
Separation of Insect Trajectories in Dynamic Vision Sensor Data
Juliane Arning, Christoph Dalitz
a
and Regina Pohle-Fr
¨
ohlich
b
Institute for Pattern Recognition, Niederrhein University of Applied Sciences, Reinarzstr. 49, Krefeld, Germany
Keywords:
Event Camera, Clustering, Insect Monitoring.
Abstract:
We present a method to separate insect flight trajectories in dynamic vision sensor data and for their math-
ematical description by smooth curves. The method consists of four steps: Pre-processing, clustering, post-
processing, and curve fitting. As the time and space coordinates use different scales, we have rescaled the
dimensions with data-based scale factors. For clustering, we have compared DBSCAN and MST-based clus-
tering, and both suffered from undersegmentation. A suitable post-processing was introduced to fix this. Curve
fitting was done with a non-parametric LOWESS smoother. The runtime of our method is sufficiently fast to
be applied in real-time insect monitoring. The data used for evaluation only had two spatial dimensions, but
the method can be applied to data with three spatial dimensions, too.
1 INTRODUCTION
The “Krefeld Study” from 1989 to 2016 (Hallmann
et al., 2017) has shown a decline in flying in-
sect biomass of more than 75 percent, which has
raised considerable interest in insect monitoring. The
Krefeld Study utilised Malaise traps. These kill the
trapped insects, which are subsequently manually
classified and weighted in a tedious process, and it
would be desirable to have another method available
that is nonlethal and less time consuming.
The BeeVision project (Pohle-Fr
¨
ohlich and
Bolten, 2023) addresses this problem and explores
the use of Dynamic Vision Sensors (DVS) for
monitoring flying insects. These sensors detect local
variations in brightness and trigger events only for
brightness variations greater than some threshold.
Position, time, and the sign of the brightness change
are recorded for each event. Unlike traditional frame
based cameras, DVS operate almost continuously
and do not yield frames, but point clouds. This has
the advantage of a much higher time resolution and
a smaller data size, but it requires special algorithms
for identifying and separating insect tracks in point
clouds.
Insect tracks occur in DVS point clouds as piece-
wise continuous dense strips. These can be inter-
rupted by gaps due to occlusion by other objects.
Moreover, rest periods on, e.g., flowers lead to in-
a
https://orcid.org/0000-0002-7004-5584
b
https://orcid.org/0000-0002-4655-6851
terruptions, too, because dynamic vision sensors only
see moving objects. Starts and landings on plants can
also lead to sharp turns in the flight path, so that the
flight trajectories are not necessarily smooth.
The problem thus consists in partitioning a point
cloud into an unknown number of clusters repre-
senting insect tracks and another cluster represent-
ing noise. This is similar to the problem of parti-
cle track identification in Time Projection Chambers
(Dalitz et al., 2019a). Although there are algorithms
like CLUE (Rovere et al., 2020) or TriplClust (Dalitz
et al., 2019b) that have been devised for this partic-
ular use case, we have not been successful in apply-
ing them to our data. CLUE requires an energy for
each event, which was lacking in our data and there-
fore had to be replaced by some dummy value, and
we found no parameter settings for the implementa-
tion CLUEstering
1
that worked in our use case. The
use of TriplClust was not feasible due to its high
time and space complexity: For instance, a noise
free DVS recording of approximately 160 seconds re-
quired about 126 GB of memory in TriplClust.
We therefore resorted to density based clustering
methods like DBSCAN (Ester et al., 1996) and MST
splitting (Zahn, 1971). These showed deficiencies in
our use case, too, especially in challenging scenar-
ios, such as when insect tracks cross each other. In
the present report, we discuss how these shortcomings
can be overcome, with special attention on improving
the runtime in order to enable processing in real-time.
1
https://github.com/cms-patatrack/CLUEstering
Arning, J., Dalitz, C. and Pohle-Fröhlich, R.
Separation of Insect Trajectories in Dynamic Vision Sensor Data.
DOI: 10.5220/0013140500003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
69-77
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
69
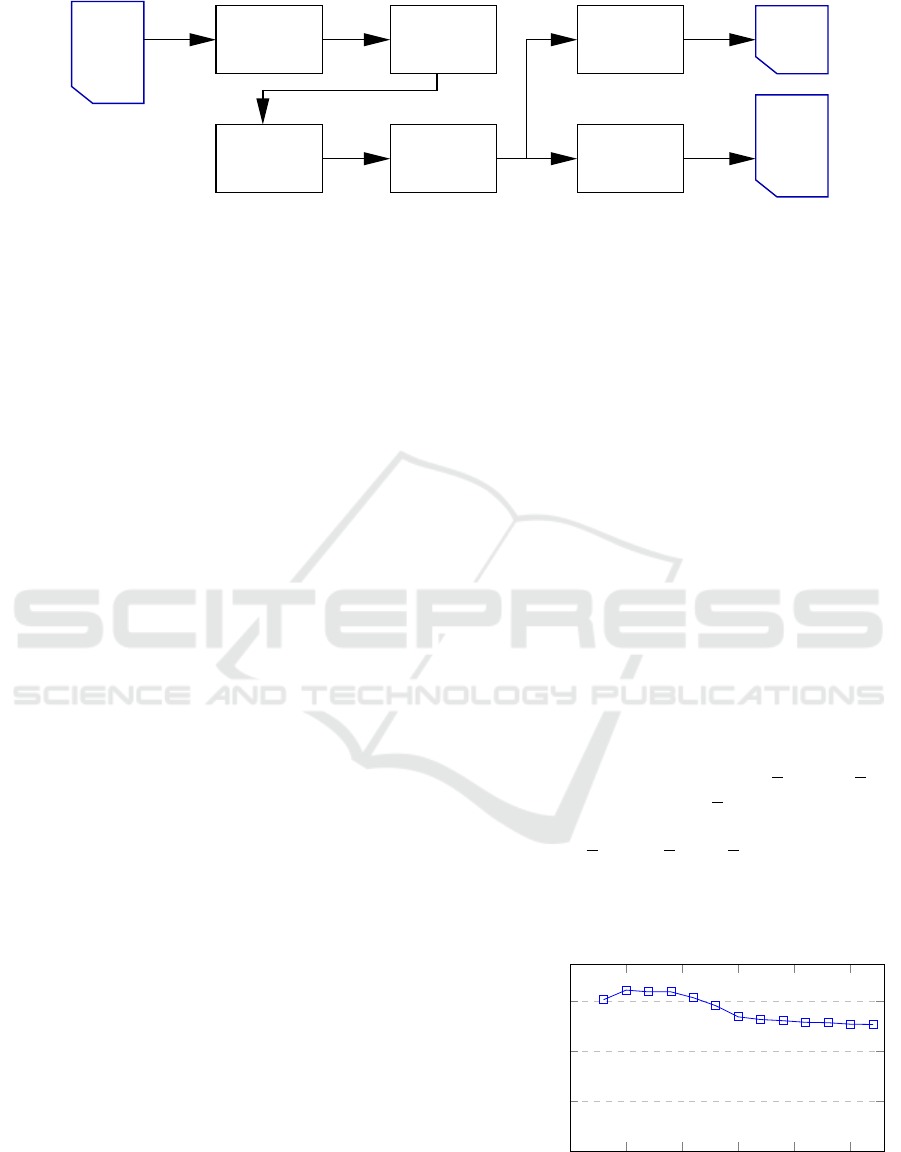
Scaling
Clustering
or MST)
(DBSCAN
Subsampling
LOWESS
Remapping
unla−
belled
points
paths
flight
label−
led
points
Fix merged
clusters
Figure 1: Processing pipeline of the segmentation method.
