
Generative Adversarial Network for Image Reconstruction from
Human Brain Activity
Tim Tanner
1a
and Vassilis Cutsuridis
1,2 b
1
School of Computer Science, University of Lincoln, Lincoln, U.K.
2
School of Engineering, Computing and Mathematics, University of Plymouth, Plymouth, U.K.
Keywords: Visual Perception, EEG, AC-GAN, Thought Decoding, Image Generation, Classification, Embedding,
Modulation Layer.
Abstract: Decoding of brain activity with machine learning has enabled the reconstruction of thoughts, memories and
dreams. In this study, we designed a methodology for reconstructing visual stimuli (digits) from human brain
activity recorded during passive visual viewing. Using the MindBigData EEG dataset, we preprocessed the
signals and cleaned them from noise, muscular artifacts and eye blinks. Using the Common Average
Reference (CAR) method and past studies’ results we reduced the available electrodes from 14 to 4 keeping
only those containing discriminative features associated with the visual stimulus. A convolutional neural
network (CNN) was then trained to encode the signals and classify the images. A 92% classification
performance was achieved post-CAR. Three variations of an auxiliary conditional generative adversarial
network (AC-GAN) were evaluated for decoding the latent feature vector with its class embedding and
generating black-and-white images of digits. Our objective was to create an image similar to the presented
stimulus through the previously trained GANs. An average 65% reconstruction score was achieved by the
AC-GAN without a modulation layer, a 60% by the AC-GAN with modulation layer and multiplication, and
a 63% by the AC-GAN with modulation and concatenation. Rapid advances in generative modeling promise
further improvements in reconstruction performance.
1 INTRODUCTION
In our everyday lives we are bombarded with visual
stimuli for which we form visual memories of them
and of their between associations. A defining feature
of visual cognition is our ability to imagine these
stored memories even in the absence of any stimulus,
allowing us to escape from the limitations of our
current perspective into a limitless range of virtual
worlds (Fulford et al., 2018). But how does our brain
store and retrieve visual memories even in the
presence/absence of a visual stimulus? Which brain
areas participate in this visualization process? Can we
decode our brain signals and reconstruct these visual
representations? The scope of this study is to address
these questions. Utilizing the MindBigData dataset of
brain signals via EEG during passive visual viewing
of images of digits (0-9) from the MNIST dataset we
employed an encoder (a CNN) to extract the latent
feature vector from these brain signals and a decoder
a
https://orcid.org/0009-0009-5908-6869
b
https://orcid.org/0000-0001-9005-0260
(a GAN) to reconstruct the images viewed while these
brain signals were recorded. The quality of the
reconstructed images was then compared to the
ground truth (MNIST images) using two performance
metrics (the Dice and Structural Similarity Index
scores) and inferences were drawn.
2 RELATED WORK
The reconstruction of high-quality perceived or
imagined images from brain signals always laid in the
realm of science fiction. Recent advancements in
generative AI (GANs) have allowed scientists to turn
the image reconstruction from brain signals into a
reality. Below we provide a brief overview of some
of these attempts. Kavasidis and colleagues (2017)
employed an encoder (an LSTM layer), which aimed
to identify a latent feature space for brain signal
classification, and a decoder (a VAE or a GAN),
868
Tanner, T. and Cutsuridis, V.
Generative Adversarial Network for Image Reconstruction from Human Brain Activity.
DOI: 10.5220/0013149300003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 868-877
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

which turned the learned feature into images using a
deconvolution approach. Their encoder reached an
86% classification accuracy, whereas their decoder
achieved a below average image reconstruction
quality. Tirupattur and colleagues (2018) employed
an 1D-CNN and modified DC-GAN to generate
images from thought using EEG signals with average
generation success. Jiao et al. (2019) the EEG data
were first converted into EEG Map image, which was
further encoded using visual CNN. The brain signal
classification accuracy was approximately 93%. The
encoded signal was then used with a GAN to
reconstruct high quality perceived images. Recently,
Khare et al. (2022) developed a pipeline consisting of
a feature extractor based on a mixture of LSTM and
gated recurrent neural network layers in order to
extract important visual features from the EEG data
regarding the type and structure of visual stimuli. The
mapping between the extracted feature vector and the
corresponding image was learned using Conditional
Progressive GAN (cProGAN). After training and
testing on a publicly available dataset, their EEG
classifier was able to achieve 98.8% test accuracy,
and cProGAN achieved an inception score (IS) of
5.15 surpassing the previous best 5.07 IS.
