
The Devil Lies in the Details for Species Coexistence Stability in Ablated
and Unablated Five-Species Evolutionary Spatial Cyclic Games
Dave Cliff
a
School of Engineering Mathematics and Technology, University of Bristol, Bristol BS8 1UB, U.K.
Keywords:
Agent-Based Mode Biodiversity, Cyclic Competition, Asymmetric Interaction, Species Coexistence,
Evolutionary Spatial Games, Rock-Paper-Scissors-Lizard-Spock, Replication.
Abstract:
I present exploration of key results from the series of ablated five-species “Rock-Paper-Scissors-Lizard-Spock”
minimal agent-based evolutionary models of biodiversity introduced by Zhong, Zhang, Li, Dai, & Yang in their
2022 paper “Species coexistence in spatial cyclic game of five species” (Chaos, Solitons and Fractals, 156:
111806). At the heart of Zhong et al.’s model of ecosystems coexistence is the Elementary Step (ES) algorithm
in which one or two neighboring agents are chosen at random to engage in one or more interactions selected
at random from the set {COMPETE, REPRODUCE, MOVE}. Minor revisions to the ES algorithm have recently
been introduced to make it more computationally efficient in space and in time, and one contribution of this
paper is to demonstrate that switching to this “Revised ES” (RES) has the unexpected effect of totally changing
the outcomes of Zhong et al.’s simulation experiments. I present analysis of the RES-based experiments which
shows that the key difference is that in RES the likelihood of an agent moving is decoupled from the likelihood
of an agent reproducing or competing, whereas in the original ES the likelihoods of the three possible actions
are interdependently coupled. The fact that such relatively minor changes to the ES algorithm result in such
major changes in the experiment outcomes casts significant doubt on the extent to which results such as those
from Zhong et al.’s original experiments can be trusted as truly representative of the real-world biological
systems that they are supposedly intended to model. Python source-code available on GitHub can be used to
replicate the results presented here.
1 INTRODUCTION
1.1 Evolutionary Spatial Cyclic Games
There is a long tradition of empirical bioinformatics
research exploring issues in ecosystems biodiversity,
and specifically in the population dynamics of species
co-existence, by running sets of computer simulation
experiments of minimal models of multiple interact-
ing species in what are known as Evolutionary Spatial
Cyclic Games (ESCGs). In these simulated systems,
each individual agent (i.e., each modelled organism)
in the population can move (i.e., each agent i occu-
pies a specific position in space at time t, denoted
by ⃗p
i
(t), and it may move to some different location
later in time such that ⃗p
i
(t + ∆
t
) ̸= ⃗p
i
(t)). Neighbor-
ing agents in these models may compete in pairwise
fight-to-the-death predator/prey interactions, in which
the loser dies leaving an empty space; and any empty
a
https://orcid.org/0000-0003-3822-9364
space may subsequently be filled with a new agent,
created by reproduction, asexually spawning a clone
of a neighboring living agent into the empty spot.
Inspired by the work of (May and Leonard, 1975),
the complexities of real-world predator-prey inter-
actions are reduced in these models to very simple
two-player competitive games that require almost no
time to simulate, and to give interesting coevolution-
ary population dynamics the game is usually cyclic,
i.e. it has intransitive dominance relationships. The
most well-known of such cyclic games is the hand-
gesture game Rock-Paper-Scissors (RPS): Rock beats
Scissors; Scissors beats Paper; and Paper beats Rock.
Using RPS as the predator-prey game allows for the
study of three-species model ecosystems (i.e., where
there are three species denoted by s
R
, s
P
, and s
S
, with
individuals in species s
R
always playing Rock, etc).
Despite their abstract simplicity, RPS-based
ESCGs have been useful in explaining aspects of
real-world biological systems in which such cyclic
dominance relationships do occur and are sustained:
Cliff, D.
The Devil Lies in the Details for Species Coexistence Stability in Ablated and Unablated Five-Species Evolutionary Spatial Cyclic Games.
DOI: 10.5220/0013132500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 545-554
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
545

Figure 1: Dominance network digraphs for Rock-Paper-
Scissors (RPS: left) and for Rock-Paper-Scissors-Lizard-
Spock (RPSLS: right). In both digraphs, the N
S
different
species S
i
are denoted by nodes labelled with index i ∈
{1,...,N
S
−1}, with arcs (directed edges) running from the
dominator (“predator”) species to the dominated (“prey”’)
species. The rules of RPSLS are: scissors cut paper; pa-
per covers rock; rock blunts scissors; scissors decapitates
lizard; lizard eats paper; paper disproves Spock; Spock va-
porizes rock; rock crushes lizard; lizard poisons Spock; and
Spock smashes scissors.
for examples, see the various papers cited in (Nag
Chowdhury et al., 2023).
1
An extension of RPS
to a five-gesture cyclic game known as Rock-Paper-
Scissors-Lizard-Spock (RPSLS) was introduced by
(Kass and Bryla, 1998) and that game has since been
widely used to study five-species model ecosystems.
Groundbreaking papers in this field of study were
published over the period 2006-2008: see, for exam-
ple, (Laird and Schamp, 2006; Reichenbach et al.,
2007a; Reichenbach et al., 2007b; Laird and Schamp,
2008; Reichenbach et al., 2008)
This current paper presents results from ESCG ex-
periments where one or more of the dominance re-
lationships in the RPSLS game have been removed,
giving rise to systems that I refer to as ESCGs with
ablated dominance networks.
1.2 Ablated Dominance Networks
Let N
S
denote the number of species in the ESCG
model. The intransitive predator/prey dominance re-
lationships of the N
S
=3 RPS and N
S
=5 RPSLS games
are illustrated as directed graphs (or digraphs) in Fig-
ure 1. Such digraphs are referred to here as domi-
nance networks.
In a recent paper, (Zhong et al., 2022) explored the
effects of selectively ablating one or more relation-
ships from the RPSLS dominance network. That is,
Zhong et al. deleted one or more of the arcs (directed
edges) from the RPSLS digraph, and then observed
the frequency distribution of asymptotic experiment
outcomes from large numbers of independent and
1
These include (Jackson and Buss, 1975; Sinervo and
Lively, 1996; Durrett and Levin, 1998; Elowitz and Leibler,
2000; Kerr et al., 2002; Gilg et al., 2003; Lankau and
Strauss, 2007; Nahum et al., 2011) and (Guill et al., 2011).
identically distributed (IDD) runs of the ESCG sim-
ulator using the ablated dominance network. Here,
“asymptotic” is interpreted in its usual sense, of the
response of the system as time t→∞, and the ex-
periment outcomes of interest are the number of co-
existing species at the end of the experiment. If we
let T denote the pre-specified experiment end-time
(where T is assumed to be sufficiently big that the
system does indeed settle to its asymptotic state), and
if we let n
s
(t) denote the number of species in the
system still surviving at time t, then the key observa-
tions in Zhong et al.’s study was the set of frequencies
F(n
s
(T )=c) : c ∈ {0,1,.. .N
S
}.