To this end, we utilised octree subsampling and time
rescaling, modifications to the clustering algorithms,
and a special post-processing for dealing with under-
segmentation that is inherent to the clustering algo-
rithms. After mapping the subsampled clusters back
to the original data, the flight trajectories can be inter-
polated with a LOWESS (Cleveland, 1979) multivari-
ate regression based on the time as a single predictor.
All of these methods are described in section 2.
We evaluated our method on a series of DVS
recordings provided by the BeeVision project, where
insect events had already been segmented from other
events with a lightweight U-Net (Pohle-Fr
¨
ohlich
et al., 2024). This enabled us to evaluate the algo-
rithm both with and without the presence of back-
ground noise stemming from shaking leaves, varying
illumination due to clouds, or other random effects.
The evaluation is described in section 3.
2 SEGMENTATION METHOD
The segmentation algorithm takes an unlabelled point
cloud with time and space coordinates as input and
returns two output lists: For each point, a cluster label
or a noise label is computed, and for every non-noise
cluster a fitted flight path is computed and sampled
at equidistant timestamps. The processing pipeline
of the algorithm is shown in Figure 1. It consists of
the following steps: Pre-processing, clustering, post-
processing, fitting of flight paths using the LOWESS
method, and remapping the labels to the original point
cloud. Additionally, after reading of the input CSV-
file, the timestamp is scaled by a scaling factor that
was determined for representative data.
During pre-processing, the data is subsampled and
outliers are removed. This smaller point cloud is then
clustered. The clustering has a tendency to underseg-
ment nearby insect tracks. These tracks are therefore
corrected in the post-processing step. Afterwards,
curves representing flight paths are fitted and the spa-
tial curve position is computed for equidistant times-
tamps. Since the clustering was done on only the
subsampled and filtered data, labels must be prop-
agated to the points that were removed during pre-
processing. To this end, every unlabelled point re-
ceives the label of the nearest labelled point. The in-
dividual steps are described in detail in the following
subsections.
2.1 Pre-Processing
The timestamp dimension t is of a different unit (mi-
croseconds) and thus differs several orders of mag-
nitude from the spatial dimensions (pixel). With the
original timescale of microseconds, the distances in
the temporal dimension thus dominate the distances
of the spatial dimensions, which has a serious nega-
tive impact on the performance of DBSCAN and the
MST-based clustering methods.
We therefore scale the time with a data based scal-
ing factor. As the DVS recording scenario is the same
for all recorded data, it was sufficient to estimate an
appropriate scaling factor only once from a represen-
tative subset of the data. We have chosen the scaling
factor s such that the mean distances to the k-th near-
est neighbour in the spatial direction d
x
(k) and d
y
(k)
is equal to the mean distance d
t
(k) in the t direction,
i.e.