3 MATERIALS AND METHODS
3.1 EEG Data
The EEG used in our study was part of the
MindBigData dataset of Vivancos and Cuesta (2022).
Briefly, a human participant was seated in front of a
65’’ TV screen viewing for 2 seconds a single digit
from 0 to 9. Each digit was presented in white font
over full black background (see Fig. 1). The order of
presentation of the digits was random with a black
screen in between them. The brain activity of the
human participant was recorded using a 14-channel
(AF3, AF4, F7, F8, F3, F4, FC5, FC6, T7, T8, P7, P8,
O1 & O2) Emotiv EPOC device (see Fig. 2a) at an
average sampling rate of 128Hz resulting in ~52000
brain signals (see Table 1).
Figure 1: Idealized MNIST stimuli (digits from 0 to 9).
Figure 2: (a) Emotiv EPOC electrode positions (Stytsenko
et al., 2011). (b) Four electrode positions used in our study.
3.2 Algorithmic Pipeline
Our high-level algorithmic pipeline is depicted in Fig.
3. Every step in the pipeline is described in detail in
the subsequent sections.
Figure 3: Algorithmic pipeline (from EEG data collection
to image reconstruction).
3.2.1 Signal Preprocessing
EEG signals are noisy and full of artifacts. A zero-
phase AC notch filter at 50Hz with a 1hz band was
applied to the raw EEG data to remove any effects
from line noise potentially created by interference
from the mains 50hz circuits. Since the oscillation
signals are typically in the range of µVs, AC
fluctuations can completely overwhelm these signals.
Next, a 5th order Butterworth, non-causal bandpass
filter was applied using a 0.4hz low-pass and 60Hz
high-pass band limits to keep only those frequencies
relevant to visual perception (0.4-60Hz). A -6db
cutoff frequency was applied to both band limits and
a Hamming window with 0.0194 passband ripple was
used. Next, the functionality of the MNE package
(Gramfort et al. 2013) was utilized to remove epochs
contaminated with muscular artifacts and eye blinks.
A maximum 100µV peak-to-peak threshold was set
based on previous studies (Sanei and Chambers
2013). Epochs which exceeded this threshold level
were excluded from further analysis (see Table 1).
Generative Adversarial Network for Image Reconstruction from Human Brain Activity
869

Table 1: EEG signals per class (digit).
Class
label
Raw EEG
signals
from MBD
EEG signals
after MNE
thresholdin
g
EEG signals
after CARs
0 5212 3845 1180
1 5080 3798 1090
2 5196 3809 1130
3 5294 3950 1100
4 5079 3765 1100
5 5256 3916 1180
6 5218 3868 1170
7 5069 3780 1195
8 5241 3861 1115
9 5250 3917 1150
Total 51895 38509 11410
Because the Emotiv EPOC device uses no
reference signal, we employed the CAR method
(Mishra et al., 2021), which required to first calculate
the average signal from all 14 channels for each digit,
and then calculate the correlation coefficient ρ
between each channel signal and the mean signal for
each digit. The value of correlation coefficient is in
between -1 to +1. If the value of the correlation
coefficient was high, that meant the channel signal
was closer to the mean signal and thus it could be
considered as less noisy. We selected only those
signals that were having a correlation coefficient
greater than a certain threshold (ρ > 0.9) (see Table
1). Finally, the signal-to-noise ratio (SNR) was
calculated for all digit signals before CAR and after
CAR to assess the effectiveness of the CAR method
(see Table 2).
Table 2: Final selected EEG signals per class at the end of
the preprocessing pipeline. SNR measure of effectiveness
of filtering, epoch rejection and correlation.