Zhong et al. studied the effect of varying the mo-
bility parameter in the ESCG (denoted by M, and ex-
plained in more detail in Section 2), which deter-
mines how mobile the individual agents are, and for
each ablated dominance network they plotted graphs
of F(n
s
(T )=c) as a function of M. Zhong et al. re-
ported a diversity of experiment outcomes, depending
on exactly which arc or arcs had been ablated from the
RPSLS dominance network. Section 2 summarises
the work of Zhong et al. in more detail.
In (Cliff, 2024b) I presented results which repli-
cated and confirmed Zhong et al.’s results, before
questioning whether they had chosen a sufficiently
large valure of T for their simulations to truly con-
verge on asymptotic states. In this paper I go
on to fully explore the effects of re-running Zhong
et al.’s experiments with a slightly different style
of ESCG simulation recently introduced in (Cliff,
2024a), which involves what are prima facie only
very minor adjustments to the core algorithm in the
ESCG simulation, the so-called Elementary Step (ES).
The initial motivation for altering the ES was to im-
prove its efficiency in space and in time. The revised
algorithm and associated initial results were first pub-
lished in (Cliff, 2024a), a paper presented at an in-
ternational conference on modeling and simulation
methods, where the focus was on the computational
efficiency increases (reductions in execution time for
the simulations) and where I did not seek to explore
the effects of using the revised ES algorithm across
the full range of ablated dominance-network species
co-existence experiments reported by Zhong et al..
My new results and analysis presented here show
that switching from the “Original” Elementary Step
(OES) used by (Zhong et al., 2022) to the “Revised”
Elementary Step (RES) introduced in (Cliff, 2024a)
has a major effect on every single one of the key ex-
periment outcomes reported in (Zhong et al., 2022);
that is, for all of Zhong et al.’s key results, I demon-
strated in (Cliff, 2024b) that they can be replicated
very well with OES and then in this paper, when I
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
546

switch the simulator to instead use RES, I get to-
tally different outcomes – identifying and explaining
that difference is the key contribution of this paper.
The Python source-code initially developed for the
research reported in (Cliff, 2024a) is freely-available
public-domain open-source software on GitHub,
2
.
I give further details of the OES-based ESCG as
used by Zhong et al. (and many other researchers) in
Section 2, and then explain the differences between
OES and RES in Section 3. I discuss differences be-
tween OES and RES results in Section 4. Then in
Section 5 I explain the root cause of those differences
and discuss the extent to which they cast doubts on
just how far results from OES can be trusted as being
representative of the real-world biosystems that they
are intended to be informative models of.
2 THE ESCG ALGORITHM
Zhong et al. used the de facto standard algorithm for
their Evolutionary Spatial Cyclic Game (ESCG), the
core elements of which were established in (Reichen-
bach et al., 2007a).
The position ⃗p
i
of each individual agent i in the
population is represented by the values on a Cartesian
lattice l of some dimensionality, and almost all work
in this field studies systems where the lattice is a 2-
dimensional square of side-length L, so the maximum
total population size is N=L×L, where the position ⃗p
i
of an individual agent i can be represented by an (x,y)
row-column co-ordinate pair indexing into the lattice
as l(x,y). Cells (or ‘positions’ or ‘locations’) in the
lattice can be empty, denoted here by
/
0, or they can be
occupied by a single individual agent. In the RPSLS-
based systems explored by (Zhong et al., 2022), the
number of species is fixed at N
S
=5, and the species
identifier indexes will here run 0,. .., N
S
− 1, so the
set of possible lattice cell-values is C
5
= {
/
0,0,. .. ,4}.
Let U(S) denote the uniform (equiprobable) ran-
dom choice of a member from set S, and let U[n
lo
,n
hi
]
denote a fresh random draw from a uniform distri-
bution over the range [n
lo
,n
hi
] ⊂ R. To initialise
the lattice, some researchers include a number of
empty cells in the initial state, but (Zhong et al.,
2022) do as many other authors do, starting in-
stead with every cell containing an agent, with each
species s
i
being chosen with probability
1
N
S
∀i such
that l(x, y)=U({0, ... ,4})∀(x, y).
The ESCG is then simulated as a simple discrete-
time system. At the core of the simulation is the El-
ementary Step (ES), the original and revised forms
2
See github.com/davecliff/ESCG Python
of which (i.e., the OES and RES) are discussed fur-
ther in Section 3. In both the OES and the RES, the
ES implements an interaction between two randomly-
chosen neighboring cells, and this requires the re-
searcher to commit to a neighborhood function and
to a specific boundary condition. The neighborhood
function is typically either the von Neumann or the
Moore. The boundary condition is typically either
periodic, where the square lattice is, topologically, a
toroidal surface; or no-flux, where the edges of the lat-
tice are hard boundaries on a ‘walled garden”. Zhong
et al. used the von Neumann Neighborhood and no-
flux boundary conditions, and so that is what I used
in the replication experiments described here.
Implementing the random selection of a cell and
its randomly chosen neighbor is trivial. Once two
neighboring cells have been chosen, they may inter-
act, and the opportunity set of possible interactions is
the same for both OES and RES:
• If both cells contain agents, they might compete
according to the rules of the ESCG’s dominance
network for species-to-species interactions. Com-
petition will result either in a draw (no change), or
the dominated ‘prey’ individual dies and the lat-
tice cell it was occupying is replaced by
/
0.
• If one cell contains an agent and the other contains
/
0 (i.e., is empty), the agent might reproduce by
cloning a copy of itself into the empty cell.
• The contents of the two cells might be swapped,
which implements a one-step move for each of the
two cells’ contents.
Which, if any, of these three interactions occurs
on any one ES is determined stochastically, and the
random choice of interaction is determined by three
system hyperparameters: µ, which primarily affects
the probability of choosing compete; σ, which primar-
ily affects the probability of choosing reproduce; and
ε which primarily affects the probability of choosing
move. Zhong et al. describe these three as “probabil-
ities” but (as is explained below in Section 3) this is
incorrect: they are relative interaction rates.
The convention in this field of research is to talk of
time-steps at two levels of granularity: the ES is the
finer-grained time-step, but it involves only a single
potential interaction between two randomly chosen
cells in what is usually a large lattice of such cells. At
a coarser-grained level, researchers talk of the Monte
Carlo Step (MCS) as being the primary unit of time,
a time-step in which all N cells in the lattice can po-
tentially experience at least one interaction: i.e., one
MCS involves N elementary steps being executed.