s · d
t
(k) =
d
x
(k) + d
y
(k)
/2 (1)
It is interesting to note that the scale factor s was quite
robust with respect to the choice of k, as can be seen
0
5
10
15
20
25
0
2
4
6
·10
−4
k
scale factor s
Figure 2: Dependency of the scaling factor s according to
Eq. (1) on the number of neighbours k.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
70
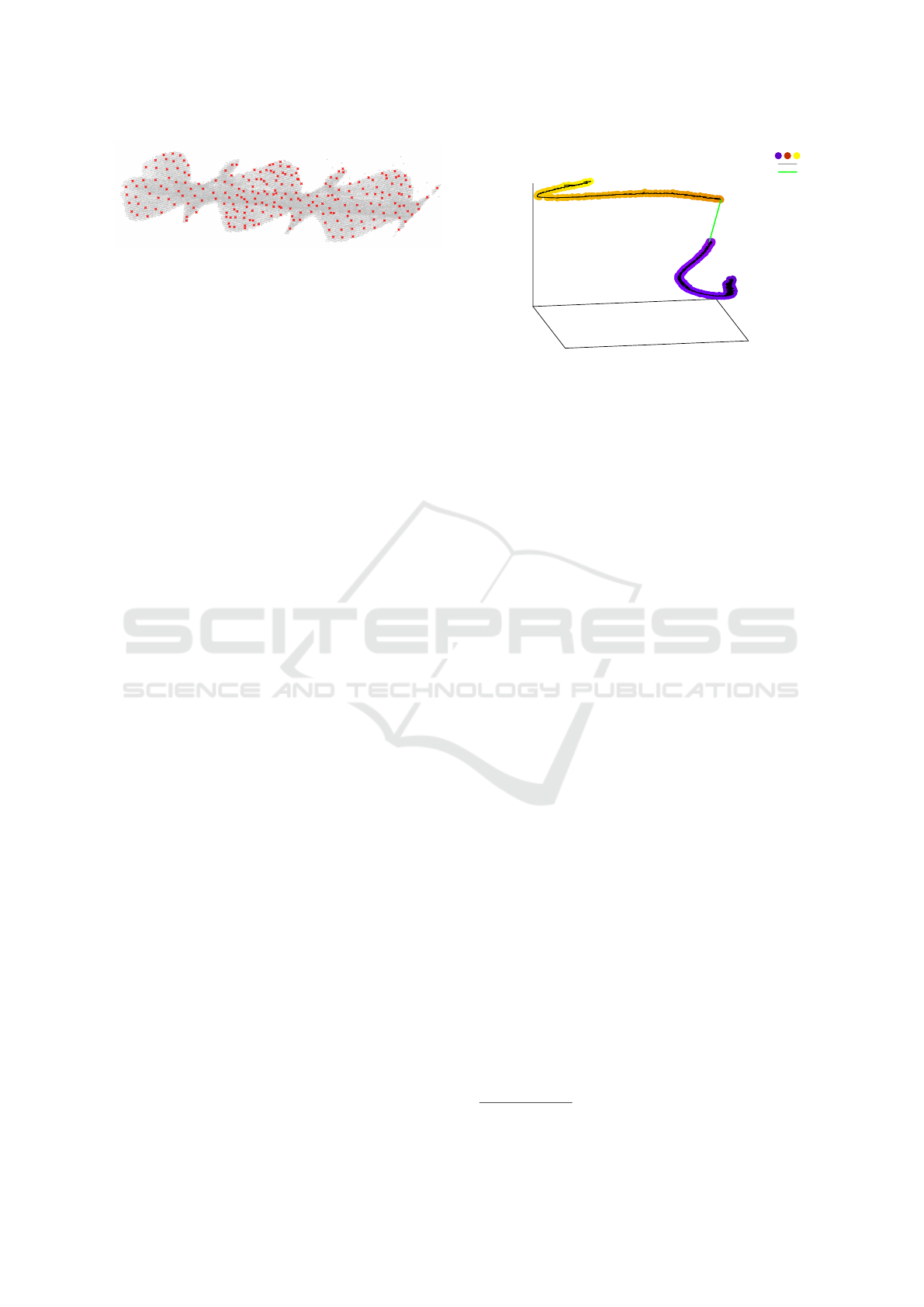
Figure 3: Subsampling: grey points are the original point
cloud and the red crosses are the subsampled points.
in Figure 2. Moreover, we have observed that either
choice in this range resulted in a similar performance
of the clustering algorithm, and we have settled on
s = 5.3 · 10
−4
µs
−1
.
After scaling the timestamps, an octree-based sub-
sampling is applied in order to reduce both data
volume and noise. Octree subsampling works by
constructing a tree structure that recursively divides
the 3D space into eight evenly-sized cubes, stop-
ping at a predefined depth (Meagher, 1982). This
process leaves the non-empty space divided into
cubes, and for each cube a representative point is se-
lected. These representative points form the subsam-
pled point cloud. To make the filtering effect inde-
pendent from the size of the point cloud, we chose the
side length of the cube at the lowest level of the octree
as the stopping criterion instead of the depth. A larger
value causes more filtering and a smaller value less fil-
tering. In our use case, the full octree data structure
was not needed, because no nearest-neighbour queries
are done on the full point cloud, only on the subsam-
pled data. We therefore divide the space directly into
cubes.
There are different options to select the represen-
tative point, e.g., the point closest to the centroid of
the cube, or the point closest to the centroid of the
points in the cube. This, however, has only little ef-
fect on the results. A result of the subsampling step
is shown in Figure 3. The octree subsampling has the
positive side-effect that it can also be used for remov-
ing noise in sparse regions simply by skipping cubes
with fewer points than a defined threshold. This sim-
ple noise reduction step does not increase the run-
time; it only requires some additional space since the
skipped points have to be memorised in order to ob-
tain the noise label in the remapping step.
2.2 Clustering
As neither the number of insect tracks, nor their shape
is known, we cannot use clustering algorithms like
k-means that look for spherical clusters and a given
number of clusters. A characteristic of insect flight
trajectories is that they typically are represented by re-
gions with a higher point density and are separated by
less dense regions. It is thus natural to use a clustering
points
edges
inconsistentedges
x
y
t
Figure 4: The MST of two clusters with an “inconsistent
edge” highlighted in green and the points coloured by clus-
ter.
method based on closeness of points and thus a local
density. We have implemented both DBSCAN and
MST-based clustering as alternative options. Both are
clustering algorithms that can detect clusters of arbi-
trary shape and do not require the number of clusters
beforehand.
MST-based clustering algorithms build a weighted
graph with weights corresponding to the Euclidean
distance between node points, and remove “inconsis-
tent edges” from the Minimum Spanning Tree (MST)
of this graph (Zahn, 1971). Edges are considered “in-
consistent” if they have a weight that is above some
global or local distance threshold. This splits the MST
into connected components which represent the clus-
ters. The MST-based clustering method is illustrated
in Figure 4 with the MST of two insect tracks. The
inconsistent edge highlighted in green is removed,
which results in two connected components repre-
senting insect tracks. These can be easily identified
using graph traversal techniques such as breadth-first
or depth-first search.
In general, building an MST for N nodes has run-
time complexity O(N
2
), but as our distance is Eu-
clidean, it is not necessary to build the complete
distance matrix and the runtime can be reduced to
O(N log N) (March et al., 2010). Moreover, we could
rely on a fast Open Source C++ implementation by
Andrii Borziak
2
.