SNR
Class
label
Before
CAR
After CAR
(ρ>0.9)
Final selected
EEG signals
0 0.378 2.126 203
1 0.521 2.213 204
2 0.374 2.131 191
3 0.352 2.239 193
4 0.323 2.191 193
5 0.312 2.068 196
6 0.474 2.139 197
7 0.299 1.983 194
8 0.382 2.155 195
9 0.280 2.077 192
Total 1958
As Mishra and colleagues (2021) have shown T7,
P7, T8 and P8 channels are the only electrodes
containing discriminative features associated with the
visual stimulus. Based on their prediction we used
only these four-channel data (see Fig. 2b) for further
processing. These data consisted of 256-time samples
and the associated class label, which were then
resampled using a sliding window of 32 samples with
a 4-sample overlap. This resulted in each channel
being split into 9 x 32 matrix x 4 channels per class
label. The result of this segmentation was to increase
the number of training examples.
The resulting data were then split into 80% for
training and 20% for testing. The 80% training data
were further split into 75% for training and 25% for
validation.
3.2.2 Encoding and Classification
For the encoding and classification of the EEG data,
a CNN (see Supplementary Table 1 for CNN design)
was used consisting of sets of 2D convolutional layers
where the kernel size was changed so that both time
and channel axis were passed through the convolution
together keeping any relation within the same set of
filters. To achieve this the first layer had its kernel
size oriented to the time axis, the second layer
changed its orientation to the channel axis after which
a max pooling layer was used to concentrate the key
activations. A further two 2D convolutional layers
were used to increase the filters with the intention of
producing a more detailed set of filters to identify the
smaller interactions. The output from the
convolutional block was then flattened before passing
onto a set of fully connected (FC) layers. Batch
normalization was used before and after the
convolutional block to help with regularization. The
FC block took the output vector from the
convolutional block and reduced the latent dimension
down to the final output dimension of 128, using a
10% dropout between each FC layer again to help
with regularization and stopped potential over
training. The final FC layer was also batch
normalised; and used as the latent space vector for
input to the GAN generator. This layer was finally
passed to the FC layer with a softmax activation and
10 nodes to determine the classification probability
distribution.
The loss function was based on categorical cross-
entropy, as the class labels were one-hot encoded
before being used. Optimization of the loss function
