesion image
classification. Bioengineering, 10.
et al., F. D. (2017). Segmentation and classification of
melanoma and benign skin lesions. Optik, 140:749–
761.
et al., G. (2019). Atlante di dermoscopia. Piccin.
et al., V. R. (2021). Metabolomic analysis of actinic ker-
atosis and scc suggests a grade-independent model of
squamous cancerization. Cancers, 13.
Hafner, C. and Vogt, T. (2008). Seborrheic keratosis.
Khan, H., Yadav, A., Santiago, R., and Chaudhari,
S. (2020). Automated non-invasive diagnosis of
melanoma skin cancer using dermo-scopic images.
ITM Web of Conferences, 32:03029.
Lara, M. A. R., Kowalczuk, M. V. R., Eliceche, M. L.,
Ferraresso, M. G., Luna, D. R., Benitez, S. E., and
Mazzuoccolo, L. D. (2023). A dataset of skin lesion
images collected in argentina for the evaluation of ai
tools in this population. Scientific Data, 10.
Messadi, M., Cherifi, H., and Bessaid, A. (2021). Segmen-
tation and abcd rule extraction for skin tumors classi-
fication.
Oliveira, R. B., Filho, M. E., Ma, Z., Papa, J. P., Pereira,
A. S., and Tavares, J. M. R. (2016). Computational
methods for the image segmentation of pigmented
skin lesions: A review.
Otsu, N. (1979). A threshold selection method from gray-
level histograms. IEEE Transactions on Systems,
Man, and Cybernetics, 9(1):62–66.
Rao, B. K. and Ahn, C. S. (2012). Dermatoscopy for
melanoma and pigmented lesions.
Reddy, D. A., Roy, S., Kumar, S., and Tripathi, R. (2022). A
scheme for effective skin disease detection using opti-
mized region growing segmentation and autoencoder
based classification. In Procedia Computer Science,
volume 218, pages 274–282. Elsevier B.V.
Rojas, K. D., Perez, M. E., Marchetti, M. A., Nichols, A. J.,
Penedo, F. J., and Jaimes, N. (2022). Skin cancer:
Primary, secondary, and tertiary prevention. part ii.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-net:
Convolutional networks for biomedical image seg-
mentation. CoRR, abs/1505.04597.
Seeja, R. D. and Suresh, A. (2019). Deep learning
based skin lesion segmentation and classification of
melanoma using support vector machine (svm). Asian
Pacific Journal of Cancer Prevention, 20:1555–1561.
Society, C. C. (2019). Canadian cancer society.survival
statistics for non-melanoma skin cancer. [Accessed
12 August 2019].
Tong, X., Wei, J., Sun, B., Su, S., Zuo, Z., and Wu, P.
(2021). Ascu-net: Attention gate, spatial and channel
attention u-net for skin lesion segmentation. Diagnos-
tics, 11.
Vandaele, R., Nervo, G. A., and Gevaert, O. (2020). Topo-
logical image modification for object detection and
topological image processing of skin lesions. Scien-
tific Reports, 10.
Wang, Y., School, T. H., Sun, S., and School, J. Y. T. H.
(2018). Skin lesion segmentation using atrous convo-
lution via deeplab v3.
Xu, L., Jackowski, M., Goshtasby, A., Roseman, D., Bines,
S., Yu, C., Dhawan, A., and Huntley, A. (1999). Seg-
mentation of skin cancer images. Image and Vision
Computing, 17(1):65–74.
Yuan, Y., Chao, M., and Lo, Y. C. (2017). Automatic
skin lesion segmentation using deep fully convolu-
tional networks with jaccard distance. IEEE Trans-
actions on Medical Imaging, 36:1876–1886.
Zafar, M., Amin, J., Sharif, M., Anjum, M. A., Mal-
lah, G. A., and Kadry, S. (2023). Deeplabv3+-based
segmentation and best features selection using slime
mould algorithm for multi-class skin lesion classifica-
tion. Mathematics, 11.
A Real-World Segmentation Model for Melanocytic and Nonmelanocytic Dermoscopic Images
323