Due to a considerable spread in point density be-
tween different tracks, local thresholds for identifying
inconsistent edges resulted in undersegmentation. We
therefore used a global threshold for removing incon-
sistent edges, which was chosen in dependence of the
length d of the pre-processing octree cube diagonal as
5 · d. However this threshold is chosen, there are two
unavoidable problems: Splits due to gaps in partially
occluded tracks, and merges of tracks coming close to
2
https://github.com/AndrewB330/EuclideanMST
Separation of Insect Trajectories in Dynamic Vision Sensor Data
71
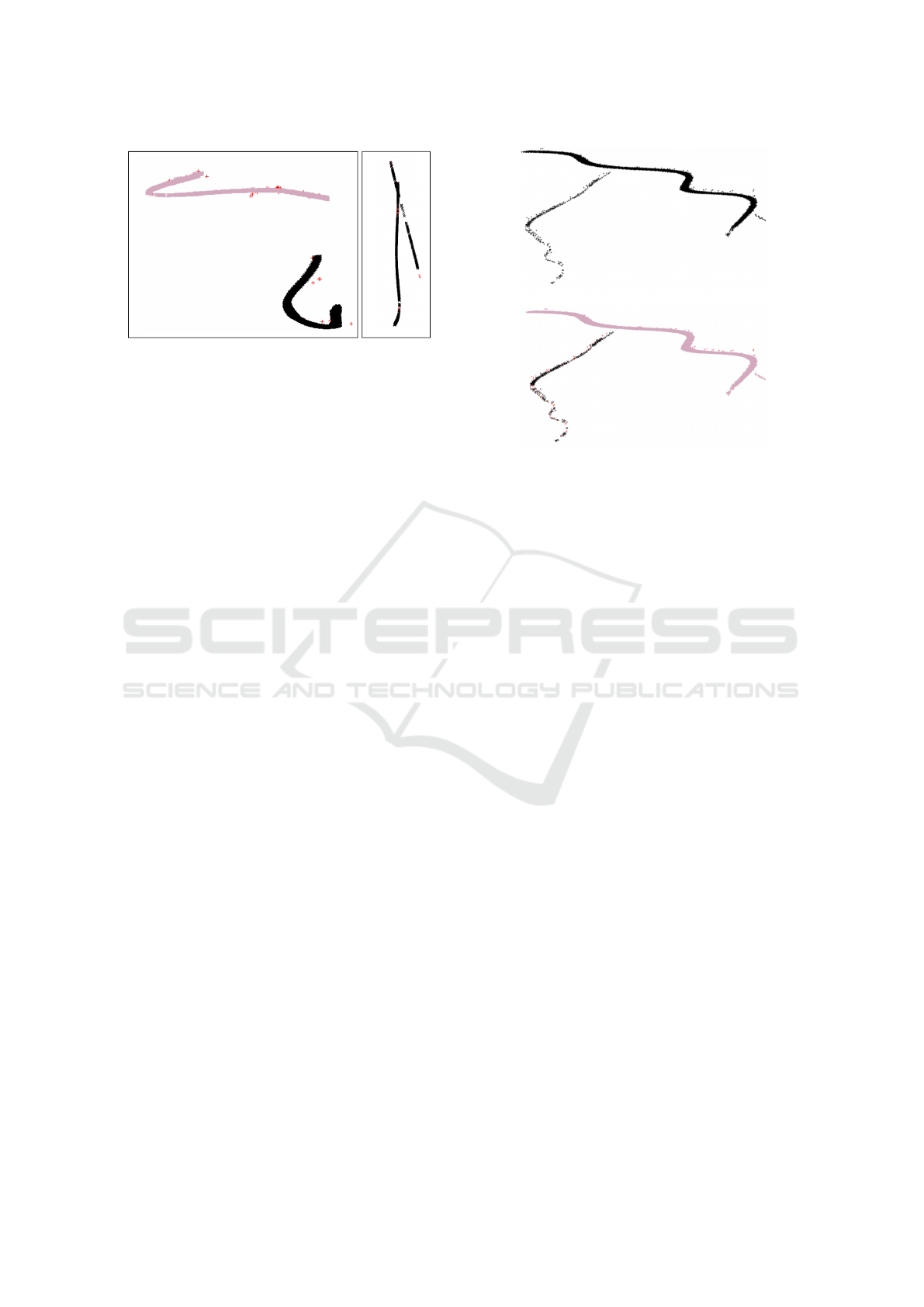
Figure 5: MST-based segmentation works in the example
on the left, and fails in the example on the right. The same
occurs for DBSCAN.
each other. An example can be seen in Figure 5.
The MST-based clustering also offers a simple
method identify falsely recognised clusters in dense
regions that actually represent noise. When the edges
are traversed in the MST, the mean edge weight can
be calculated. Clusters with a small mean edge weight
can be removed as they often consist of noise. This
method adds more robustness against very strong
noise, which is common in DVS-recordings and can-
not be removed by the filtering in the subsampling
step.
DBSCAN (Ester et al., 1996) is a density-based
clustering algorithm, that defines clusters as regions
of high density that are separated by regions of lower
density. DBSCAN has two parameters, eps and
minPts. Every point with at least minPts neighbours
within a radius of eps is a core point. Whenever an
unlabelled core point is found, a new cluster is ini-
tialised and the core point is added to it. The new
cluster is iteratively propagated to core points within
radius eps. This process is iterated until all core points
and their neighbours are assigned to clusters. Points
that are not neighbours of a core point are marked as
outliers.
Schubert et al. (Schubert et al., 2017) suggested
simple heuristics for choosing the parameters. For
minPts, they suggested minPts = 2· #Dimensions = 6,
which we have adopted. We have not utilised their
suggestion to analyse a ’k-dist plot’ to determine an
appropriate eps, because the octree cell size of our
subsampling process already limits the possible max-
imum point density. We therefore set eps equal to a
multiple of the diagonal of the cube, which is the fur-
thest distance between two points in a dense insect
track should be separated. After experimentally test-
ing different values for the multiplication factor, we
settled on eps = 6 · d, where d is the diagonal length
of a cell in the pre-processing octree.
Like MST-based clustering, DBSCAN cannot
separate tracks when insect tracks come close to each
(a) After clustering.
(b) After post-processing.
Figure 6: Example for two merged tracks that are correctly
split up during post-processing.
other (see Figure 5), because both algorithms are
purely based on point distances. To correct this prob-
lem, a post-processing step is necessary.