was achieved using adaptive moment estimation
(Adam) method with a starting rate of 0.001 and
momentum 0.8. The final two layers are outputs from
the network, predicted label, and encoded EEG latent
space vector. The network was trained for a
maximum of 150 epochs, with a batch size of 128.
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
870
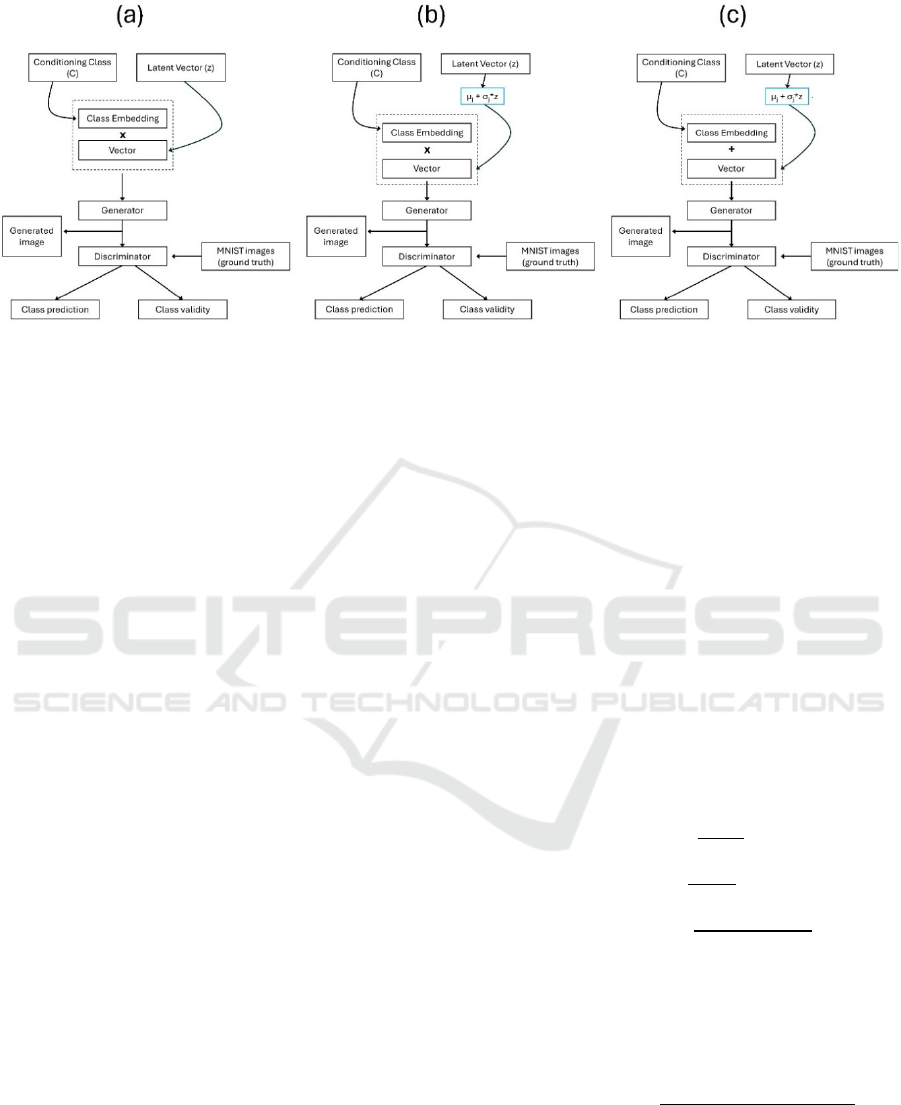
Figure 4: AC-GAN model architectures. (a) AC-GAN without a modulation layer. The conditional class embedding is
multiplied with latent vector (z). (b) AC-GAN with a modulation layer. The conditional class embedding is multiplied with
the modulated vector (µ
z
+ σ
z
• z). (c) AC-GAN with a modulation layer. The conditional class embedding is concatenated
with the modulated vector (µ
z
+ σ
z
• z).
3.2.3 Decoding and Reconstruction
For decoding and image generation, a AC-GAN
(Odena et al. 2017) was used. In the original AC-
GAN, every generated sample has a corresponding
conditioning class label, c in addition to the noise z.
In our study the noise z is replaced by the latent vector
Latent(z) of the encoder. The generator (see
Supplementary Table 2) then uses both the Latent(z)
and the Conditioning label c to generate images X
fake
= G(c; Latent(z)). The discriminator (see
Supplementary Table 3) gives both a probability
distribution over sources and a probability
distribution over the class labels, P(S j X); P(C j X) =
D(X). The objective function has two parts: the log-
likelihood of the correct source, L
S
, and the log-
likelihood of the correct class, L
C
.
𝐿
= 𝐸[𝑙𝑜𝑔 𝑃(𝑆 = 𝑟𝑒𝑎𝑙 | 𝑋
)] + 𝐸[𝑙𝑜𝑔 𝑃(𝑆 =
𝑓𝑎𝑘𝑒 | 𝑋
)] (1)
𝐿
= 𝐸[𝑙𝑜𝑔 𝑃(𝐶 = 𝑐 | 𝑋
)] + 𝐸[𝑙𝑜𝑔 𝑃(𝐶 =
𝑐 | 𝑋
)] (2)
D is trained to maximize L
S
+ L
C
while G is trained
to maximize L
C
- L
S
.
In some simulations, we added an extra layer
(modulation layer) before the generator, which
allowed the modulation of the latent feature space as
a weighted deterministic function of µ
z
and σ
z
described in (Gurumurthy et al., 2017):
𝜇
+𝜎
∗𝑧 (3)
Our objective was then to learn µ
z
, σ
z
, and the AC-
GAN parameters in order to maximize p
data
=
G((µ
z
+σ
z
*Latent(z))|Latent(z)). During the training
of the AC-GAN on the EEG data, we explored three
different architectures of it (see Fig. 4): (a) No
modulation layer, (b) Multiplication of the
modulation layer with the conditional class
embedding, and (c) Concatenation of the modulation
layer with the conditional class embedding. The
training was left to run for 2000 epochs after which
time image generation was stabilized. An Adam
optimizer with learning rate of 0.0002, beta = 0.5 and
decay of 1e-6 was used for both the generator and
discriminator.