In all the experiments they reported in detail,
Zhong et al. used L=200 and (in common with many
The Devil Lies in the Details for Species Coexistence Stability in Ablated and Unablated Five-Species Evolutionary Spatial Cyclic Games
547

other researchers) they set µ=σ=1.0 for all experi-
ments, stating that they did so without loss of gener-
ality. Following the de facto standard procedure es-
tablished in many prior papers in this field, Zhong et
al. did not explicitly set values for the movement fac-
tor ε, but instead systematically varied, as the primary
independent variable in their experiments, a mobility
parameter M, which relates the movement rate ε to
the lattice size N via ε=2MN. For each of their key
experiments, Zhong et al. varied M in discrete steps
over M∈[10
−7
,10
−3
], sampling the response of the
system, its frequency distribution of final outcomes, at
each M-value. At each value of M sampled, Zhong et
al. performed N
IID
=500 independent and identically
distributed (IID) simulation experiments.
Finally, to complete the specification for one of
the RPSLS-style ESCGs simulated by (Zhong et al.,
2022), the dominance network needs to be defined. In
(Cliff, 2024b) I introduced terminology to distinguish
the various ablated RPSLS dominance networks that
Zhong et al. explored (each of which, Zhong et al. re-
ferred to solely by reference to which figure-number
in their paper the relevant network diagram appeared
in): let Z0 denote the original unablated RPSLS net-
work as illustrated in Figure 1, and then let any ver-
sion of that network with n
a
arcs ablated be referred
to as a Zn
a
network, so for example Z0 with two ab-
lated arcs would be referred to as a Z2 network; then
in the cases where Zhong et al. studied two topologi-
cally nonisomorphic networks of type Zn
a
, add qual-
ifying letters a and b as suffixes. In total, Zhong et
al. studied the dynamics of ESCGS with six different
ablated versions of Z0, referred to as Z1, Z2a, Z2b,
Z3a, Z3b, and Z4; for full details, see (Cliff, 2024b).
The ESCG algorithm used by Zhong et al. is sim-
ple to describe: the 2D lattice L is randomly initialised
at t=0, as described above, and the dominance net-
work D is set up. An outer time-step loop iterates
over each MCS, within which an inner loop iterates
N times over the Elementary Step (ES) procedure
ELSTEP. After each MCS time-step, the population
density of each of the N
S
species (denoted by ρ
i
for
i ∈ {0,.. .,N
S
− 1}), along with the density of empty
cells (denoted by ρ
/
0
) is recorded, optionally along
with the state of the lattice at that MCS. At the end
of each IID simulation run, the time-series of densi-
ties ρ
/
0
and ρ
i
∀i are written to an output file, and the
set of frequency distributions of experiment outcomes
(i.e., F(n
s
(T )=c) for c ∈ {1,. .., 5}) are updated. The
final Fs for the set of N
IID
simulations is, in the cur-
rent context, the primary data of interested returned
by the ESCG.
The elementary step (ES) algorithm ELSTEP is
similarly straightforward. It is trivial to randomly
choose a cell in the lattice, and to randomly choose
a neighbor of that cell given the pre-defined neigh-
borhood function and boundary conditions. Imple-
menting the three individual cell-to-cell interaction
procedures COMPETE, REPRODUCE, and MOVE is
similarly unproblematic. However, to better facili-
tate the comparison between OES and RES, there is
some subtlety required in describing the method for
randomly selecting the interaction(s) to be enacted on
any one elementary step: here we’ll assume that EL-
STEP can call a function GET3PR which returns three
interaction probabilities P
∗
µ
,P
∗
σ
, and P
∗
ε
that respec-
tively set the likelihood of ELSTEP calling COMPETE,
REPRODUCE, and MOVE. This can be simply imple-
mented by generating three random real values r
1
to
r
3
each via U[0,1], and then executing COMPETE if
r
1
< P
∗
µ
; executing REPRODUCE if r
2
< P
∗
σ
; and exe-
cuting MOVE if r
3
< P
∗
ε
. For OES, GET3PR always
returns two of the three probabilities as zero, and the
other probability as one – this ensures that only one
of the three possible interactions occurs on any one
elementary step, which is the convention introduced
in the seminal papers of (Reichenbach et al., 2007a);
a convention that has been continued in all published
literature that replicates and builds upon those works.
The calculation of the three interaction probabili-
ties is where the revised elementary step (RES) differs
from the original elementary step (OES), and that is-
sue is discussed in more detail in the next section.
3 OES vs. RES
The difference between the OES, used by (Zhong
et al., 2022) and many others,
3
and the RES, intro-
duced in (Cliff, 2024a), boils down to how the three
probabilities used in ELSTEP, P
∗
µ
,P
∗
σ
, and P
∗
ε
, are cal-
culated from the interaction rates µ, σ, and ε. Be-
cause we’re repeatedly dealing here with triplets of
values, let
⃗
R = (µ,σ, ε) denote the vector of inter-
action rates; let
⃗
P
∗
(t
ES
) = (P
∗
µ
(t
ES
),P
∗
σ
(t
ES
),P
∗
ε
(t
ES
))
denote the vector of interaction probabilities at time
t
ES
, measured here at the micro-level of the count
of individual elementary steps, rather than the macro
level of MCS-number; and let
⃗
P = (P
µ
,P
σ
,P
ε
) denote
3
The core of OES can be traced back to (Gillespie,
1976), cited approvingly in the landmark paper of (Reichen-
bach et al., 2007a). Other authors who have reported re-
sults from OES-based studies include (Nagatani et al., 2018;
Kabir and Tanimoto, 2021; Mood and Park, 2021; Park,
2021; Avelino et al., 2022; Bazeia et al., 2022; Menezes
et al., 2022a; Menezes et al., 2022b; Zhang et al., 2022;
Park and Jang, 2023; Menezes et al., 2023); and (Kubyana
et al., 2024).
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
548

the vector of normalized probabilities, explained fur-
ther below.
The OES method, which is the de facto standard
method in ESCG modelling, can be traced back to first
usage in the landmark papers of (Laird and Schamp,
2006) and (Reichenbach et al., 2007a). In those origi-
nal sources, each of the elements of
⃗
R clearly have the
potential to take on values greater than one.
However, quite often the three interaction rates
have since been described by other authors of papers
using OES as being bounded to the range [0.0,1.0]
and are also referred to by several authors as ‘prob-
abilities’, despite the fact that they are not actually
treated as literal probabilities: for example, µ = 0.5
does not necessarily mean there is a 50% chance of
COMPETE being the next interaction: it might do, and
it might not, depending on the contemporaneous val-
ues of σ, and ε. This is because the three interac-
tion rates are first subjected to a normalization pro-
cess that divides each of the three by the sum of the
three, giving three “normalized probabilities” P
µ
,P
σ
,
and P
ε
, the elements of
⃗
P, which are each within [0,1]
and which collectively sum to one.