2.3 Post-Processing
To separate touching insect tracks that were erro-
neously merged into the same cluster, we imple-
mented a post-processing that consists of three steps.
First, possibly merged clusters are identified and, sec-
ondly, these clusters are split up at branching points
in their Minimum Spanning Tree (MST). As the split-
ting leads to oversegmentation, that is corrected in the
third step by merging the most appropriate branches
based on their direction and time alignment. An ex-
ample can be seen in Figure 6.
2.3.1 Identifying Merged Clusters
To minimise runtime and prevent over-segmentation,
we first detect merged clusters using a simple heuris-
tic. This approach is based on the premise that when
insect tracks intersect, there should be simultaneous
activity in different spatial regions at the same time.
As an indicator for this phenomenon, we compute the
spatial Euclidean distance between consecutive time
points for each cluster, which results in an array of
n − 1-values which we call ∆s. The computation is
facilitated by the nature of the DVS stream, which
records events according to their time order, so that
consecutive points occur at similar times.
If the standard deviation of the array sd(∆s) ex-
ceeds a threshold θ
sd
, the cluster is flagged for further
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
72
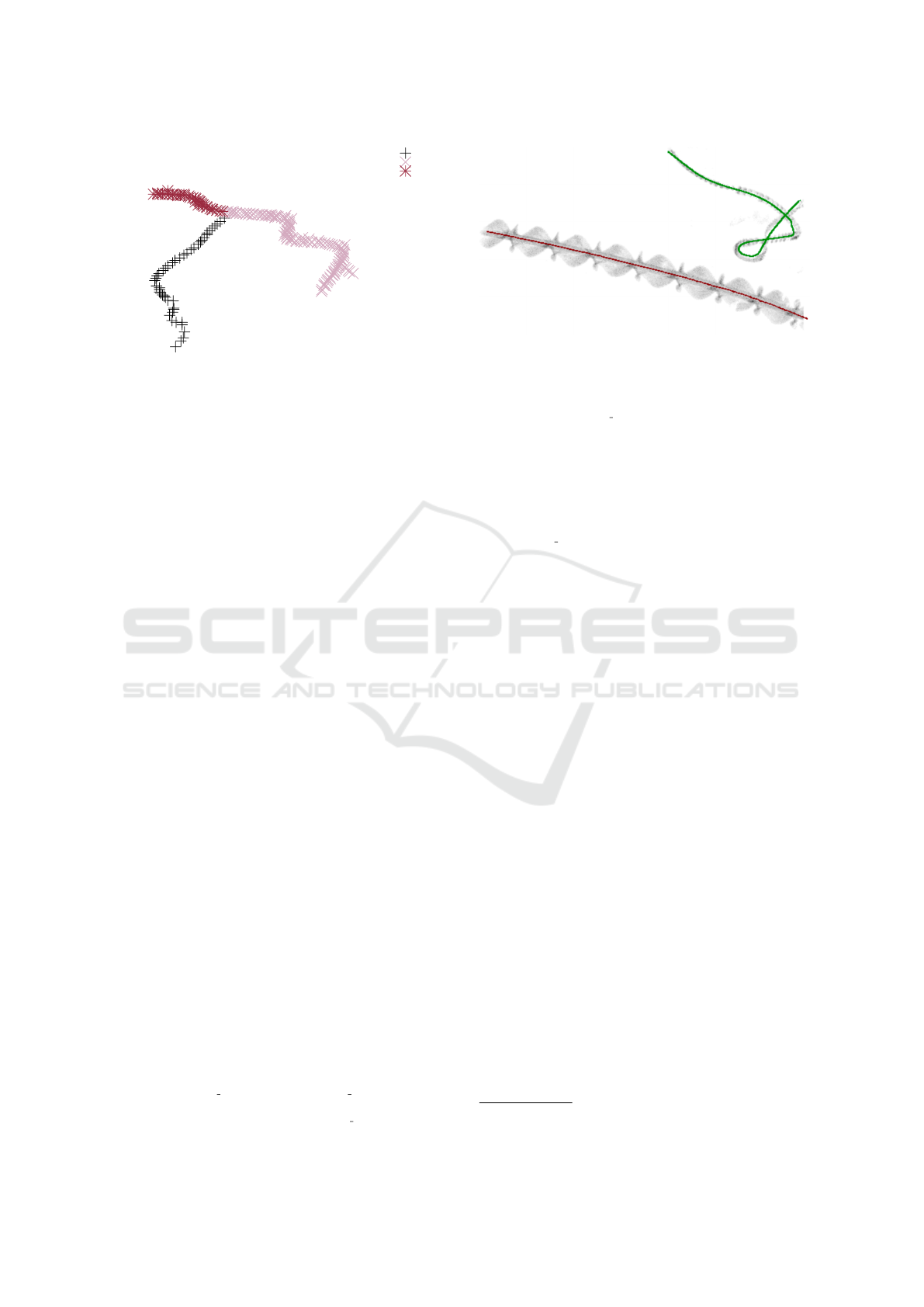
curve0
curve1
curve2
Figure 7: Example for oversegmentation due to splitting at
crossing points.
post-processing steps. In the longest recording and
for the threshold value θ
sd
= 22.36 that worked best
in our case, 16 out of 598 clusters met this criterion,
including all 12 merged tracks and 4 incorrect classi-
fications.
2.3.2 Splitting Clusters
For each flagged cluster, an MST is built and nodes
with three or more branches of sufficient depth are
identified as split points. The depth threshold is cho-
sen proportional (α · n
i
) in order to adapt it to varying
cluster sizes.
At each split point, all edges except the short-
est one are deleted. This splits the cluster into con-
nected components, which are identified and num-
bered. These preliminary clusters are referred to as
segments. These must be further processed, though,
because the splitting method can result in overseg-
mentation at X-crossing points where the straight con-
tinuation cannot be preserved (see Figure 7).
2.3.3 Merging Over-Segmented Components
To correct over-segmentation due to MST splitting,
segments are merged again, if they are continuations
of each other in the direction extrapolated from the
end of the earlier segment. For every segment S
i
that occurred during splitting the clusters at branches,
another fitting segment S
j
is sought as continuation.