3.2.4 Performance Metrics
We used the following metrics for evaluating the
performances of our classifier:
𝑃𝑟𝑒𝑐𝑖𝑠𝑖𝑜𝑛 =
(4)
𝑅𝑒𝑐𝑎𝑙𝑙 =
(5)
𝐹1 𝑠𝑐𝑜𝑟𝑒 = 2 ∗
∗
(6)
where TP are the true positives, TN are the true
negatives, FP are the false positives and FN are the
false negatives.
We used the following metrics for evaluating the
performance of our image generator:
𝐷𝑖𝑐𝑒 𝑠𝑐𝑜𝑟𝑒 = 2 ∗
|∩ |
||| |
(7)
This metric evaluates the similarity between two
images, the predicted one (generated) against the
ground truth (MNIST image). Its value ranges
between 0 and 1, with 1 being the perfect overlap
Generative Adversarial Network for Image Reconstruction from Human Brain Activity
871
(100% similarity between the predicted and the
ground truth) and 0 being no overlap (0% similarity).
The Structural Similarity Index Measure (SSIM)
is a perceptual metric used to evaluate the quality of
generated images by comparing them directly to
ground truth images. Unlike traditional measures like
Mean Squared Error (MSE) that only assess pixel-by-
pixel differences, SSIM considers human visual
perception, focusing on structural information,
luminance, and contrast to assess image similarity. It
produces a score between -1 and 1, where 1 indicates
perfect similarity. SSIM is calculated by comparing
local patterns of pixel intensities normalized for
luminance and contrast, providing a more holistic
view of image quality. It is particularly useful for
evaluating image generation tasks, such as super-
resolution, denoising, and inpainting, where
maintaining structural fidelity and visual quality is
crucial. By accounting for variations in structure,
texture, and lighting, SSIM offers a robust measure of
how closely generated images resemble their
corresponding ground truth, making it an effective
tool for evaluating generative models:
𝑆𝑆𝐼𝑀
(
𝑥, 𝑦
)
=(𝑙
(
𝑥, 𝑦
)
)
∗(𝑐
(
𝑥, 𝑦
)
)
∗(𝑠(𝑥,𝑦))
(8)
where
𝑙(𝑥, 𝑦) = (2𝜇𝑥 ∗ 𝜇𝑦 + 𝐶1)/(𝜇𝑥
+ 𝜇𝑦
+𝐶1)
𝑐(𝑥, 𝑦) = (2𝜎𝑥 ∗ 𝜎𝑦 + 𝐶2)/(𝜎𝑥
+ 𝜎𝑦
+𝐶2)
𝑠(𝑥, 𝑦) = (𝜎𝑥𝑦 + 𝐶3)/(𝜎𝑥 ∗ 𝜎𝑦 + 𝐶3)
where µ is the mean pixel value, σ is the standard
deviation of the pixel value and σxy is the covariance
of the pixel value. C1, C2, and C3 are constraints to
avoid division by zero with C3 = C2 /2. α, β, and γ are
positive exponents that change the components
contribution to the overall SSIM.
4 RESULTS
4.1 Classification
Our CNN model was trained on two sets of data. The
post filtered data prior to CAR correlation selection
method and the data set after final CAR correlation
selection. From Tables 6 and 7, it is evident that
accuracy improves from an approximate average 46%
in the pre-CAR selection case to an approximate
average 92% in the post-CAR selection case.
Table 3: CNN evaluation after trained with pre-CAR
selection data.
Label precision Recall F1-score
0 0.48 0.48 0.48
1 0.46 0.43 0.45
2 0.43 0.40 0.41
3 0.50 0.51 0.51
4 0.46 0.39 0.42
5 0.45 0.48 0.46
6 0.45 0.46 0.45
7 0.41 0.47 0.47
8 0.43 0.43 0.43
9 0.50 0.47 0.47
Accurac
y
0.46
Micro avg 0.46 0.46 0.46
Wei
g
hted av
g
0.46 0.46 0.46
Table 4: CNN evaluation after trained with post-CAR
selection data.