To understand this, the values for the interac-
tion rates µ, σ, and ε can be thought of in OES as
indications for what relative proportion of a three-
segment roulette-wheel is allocated to each possi-
ble interaction, and then in the OES the concep-
tual roulette-wheel is spun once, generating a ran-
dom value r(t
ES
)=U[0.0, 1.0], which is then com-
pared to the elements of
⃗
P such that the interaction
at t
ES
will be COMPETE if 0 ≤ r(t
ES
) < P
µ
; REPRO-
DUCE if P
µ
≤ r(t
ES
) < P
µ
+ P
σ
; and MOVE other-
wise (because P
ε
= 1.0 − (P
µ
+ P
σ
)). This method is
widely known as roulette selection in the literature on
evolutionary computation (see e.g. (Lloyd and Amos,
2017)). Hence, with OES,
⃗
P
∗
(t
ES
) is always either
(0,0,1) or (0,1, 0) or (1,0, 0), roulette-selected on the
basis of
⃗
P and r(t
ES
): i.e., only one randomly-chosen
type of interaction can ever happen per single OES.
The corresponding calculation in RES is much
simpler to explain because the interaction rates µ and
σ are interpreted instead as actual probabilities, de-
fined only over [0,1], and mapping directly (via the
identity function) onto P
∗
µ
and P
∗
σ
, respectively; the
movement rate ε is required to be non-negative and is
interpreted as mapping directly onto P
∗
ε
, with the sim-
ple filter that if ε>1.0 then it is capped to one. Thus
in RES there is no
⃗
P and instead an essentially direct
mapping from
⃗
R to
⃗
P
∗
(t
ES
). This means that, whereas
in OES there is a constraint that |
⃗
P
∗
(t
ES
)|=1 ∀t
ES
, in
RES instead |
⃗
P
∗
(t
ES
)| ∈ [0, 3] ∈ R – i.e., at the limit,
⃗
P
∗
(t
ES
)=(1,1,1) is perfectly well defined.
On the face of it, the change between OES and
RES is pretty minimal. Whereas OES’s roulette-
selection means that only one interaction can oc-
cur per ES, the definition of RES allows up to
three interactions to occur per ES. For example,
if µ=σ=ε=1.0 (i.e.,
⃗
R=(1,1,1)) then in OES one
of the three possible actions will be randomly
chosen, using r(t
ES
) and
⃗
P=(
1
3
,
1
3
,
1
3
), and then
⃗
P
∗
(t
ES
) ∈ {(1,0,0), (0,1,0),(0,0, 1)} depending on
r(t
ES
); whereas in RES, for the same
⃗
R we have
⃗
P
∗
(t
ES
)=
⃗
R=(1,1,1) and hence, whenever possible,
on each RES there will be a call to COMPETE, fol-
lowed by a call to REPRODUCE, followed by a call to
MOVE. This means that, when
⃗
R=(1,1,1), if possi-
ble a single agent can, in one RES, kill its neighbor-
ing opponent in COMPETE, and then REPRODUCE a
clone of itself into the
/
0-cell just vacated by the dead
opponent; and then MOVE into the neighboring cell,
swapping position with its newly-spawned offspring.
Whereas with OES the same agent would engage in
at most one randomly-chosen interaction per ES, and
so would require three consecutive ESs to achieve the
same sequence of events.
For that reason, one might expect a RES-based
replication of any of the experiments reported in
(Zhong et al., 2022) to proceed up to three times faster
than the comparable OES-based experiment – and in-
deed it was a desire for reduced run-times that moti-
vated the development of RES, as reported in (Cliff,
2024a), where further analysis and discussion of the
speed-up enabled by RES is given. Here, the focus is
not on the run-times of the simulations, but instead on
the unexpected results that I report in the next section:
the outcomes of every single one of Zhong et al.’s ab-
lation experiments is dramatically altered when the
ES is switched from OES to RES, an unexpected con-
sequence of RES that was not explored in any depth
in (Cliff, 2024a).
4 RESULTS
4.1 Original Elementary Step (OES)
As with the original results of (Zhong et al., 2022)
and also with my replication of those results reported
in (Cliff, 2024a), in all the experiments I discuss in
(Cliff, 2024b) I set T =10
5
MCS, L=200, µ=σ=1.0,
and ranged M from M
min
=10
−7
to M
max
=10
−3
, us-
ing a multiplier of 10
1/5
to give five sampled M data-
points per order of magnitude, and I ran OES-based
simulation experiments for all six of Zhong et al.’s
networks Z1, Z2a, Z2b, Z3a, Z3b, and Z4 – the com-
plete set. All my results presented in (Cliff, 2024b)
The Devil Lies in the Details for Species Coexistence Stability in Ablated and Unablated Five-Species Evolutionary Spatial Cyclic Games
549

agree very well with the original results of (Zhong
et al., 2022).
In each set of experiment outcomes the key ob-
servable feature, as highlighted by Zhong et al., is
a sudden transition part-way between M=10
−5
and
M=10
−4
from three-or-more-species coexistence be-
ing the dominant outcome, to two or fewer coexisting
species always being the outcome – i.e., a “collapse”
in ecosystems diversity. If this result is general, i.e.
if it is applicable to real-world ecosystems and is not
just an artefact of the particular details of Zhong et
al.’s ESCG modelling methods, then surely we should
expect to see it also when the only change made to the
ESCG is to switch from OES to RES – and in fact, if
anything, we should expect to see the collapse sooner
(after a shorter T ) when working with RES, because
of its potential for a three-times speed-up.
4.2 Revised Elementary Step (RES)
For my RES experiments, I swept M within the range
[5×10
−8
,1.25×10
−5
] to give the same range of P
ε
values as were used by (Zhong et al., 2022) in their
OES experiments.
To avoid what would otherwise surely be a strong
contender for the most boring set of graphs ever pub-
lished in a scientific paper, here I give only verbal de-
scriptions of the outcomes of my RES-based replica-
tions of Zhong et al.’s experiments. I only need to de-
scribe two outcomes because my results were unam-
biguously bimodal: for networks Z0, Z2b, Z3b, and
Z4, the FvsM outcome was essentially identical: the
system never deviated from F(n
s
(T )=5)=1.0 across
the entire range of M values; and for the remaining
networks (i.e., Z1, Z2a, and Z3a) the outcome was
again essentially identical, but in these three cases
the system’s asymptotic state was F(n
s
(T )=3)=1.0
across the entire range of M values. That is, in all my
RES experiments, the outcome at T was always ei-
ther three-species coexistence or five-species coexis-
tence, for every network studied by Zhong et al.: that
is, once you switch to RES, the collapse in ecosystems
diversity seen every time when using OES simply dis-
appears.