Both the time order of both segments is considered,
as well as the direction of the segments around the
ends with the closest timestamps.
Only segments S
j
with timestamps greater than S
i
,
within a given tolerance and with a sufficiently small
gap, are considered. The time difference between the
last point of S
i
(t
e
) and the first point of S
j
(t
s
) must
satisfy:
−0.5 · θ
t
tolerance
< t
s
− t
e
< θ
t tolerance
(2)
This allows for gaps of the length θ
t tolerance
and an
Figure 8: Curves ⃗x(t) fitted with LOWESS for two insect
tracks. The fitted curves are coloured and the DVS points
are shown in grey.
overlap of half of θ
t tolerance
. The gap length has to
be greater than the tolerance of overlap because one
insect often covers the other. This leads to large gaps
in the track. A time overlap that large, however, did
not occur in our data, which is why 0.5 was somewhat
arbitrarily chosen to reduce the allowed overlap. A
threshold that worked in our case was, in the original
time unit, θ
t tolerance
= 35ms.
For approximating the outgoing and incoming di-
rection, ⃗v
in
and ⃗v
out
of the tracks, the first PCA com-
ponent of the beginning and end of every segment is
calculated. The size of the beginning and end is de-
termined by a proportion of the size of the segment.
The similarity of the directions is measured with
the cosine similarity, and a segment S
j
is only consid-
ered as a continuation of segment S
i
, if
cos(⃗v
in
j
,⃗v
out
i
) > θ
mincos
= 0.7 (3)
Moreover, there is the constraint that each segment
cannot be used more than once as a continuation of
another segment. If more than one segment fulfils all
conditions, the segment with the maximal cosine sim-
ilarity cos(⃗v
in
j
,⃗v
out
i
) is chosen as the continuation.
2.4 Curve Fitting
For visualisation or for an analysis of insect flight
patterns, it is useful to represent each track as a spa-
tial curve ⃗x(t) that yields a position for every time t.
As each point in our point cloud already has a time
stamp, this time coordinate can be used as a predic-
tor in a local regression model with the space coor-
dinates as a multivariate response variable. An es-
tablished method for local regression is Cleveland’s
Locally Weighted Estimated Scatterplot Smoothing
(LOWESS) (Cleveland, 1979). We used the Open
Source C++ implementation CppWeightedLowess
3
by
Aaron Lun.
3
https://github.com/LTLA/CppWeightedLowess
Separation of Insect Trajectories in Dynamic Vision Sensor Data
73

Figure 9: Example result showing the clustering of the points into noise (red) and different insect tracks (other colours).
For each time value t, LOWESS fits a second or-
der polynomial locally through the r neighbouring
points in the predictor space, which is the time in our
case. Apart from only using the nearest neighbours,
the points are also inversely weighted according to
their distance in the predictor space. This restricts the
regression further to the local shape. From a set of
sample points, LOWESS predicts a response ⃗x(t) for
arbitrary predictor values t within the sample range,
but the resulting curve is non-parametric. We there-
fore compute the predicted x and y coordinates for
equally spaced timestamps and store both together as
the result of the curve fitting.
The result of LOWESS depends on the number
of neighbours that are used for local fitting, which
is determined by a parameter f , also known as span.
A smaller value for f produces a less smooth curve,
whereas a larger value result in a smoother curve. f
can be given as a proportion of the entire set, e.g.
10%, or as a fixed number of neighbours. A curve
tangential to the flight direction is only obtained, if
the neighbourhood extends much more in the flight
direction than perpendicular to it. Unfortunately, this
means that a fixed number of neighbours does not
work for fitting insect tracks in general, because in-
sects vary considerable in size and wing beat rate,
which means that the number of DVS events per time
interval can vary considerably. Therefore, we chose
0.2 as value for f , i.e. 20% of the points per cluster
were used to fit each value. As can be seen in Figure
8. This may result in overly smoothed curves, how-
ever, in cases of very long clusters.
3 RESULTS
We have evaluated our algorithm on eight recordings
of insect flights on a meadow that were captured in the
context of the BeeVision project (Pohle-Fr
¨
ohlich and
Bolten, 2023). The data was recorded with a Proph-
esee EVK3 Gen4.1 event camera (1280 × 720 resolu-
tion), where time was recorded in microseconds. The
length varied between 16 and 160 seconds, compris-
ing 308MB and about 10
7
events. The data points had
already been segmented with a U-Net (Pohle-Fr
¨
ohlich
et al., 2024), and the time was scaled to milliseconds.
To obtain ground truth data, the files were man-
ually labelled, assigning each insect track a unique
label and an additional label for points considered to
be noise. This was done with the Semantic Segmenta-
tion Editor
4
that allows for 3D point cloud labelling.
To assess the ground truth labelling, a sample of the
test data was labelled manually again a few months
apart from the first pass. This provided a reference
value for the accuracy of the labelling.
To evaluate the post-processing, 19 merges of sev-
eral tracks into one cluster were identified in the
dataset and isolated with some surrounding clusters.
To assess the robustness of our algorithm with respect
to noise, both the original dataset with noise and the
dataset without noise were used.
3.1 Evaluation Criteria
There are two categories of evaluation criteria for the
evaluation of clusterings, internal and external in-
dices (Hassan et al., 2024). Internal indices, like the
Silhouette Index and Calinski-Harabasz Index, assess
clusters based on properties such as intra- and inter-
cluster variance, but they assume clusters are spheri-
cal and are thus not suitable for insect tracks.
External metrics compare the clustering results
with ground truth data. These metrics evaluate clus-
terings by counting whether pairs of points fall in the
4
https://github.com/Hitachi-Automotive-And-Industr
y-Lab/semantic-segmentation-editor
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
74

Figure 10: Example for the result of noise removal: red points are noise, the time is on the x-axis and the y-axis is the y-
coordinate.
same or different clusters in the respective cluster-
ings. From these counts, the (adjusted) Rand Index
and the Jaccard Score are computed (Hubert and Ara-
bie, 1985). The Rand Index measures overall similar-
ity, ranging from 0 (no matching pairs) to 1 (identical
clusterings), while the adjusted Rand Index corrects
for chance. The Jaccard Score is similar but only
focuses on pairs that are in the same cluster in both
ground truth and predicted clustering. This has the
effect that the Jaccard Score is always smaller than
the Rand Index.