Label precision Recall F1-score
0 0.92 0.88 0.90
1 0.93 0.90 0.92
2 0.91 0.87 0.89
3 0.94 0.94 0.94
4 0.91 0.93 0.92
5 0.89 0.91 0.90
6 0.92 0.89 0.90
7 0.90 0.96 0.93
8 0.93 0.95 0.94
9 0.92 0.93 0.93
Accurac
y
0.92
Micro av
g
0.92 0.92 0.92
Wei
g
hted av
g
0.92 0.92 0.92
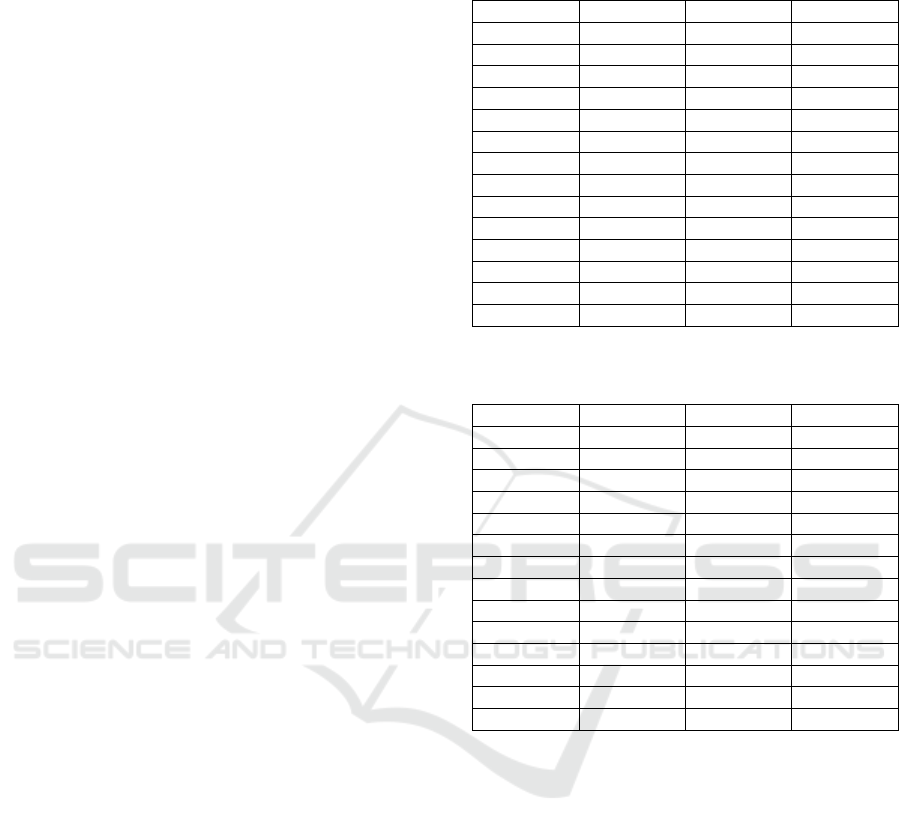
Figures 5 and 6 show the confusion matrix
between class predictions, and the t-SNE map of class
separability, respectively, before the application of
the CAR selection method. Both figures clearly show
the extent of class confusion which is caused by the
remaining noise in the signals prior to extracting the
data which had a high correlation threshold.
In contrast, figures 7 and 8 show the confusion
matrix between class predictions, and the t-SNE map
of class separability, respectively, after the
application of the CAR selection method. It is clearly
evident that the classes are mostly separable and very
little confusion between them is present.
4.2 Image Reconstruction
From Figure 9 it is clear that the AC-GAN with no
modulation layer is able to reconstruct well all digits
with the exception of ‘2’, which confuses it with ‘0’.
The SSIM score for ‘2’ is 0.098 (see Table 5).