And yet that switch, from roulette-selecting a sin-
gle interaction in OES to probabilistically selecting
one, two, or three interactions in RES, seems only a
very small change, a mere implementation detail in-
tended to speed things up a bit – the set of possible
interactions remains unchanged, so why does RES
result in such different outcomes to OES? And, un-
less it can be convincingly argued that OES is some-
how a better representation, a better abstraction, of the
natural world than is RES, then surely this fragility,
these major changes in outcomes resulting from mi-
nor changes to the internal mechanisms, casts signif-
icant doubt on the generality and applicability of the
OES-based results published by (Zhong et al., 2022),
and indeed of the results in any other published pa-
pers (and there are many) that show results from OES-
based ESCGs. This discrepancy in the results is dis-
cussed in more detail in the following section.
5 DISCUSSION
To understand why the RES outcomes differ so
markedly from the OES ones, it is necessary to go
back and consider the step in OES where the three
system hyperparameters, the interaction rates µ,σ,
and ε, are ‘normalized’ to give internal probabilities
P
µ
,P
σ
, and P
ε
prior to the roulette-selection of a sin-
gle action for that ES. The normalization process in-
tertwines the three rates, and they cease to be indepen-
dent, and yet this seems to be rarely if ever acknowl-
edged in the literature.
For example, sweeping M across a range such as
the [10
−7
,10
−3
] used by Zhong et al., is described
by Zhong et al. (and by many other authors who also
present results from parameter-sweeps on M, this be-
ing a common way of exploring ESCGs) as systemat-
ically varying mobility, i.e. treating M as the indepen-
dent parameter in the experiments. While altering M
definitely does alter the likelihood of movement for
the agents in the ESCG, in OES it also alters the like-
lihood of competition, and of reproduction. Specif-
ically, in OES, increasing mobility M increases the
probability of movement P
ε
, but it can also decrease
the probabilities of competition (P
µ
) and reproduction
(P
σ
). Figure 2 shows the changes in values of the
three probabilities over the range of M values used
by (Zhong et al., 2022), all at L=200, for various val-
ues of µ and σ including the µ=σ=1.0 case used by
(Zhong et al., 2022).
As can be seen from the upper left graph in Fig-
ure 2, the µ=σ=1.0 case, as M is increased P
ε
does
indeed rise as intended, but as it rises so P
µ
and P
σ
fall, ending up very close to zero once M≈10
−3
.
Clearly increasing M not only increases mobility, but
also greatly reduces the frequency of competition
and of reproduction: at M=10
−3
,P
ε
=0.976 while
P
µ
=P
σ
=0.012; so movement is roughly 80 times
more likely than either reproduction or competition.
In contrast, when using RES, increasing M does in-
deed increase P
ε
, but it has no effect on P
µ
or P
σ
be-
cause the interaction probabilities are independent.
To illustrate this key difference between OES and
RES, Figure 3 shows the unit cube in R
3
formed by
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
550
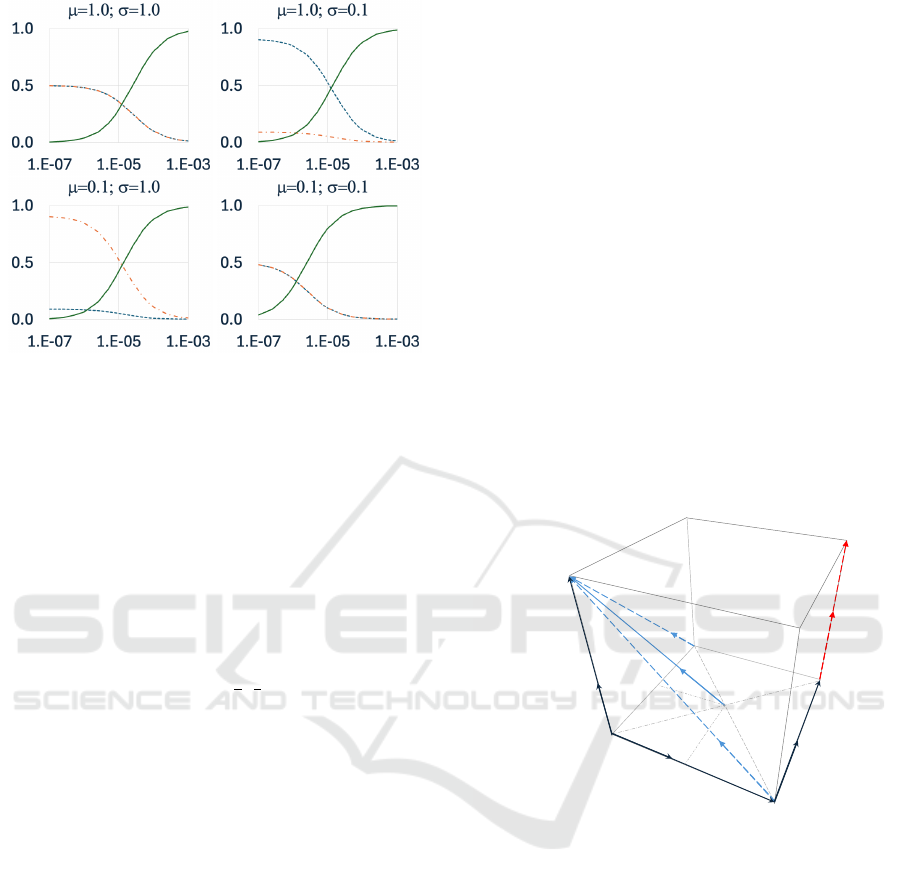
Figure 2: Graphs of the normalized internal probabilities
P
µ
, P
σ
, and P
ε
for L=200 as calculated in the OES for
M∈[10
−7
,10
−3
], the range used by (Zhong et al., 2022), for
the values of µ and σ shown in each graph title. Each graph
shows plots for P
µ
(dotted line), P
σ
(dash-dot line), and P
ε
(solid line). In all four graphs the horizontal axis is M while
the vertical axis is probability. The upper left graph shows
results from µ=σ=1.0; upper right shows results from
µ=1.0,σ=0.1; lower left shows results from µ=0.1, σ=1.0;
and lower right shows results from µ=σ=0.1.
(P
µ
,P
σ
,P
ε
)-space – a cube that I’ll refer to as “P-
space” hereafter. In Zhong et al.’s OES-based ex-
periments with µ=σ=1.0, As M is swept from 10
−7
to 10
−3
, the three interaction probabilities shift from
their initial point close to (
1
2
,
1
2
,0), and move with
nonlinearly varying speed along a straight line in P-
space, to their final point at approximately (0,0, 1) –
this is represented by the solid blue arrowed line in
Figure 3. (At L=200, as used by Zhong et al., the ac-
tual start-point is (0.498,0.498, 0.004) and the actual
end-point is (0.01,0.01,0.98)).