Since these metrics are pair-based, larger clusters
(or tracks) with more point pairs will have a greater
impact on the evaluation results. Moreover, although
their range is [0,1], it is not clear which values actu-
ally represent “good” results. We therefore compared
the two manual ground truth clusterings of the same
data set and obtained a Jaccard-Score of about 0.86
and an adjusted Rand-Score of 0.92. This means that
results around these values can be considered as good
results for our algorithm.
3.2 Clustering
As can be seen from the results in Table 1, in the ab-
sence of noise, the MST-based clustering and DB-
SCAN both have an excellent performance that is
comparable to a human. In the presence of noise,
however, DBSCAN performs considerably poorer.
This is because the MST-based clustering removes
more less dense clusters than DBSCAN. An exam-
ple of its ability to remove noise is seen in Figure 10,
where the insect track is mostly separated from the
noise with only some small artefacts.
According to Table 1, it seems that the post-
processing has almost no effect on the quality of
the segmentation. This is misleading, however, be-
cause the pairwise quality indices are dominated by
the large clusters and not very sensitive to differences
in small clusters. Therefore, we did another test with
only those clusters that were erroneously merged by
Table 1: Jaccard score J and adjusted Rand index R
ad j
for
the two clustering algorithms with and without noise or with
and without post-processing.
post- MST DBSCAN
noise proc. R
ad j
J R
ad j
J
no no 0.921 0.857 0.919 0.854
no yes 0.922 0.858 0.922 0.858
yes no 0.759 0.656 0.598 0.470
yes yes 0.765 0.662 0.589 0.467
the clustering algorithms. Out of 19 merged cluster,
the post-processing split 11 correctly, and 3 were cor-
rectly split, but not perfectly rejoined. The same test
for non-merged clusters yielded no incorrect splits.
Noise removal worked best with uniformly dis-
tributed noise but had problems when noise is con-
centrated in specific areas, e.g., due to ambient ef-
fects like wind. For the full original noisy record-
ing with points removed only when are the only point
in an octree cube or when they end up in a cluster
of only one point, 70.8% of all noise points are cor-
rectly removed, while 1.3% of insect points were in-
correctly removed. This criterion is thus too strict for
raw noisy data. Weaker thresholds are required for
noisy datasets, although more aggressive filtering dis-
proportionally affects small insect tracks. More filter-
ing will also remove points at the edges of the insect
tracks, which could impact the insect classification
downstream. With simulated uniformly distributed
noise consisting of about 197% of the original point
cloud size, 99.98% of noise points are removed, while
only 5.78% of insect points are affected.
3.3 Runtime
When the method is utilised for actual real-life in-
sect monitoring, it is crucial that the processing oc-
curs in real time, i.e., the runtime must be less than
the recording time of the DVS sensor plus other pre-
processing time possibly needed, e.g. for computing
3D spatial information from stereo recordings (Pohle-
Fr
¨
ohlich et al., 2024). Figure 11 shows how the aver-
Separation of Insect Trajectories in Dynamic Vision Sensor Data
75
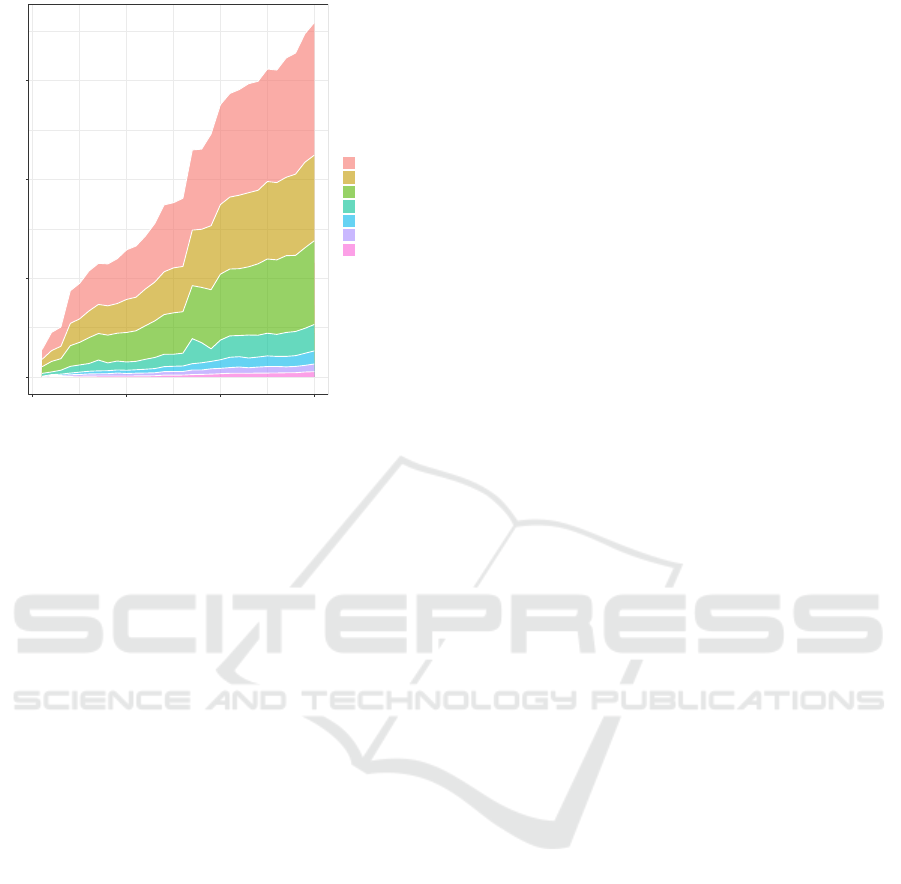
0
2
4
6
0 100 200 300
Duration of the DVS−recording [s]
Runtime [s]
group
Writing
Reading
Pre−processing
Remapping
Clustering
LOWESS
Post−processing
Figure 11: The runtime plotted against the length of the
DVS-recording and separated into the individual steps of
the processing pipeline.
age runtime varies with the DVS-recording duration
and how it is distributed among the different process-
ing steps. The runtime was measured on an AMD
Ryzen 7 5700U CPU running Ubuntu 20.04 and aver-
aged over 100 runs. As can be seen from Figure 11,
the processing runtime is always considerably shorter
than the duration of the recording.