However in RES, sweeping M over the corre-
sponding range of values, P
µ
=P
σ
=1.0;∀M, and the
only probability varied as M increases is P
ε
which
rises along the straight line in P-space from its ini-
tial point close to (1,1,0) to its final point close to
(1,1,1), which is illustrated by the vertical dashed red
arrowed line in Figure 3.
Because the three interaction probabilities are in-
dependent in RES, one can readily set P
µ
and P
σ
to be
any pair of values in [0, 1]
2
⊂ R
2
and then indepen-
dently sweep the mobility M across some range which
maps P
ε
to varying over [0,1] ⊂ R: that is, in RES it
is trivially easy to set up an experiment where the ini-
tial point in P-space is anywhere on the floor-plane of
the P-space unit cube, and for P
ε
to then range from
zero to one while P
µ
and P
σ
are held constant, giving
a vertical line in P-space. And we need not stop there:
in principle, experiments can be run with RES where
across the duration of the experiment the point in P-
space moves along some arbitrarily-oriented straight
line or curve – there is nothing in RES that limits P
ε
to being the sole independent variable, nothing that
requires only vertical lines in P-space.
However with OES the experimenter’s options are
significantly more constrained. To illustrate why this
is so, refer back to Figure 2: the upper left graph
shows the three interaction probabilities versus M for
the case where µ=σ=1.0, and the lower right graph
shows the same three probabilities for µ=σ=0.1; de-
spite the factor of ten difference in the values of µ
and σ between these two graphs, the traces shown
on the graphs are qualitatively very similar: in both
cases, P
µ
and P
σ
both start at ≈0.5 and then decay
sigmoidally to ≈0.0, while P
ε
rises sigmoidally from
≈0.0 to ≈1.0.
To understand this, consider first the location of
the start-point in P-space for an OES-based experi-
ment where M is swept from some very small initial
value (like 10
−7
) to some larger final value (such as
P
µ
P
s
P
e
(0,0,0)
(1,0,0)
(1,1,0)
(1,1,1)
(0,0,1)
(0,1,1)
(P
µ
, P
s
, P
e
)
(0,1,0)
Figure 3: Perspective plot of (P
µ
,P
σ
,P
ε
)-space, referred to
here as “P-space”, which is the unit cube [0,1]
3
⊂R
3
. The
floor-plane of the cube is the (P
µ
,P
σ
,0) unit square with,
from this viewpoint, the P
µ
axis running from the origin to
lower-right, and the P
σ
axis running from the origin to up-
per right; the vertical axis is P
ε
. For clarity, seven of the
eight corner vertices are labelled with their coordinates; the
one unlabelled vertex, nearest the viewpoint, is (1,0,1). The
solid blue arrowed line from (0.5,0.5,0.0) to (0,0, 1) shows
the straight-line path of P
ε
values explored by Zhong et al.
in their OES-based experiments as they swept M over the
range [10
−7
,10
−3
] while keeping µ=σ=1.0. The dashed
red arrowed line from (1,1,0) to (1,1,1) shows the straight-
line path of P
ε
values explored when using RES, with M
varied over the corresponding range. The dashed blue ar-
rowed lines starting at (1, 0, 0) and at (0, 1, 0), both ending
at (0,0,1), mark the two vertical-plane edges of the OES
unit 3-simplex: its horizontal edge is the line P
σ
= 1 − P
µ
,
joining (1,0,0) to (0,1,0): the significance of the OES unit
3-simplex is discussed further in the text.
The Devil Lies in the Details for Species Coexistence Stability in Ablated and Unablated Five-Species Evolutionary Spatial Cyclic Games
551

10
−3
). From the way that the three interaction prob-
abilities are calculated as functions of the three in-
teraction rates in the “normalization” step, whenever
µ=σ, we have P
µ
=P
σ
=
µ
2µ+ε
. Given that ε=2MN,
when M≪
1
2N
we have ε≈0 and hence P
µ
=P
σ
≈
µ
2µ
=
1
2
,
for any case when µ=σ. Thus it is intuitively obvi-
ous that with OES the start-point on the (P
µ
,P
σ
,0)
ground-plane of the P-space unit cube is determined
by the ratio of µ to σ: when µ=σ=c the ratio is 1.0
and the start-point is very close to (0.5, 0.5, 0.0), re-
gardless of the actual value of c; then, as σ/µ → 0,
the start-point moves ever closer to (1,0, 0); and as
σ/µ → ∞ the start-point moves ever closer to (0, 1,0).
And, in the two limit cases, when µ=0 we have P
µ
=0
but P
σ
=σ/(σ+ε) → 1.0 as ε → 0, giving a start-point
of ≈(0, 1,0); and (mutatis mutandis) when σ=0 so
P
µ
→ 1.0 as ε → 0, giving a start-point of ≈(1, 0,0).
The key thing to note here is that, for any plausible
values of µ and σ, and sufficiently low initial value of
M, the start point is always on (or very close to) the
line P
σ
=1 − P
µ
, joining (1,0,0) to (0, 1,0).
Next consider the OES end-point in P-space: this
is easy, it’s always going to be at (or very close to)
(0,0,1), for any plausible values of µ and σ, and suf-
ficiently high final value of M (i.e., for M ≫
1
2N
).
And, taken together, these constraints on the P-
space start-points and end-points for OES-based M-
sweep experiments mean that the set of possible
points sampled in P-space is tightly constrained: the
OES-based experiments are limited to the set of points
on (or very near) the unit 3-simplex (i.e., the planar
triangle defined by the points (1,0, 0), (0,1, 0), and
(0,0,1)); thus, OES experiments are simply not set
up to explore the response of the system in any area
of P-space outside of that three-sided planar area.
To emphasize the importance of this point, let us
consider the expected qualitative asymptotic experi-
ment outcomes at each of the seven corner-vertices of
P-space for which at least one of the interaction prob-
abilities is nonzero, assuming that the ESCG is run
for its duration with the three interaction probabilities
held at the vertex:
• (0, 0, 1): the agents can move, but they never
compete or reproduce, so the whole population of
agents wander aimlessly around the lattice, never in-
teracting with one another. This is a dynamic equi-
librium, but population-level statistics such as species
densities will remain static. There is no chance of
species extinctions with the system at this vertex.
• (0, 1, 0): agents can reproduce, but not move
or compete: any initially empty cells might be filled
by reproduction, but once there are no empty cells re-
maining the system will have converged to a stasis.
there’s no chance of extinctions at this vertex.
• (1, 0, 0): agents compete, but can’t move or re-
produce: any immediate neighbors in the initial lattice
configuration who are predators/prey will eventually
compete, resulting in the death of the prey. Once all
possible predator/prey interactions in the initial lat-
tice configuration have been resolved by competition,
the number of empty cells will have increased, but
the system will thereafter be at stasis. Species ex-
tinctions can occur when the system is at this vertex:
the likelihood of extinctions is entirely dependent on
the initial distribution of agents on the lattice at t=0.
With uniform random allocation of agents to the lat-
tice positions at t=0 the likelihood of one or more en-
tire species going extinct when the interaction prob-
abilities are held at this vertex is technically nonzero
but will be vanishingly small for any sufficiently large
value of L to be of serious interest.
• (0, 1, 1): agents reproduce and move, but don’t
compete: empty cells in the lattice at t=0 will eventu-
ally be filled by reproduction, and all agents can move
around freely; once the last empty cell is filled, the
system is thereafter at the same dynamic equilibrium
with static species densities as at vertex (0,0,1), and
so there is no chance of extinctions here either.
• (1, 0, 1): agents compete and move, but don’t
reproduce: the number of empty cells is likely to rise
over the course of the experiment as prey agents are
dominated by predators. Species extinctions are en-
tirely likely when the system is at this vertex, the like-
lihood depending on the dominance network.
• (1, 1, 0): agents compete and reproduce, but
don’t move: the number of empty cells is likely to
rise and fall, with the asymptotic outcome being zero
empty cells. Species extinctions can occur at this ver-
tex, the likelihood depending on l(0) (the state of the
lattice l at t=0) and on the dominance network.
• (1, 1, 1): the whole enchilada – agents can com-
pete, reproduce, and move. Species extinctions can
occur when the system is at this vertex, the likelihood
depending on l(0) and the dominance network.
From this it is clear that, if the ESCG is being run
as a minimal model of ecosystems biodiversity and
species co-existence, the final three vertices (1,0,1),
(1,1,0), and (1,1,1) are of primary interest, because
they are the three for which there is a reasonable
chance of species extinctions. These three vertices
are all on the top-plane of the P-space unit cube, and
because none of them are within the unit 3-simplex,
they simply can’t be explored via OES-based mod-
els. Instead, as mobility M is increased in OES-based
models, the system is taken ever-closer to the “aim-
less wandering” P-space vertex (0,0,1).
For example, a naive experimenter might start an
OES M-sweep simulation at µ=1 and σ=0, i.e. at
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
552

the (1,0,0) vertex in P-space, and then sweep M over
[10
−7
,10
−3
] in the (mistaken) expectation that this
will increase only P
ε
and eventually take the system
to the (1,0,1) P-space vertex. However the OES nor-
malization process will instead take the system on
a straight-line path toward the (0,0,1) vertex. Some
species extinctions may occur en route, and – as I
have shown in my replication of the results of (Zhong
et al., 2022) — the system will close in on (0,0,1)
but P
µ
and P
σ
can remain nonzero, so extinctions do
remain a possibility. But the OES experimenter’s re-
sults will say nothing about the asymptotic behavior
of the ESCG system at (1,0,1) in P-space: for that,
they would need to abandon OES and switch to RES.
The results from RES-based simulations pre-
sented here show that when areas of P-space that are
unreachable in OES-based simulations are instead ex-
plored via the ESCG with RES, the frequency distribu-
tions of asymptotic experiment outcomes can change
significantly. And surely that then calls into ques-
tion the generality and the applicability of the results
from OES-based simulations to the real-world sys-
tems that they are intended as models of: my RES
results present a first counter-example to the observa-
tion in OES-based systems of the sudden transition in
experiment outcomes from being n
s
(T )) ≥ 3 at low
M to n
s
(T )) ≤ 2 at higher M values: this happens as
the OES-based system converges on P-space vertex
(1,0,0) but does not happen when the RES-based sys-
tem is instead set to converge on (1,1,1).
And from that it seems reasonable to propose that
Zhong et al.’s OES-based results, repeatedly showing
collapses in ecosystems coexistence/diversity as mo-
bility increases, are mere artefacts of their decision to
use OES in their ESCG model: if they’d used RES,
they would have had no story to tell. The only pos-
sible line of counterargument that I can think of here
would be to defend use of OES on the basis that it
is somehow more faithful to ecological reality, more
biologically plausible, than RES, and hence to argue
that OES should continue to be used in preference to
RES because it is, in that sense, the better modelling
method. Now maybe I’m biased in favour of my
own creation, but I’ve tried very hard to think of how
OES could possibly be defended as somehow better
than RES, and thus far I have drawn a blank. The
two methods strike me as equally abstract, equally
(im)plausible approximations to reality, but in OES
the experimenter’s hands are tied in P-space, while
in RES the experimenter can roam freely within the
confines of that cube, which surely opens up more av-
enues for exploration than repeatedly moving from
the base to the apex of the unit simplex, which is
seemingly the only option with OES. And, in that
sense, arguably RES is a better approximation to re-
ality than OES, because in many real ecosystems it
seems intuitively unlikely that every species’ motil-
ity, fecundity, and likelihood of eating or being eaten
are interdependently bound together by one function
(as in OES), and more likely that these three aspects
of species’ existence are to some extent independently
variable (as in RES). I am of course very open to con-
versation with anyone who wants to argue the point:
discussion and debate is very welcome here.
In this paper I’ve shown results from RES-based
M-sweep experiments where only one path in P-
space, along the line from (1,1,0) to (1,1,1), is ex-
plored. An obvious direction for further work in
attempting to understand these ESCGs as models
of species coexistence and ecosystems biodiversity
would be to generate results from similar RES ex-
periments where the P-space path starts at a different
initial point but still ends at (1,1,1), and also to run
further RES-based simulations that end at (1,1,0) or
(1,0,1), the other two P-space vertices where species
extinctions can be expected to occur at the limit. The
OES-based results can then be judged in the wider
context of comparison to the array of results from the
various RES-based experiments: the extent to which
the sudden shift in n
s
(T ) outcomes is also seen, if
at all, in the RES experiments will then allow us to
make a reasoned judgement on the generality of that
shift occurring in OES-based outcomes. This is fur-
ther work that I intend to report in one or more forth-
coming papers, but at this stage it seems reasonable
to hypothesize that the M-dependent shift in experi-
ment outcomes seen in the OES systems as reported
by (Zhong et al., 2022) and replicated here is in fact
a mere artefact of the method, a quirk of the OES’s
normalization and roulette-selection process.