We also measured the speedup S due to pre-
processing, defined as the runtime without subsam-
pling divided by the runtime with subsampling. The
speedup for noise filtering was greater in unnoisy data
(S ≈ 20) than in noisy data (S ≈ 4). This is because
subsampling reduces the number of noisy points less
effectively, as many noise points occupy their own
cubes. The speedup for noisy data increases with
noise removal during subsampling. This in partic-
ular affected the MST-based clustering which had a
speedup of 30. This means that subsampling is essen-
tial for an application in real time.
4 CONCLUSIONS
We have developed a method for fast instance seg-
mentation of insect flight tracks in DVS data, treating
time as another dimension to preserve high temporal
resolution. The central part of the algorithm is a den-
sity based clustering, for which either DBSCAN or
MST-based clustering can be chosen. The MST-based
clustering was considerably more robust with respect
to noise and is thus preferable. Both algorithms, how-
ever, failed to separate close by tracks, and we have
implemented a post-processing step that remedies this
shortcoming in most situations.
Due to subsampling during pre-processing, the
method has a runtime much shorter than the DVS
recording duration and is thus applicable in real time.
Noise removal was optionally included in the subsam-
pling step and in the clustering step. These automatic
noise detection makes the method quite robust in the
presence of noise, which is important for its deploy-
ment in natural scenarios.
Although the method has an accuracy comparable
to a manual segmentation by a human, it occasionally
removes thin tracks. This makes the method currently
less effective for small insects like mosquitos. For
visualisation or further analysis, we also fit flight tra-
jectories through the returned clusters. Although our
usage of LOWESS was satisfactory, a local regres-
sion based on the number of neighbours can become
problematic for scenarios with a wide range of track
thicknesses and lengths. It would be interesting to in-
vestigate different local regression methods, e.g. by
basing the local region on fixed time intervals.
Although we tested our method only with DVS
data with two spatial dimensions, there is nothing spe-
cial in our algorithm that restricts it to 2D data. The
method can thus readily be deployed to 3D data like
that recorded by the new system developed in the Bee-
Vision project (Pohle-Fr
¨
ohlich et al., 2024), which
aims at counting insect populations over a long pe-
riod. We plan to deploy our algorithm in this project
and use it as a second step after semantic segmenta-
tion. This will then be followed by a classification
step which leads to an automatic counting of species
occurances in the data.
REFERENCES
Cleveland, W. S. (1979). Robust locally weighted regres-
sion and smoothing scatterplots. Journal of the Amer-
ican Statistical Association, 74(368):829–836.
Dalitz, C., Ayyad, Y., Wilberg, J., Aymans, L., Bazin, D.,
and Mittig, W. (2019a). Automatic trajectory recog-
nition in Active Target Time Projection Chambers
data by means of hierarchical clustering. Computer
Physics Communications, 235:159–168.
Dalitz, C., Wilberg, J., and Aymans, L. (2019b). TriplClust:
An algorithm for curve detection in 3D point clouds.
Image Processing On Line, 8:26–46.
Ester, M., Kriegel, H.-P., Sander, J., Xu, X., et al. (1996).
A density-based algorithm for discovering clusters in
large spatial databases with noise. In kdd, volume 96,
pages 226–231.
Hallmann, C. A., Sorg, M., Jongejans, E., Siepel, H.,
Hofland, N., Schwan, H., Stenmans, W., M
¨
uller,
A., Sumser, H., H
¨
orren, T., et al. (2017). More
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
76

than 75 percent decline over 27 years in total fly-
ing insect biomass in protected areas. PloS one,
12(10):e0185809.
Hassan, B. A., Tayfor, N. B., Hassan, A. A., Ahmed, A. M.,
Rashid, T. A., and Abdalla, N. N. (2024). From A-to-Z
review of clustering validation indices. Neurocomput-
ing, 601:128198.
Hubert, L. and Arabie, P. (1985). Comparing partitions.
Journal of classification, 2:193–218.
March, W. B., Ram, P., and Gray, A. G. (2010). Fast Eu-
clidean minimum spanning tree: algorithm, analysis,
and applications. In Proceedings of the 16th ACM
SIGKDD international conference on Knowledge dis-
covery and data mining, pages 603–612.
Meagher, D. (1982). Geometric modeling using octree en-
coding. Computer graphics and image processing,
19(2):129–147.
Pohle-Fr
¨
ohlich, R. and Bolten, T. (2023). Concept study for
dynamic vision sensor based insect monitoring. In In-
ternational Conference for Computer Vision and Ap-
plications (VISAPP), pages 411–418.
Pohle-Fr
¨
ohlich, R., Gebler, C., and Bolten, T. (2024).
Stereo-event-camera-technique for insect monitoring.
In International Conference for Computer Vision and
Applications (VISAPP), pages 375–384.
Rovere, M., Chen, Z., Di Pilato, A., Pantaleo, F., and Seez,
C. (2020). CLUE: A fast parallel clustering algo-
rithm for high granularity calorimeters in high-energy
physics. Frontiers in Big Data, 3:591315.
Schubert, E., Sander, J., Ester, M., Kriegel, H. P., and Xu, X.
(2017). DBSCAN revisited, revisited: why and how
you should (still) use DBSCAN. ACM Transactions
on Database Systems (TODS), 42(3):1–21.
Zahn, C. T. (1971). Graph-theoretical methods for detecting
and describing gestalt clusters. IEEE Transactions on
Computers, 100(1):68–86.
Separation of Insect Trajectories in Dynamic Vision Sensor Data
77