6 CONCLUSION
The results presented here offer the first clear and
comprehensive set of counterexamples to all the key
results presented by (Zhong et al., 2022), because
I have shown that in the RES-based ESCG experi-
ments, which I contend are just as plausible models
of real-world systems as the OES-based ones, no M-
dependent shift in outcomes (i.e., final counts of coex-
isting species) occurs. Provision of these counterex-
amples, and my analysis and explanation in terms of
P-space trajectories, are the contributions of this pa-
per. I see no reason to expect that the problematic
issues identified here with the use of OES in ESCGS
is limited only to the one paper by Zhong et al. that
I have focused on here. It seems plausible that con-
The Devil Lies in the Details for Species Coexistence Stability in Ablated and Unablated Five-Species Evolutionary Spatial Cyclic Games
553

clusions drawn in many other papers reporting results
from OES-based studies may need to be revised if,
when the same M-sweep experiments are instead run
with a RES-based ESCG, the results come out differ-
ent. Only further work and time will tell, but right
now, from what I’ve presented here, it does seem that
the devil lies in the details for this class of evolution-
ary spatial ecosystem simulation model.
REFERENCES
Avelino, P., de Oliveria, B., and Trintin, R. (2022). Parity
effects in rock-paper-scissors type models with a num-
ber of species ns ≤ 12. Chaos, Solitons and Fractals,
155(111738).
Bazeia, D., Bongestab, M., and de Oliveira, B. (2022). In-
fluence of the neighborhood on cyclic models of bio-
diversity. Physica A, 587(126547).
Cliff, D. (2024a). Never Mind The No-Ops: Faster and Less
Volatile Simulation Modelling. Proc. EMSS2024;
Full version: SSRN 4883174.
Cliff, D. (2024b). On long-term species coexistence in
five-species evolutionary spatial cyclic games. Chaos,
Solitons and Fractals, 190(115702).
Durrett, R. and Levin, S. (1998). Spatial aspects of inter-
specific competition. Theor. Pop. Biol., 53(1):30–43.
Elowitz, M. and Leibler, S. (2000). A synthetic oscilla-
tory network of transcriptional regulators. Nature,
403(6767):335–338.
Gilg, O., Hanski, I., and Sittler, B. (2003). Cyclic dynamics
in a simple vertebrate predator-prey community. Sci-
ence, 302(5646):866–868.
Gillespie, D. (1976). A general method for numerically sim-
ulating the stochastic time evolution of couple chemi-
cal reactions. J. Computational Physics, 22:403–434.
Guill, C., Drossel, B., Just, W., and Carmack, E. (2011). A
three-species model explaining cyclic dominance of
pacific salmon. J. Theor. Biol., 276(1):16–21.
Jackson, J. and Buss, L. (1975). Alleopathy and spatial
competition among coral reef invertebrates. PNAS,
72(12):5160–5163.
Kabir, K. and Tanimoto, J. (2021). The role of pairwise non-
linear evolutionary dynamics in the RPS game with
noise. App. Math. & Comp., 394(125767).
Kass, S. and Bryla, K. (1998). Rock Paper Scissors Spock
Lizard. samkass.com/theories/RPSSL.html.
Kerr, B., Riley, M., Feldman, M., and Bohannan, B. (2002).
Local dispersal promotes biodiversity in a real-life
game of RPS. Nature, 418(6894):171–174.
Kubyana, M., Landi, P., and Hui, C. (2024). Adaptive
RPS game enhances eco-evolutionary performance at
cost of dynamic stability. App. Math. & Comp.,
468(128535).
Laird, R. and Schamp, B. (2006). Competitive intransitivity
promotes species coexistence. American Naturalist,
168:182–193.
Laird, R. and Schamp, B. (2008). Does local competition
increase the coexistence of species in intransitive net-
works. Ecology, 89:237–247.
Lankau, R. and Strauss, S. (2007). Mutual feedbacks main-
tain both genetic and species diversity in a plant com-
munity. Science, 317(5844):1561–1563.
Lloyd, H. and Amos, M. (2017). Analysis of independent
roulette selection in parallel ant colony optimization.
In Proc. GECCO 2017, page 19–26. ACM Press.
May, R. and Leonard, W. (1975). Nonlinear aspects of com-
petition between species. J. App. Math., 29:243–253.
Menezes, J., Batista, S., and Rangel, E. (2022a). Spatial
organisation plasticity reduces disease infection risk
in RPS models. Biosystems, 221(104777).
Menezes, J., Rangel, E., and Moura, B. (2022b). Aggre-
gation as an antipredator strategy in the RPS model.
Ecological Informatics, 69(101606).
Menezes, J., Rodrigues, S., and Batista, S. (2023). Mobility
unevenness in rock-paper-scissors models. Ecological
Complexity, 52(101028).
Mood, M. and Park, J. (2021). The interplay of rock-
paper-scissors competition and environments medi-
ates species coexistence and intriguing dynamics.
Chaos, Solitons and Fractals, 153(111579).
Nag Chowdhury, S., Banerjee, J., Perc, M., and Ghosh, D.
(2023). Eco-evolutionary cyclic dominance among
predators, prey, and parasites. J. Theor. Biol.,
564:111446.
Nagatani, T., Ichinose, G., and Tainaka, K. (2018).
Metapopulation model for RPS game: Mutation af-
fects paradoxical impacts. J. Theor. Biol., 450(22–29).
Nahum, J., Harding, B., and Kerr, B. (2011). Evolution of
restraint in a structured rock–paper–scissors commu-
nity. PNAS, 108(2):10831–10838.
Park, J. (2021). Evolutionary dynamics in the RPS sys-
tem by changing community paradigm with popula-
tion flow. Chaos, Solitons and Fractals, 142(110424).
Park, J. and Jang, B. (2023). Role of adaptive intraspecific
competition on collective behavior in the RPS game.
Chaos, Solitons and Fractals, 171(113448).
Reichenbach, T., Mobilia, M., and Frey, E. (2007a). Mo-
bility promotes and jeopardizes biodiversity in rock-
paper-scissors games. Nature, 448(06095).
Reichenbach, T., Mobilia, M., and Frey, E. (2007b). Noise
and Correlations in a Spatial Population Model with
Cyclic Competition. Phys. Rev. Lett., 99(238105).
Reichenbach, T., Mobilia, M., and Frey, E. (2008). Self-
Organization of Mobile Populations in Cyclic Com-
petition. J. Theor. Biol., 254(2008):363–383.
Sinervo, B. and Lively, C. (1996). The rock–paper–scissors
game and the evolution of alternative male strategies.
Nature, 380(6571):240–243.
Zhang, Z., Bearup, D., Guo, G., Zhang, H., and Liao, J.
(2022). Competition modes determine ecosystem sta-
bility in RPS games. Physica A, 607(128176).
Zhong, L., Zhang, L., Li, H., Dai, Q., and Yang, J. (2022).
Species coexistence in spatial cyclic game of five
species. Chaos, Solitons, and Fractals, 156(111806).
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
554