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
872
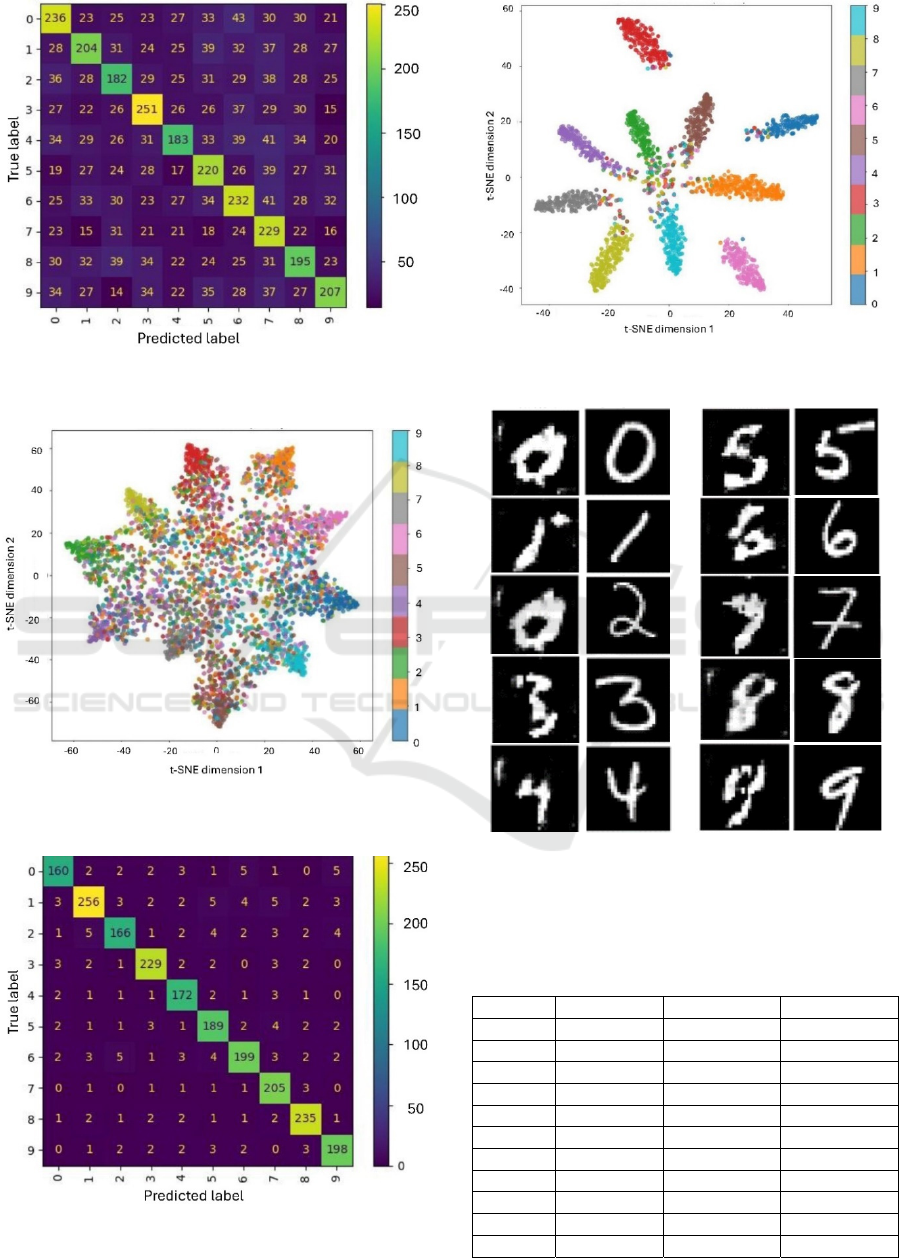
Figure 5: Class confusion matrix of CNN model trained
with pre-CAR selection data.
Figure 6: t-SNE class separability of CNN model trained
with pre-CAR selection data.
Figure 7: Class confusion matrix of CNN model trained
with post-CAR selection data.
Figure 8: t-SNE class separability of CNN model trained
with post-CAR selection data.
Figure 9: Reconstruction of digit (0-9) images (columns 1
and 3) compared to the MNIST digit (0-9) images (columns
2 and 4) by the AC-GAN without a modulation layer (see
Fig. 4a for GAN architecture)
Table 5: Summary of reconstruction evaluation results of
AC-GAN without a modulation layer.
Label Prediction Dice score SSIM score
0 TRUE 0.581 0.229
1 TRUE 0.408 0.449
2 FALSE 0.285 0.098
3 TRUE 0.437 0.293
4 TRUE 0.408 0.380
5 TRUE 0.467 0.316
6 TRUE 0.567 0.401
7 TRUE 0.198 0.139
8 TRUE 0.568 0.271
9 TRUE 0.454 0.334
Average 0.4373 0.291
Generative Adversarial Network for Image Reconstruction from Human Brain Activity
873
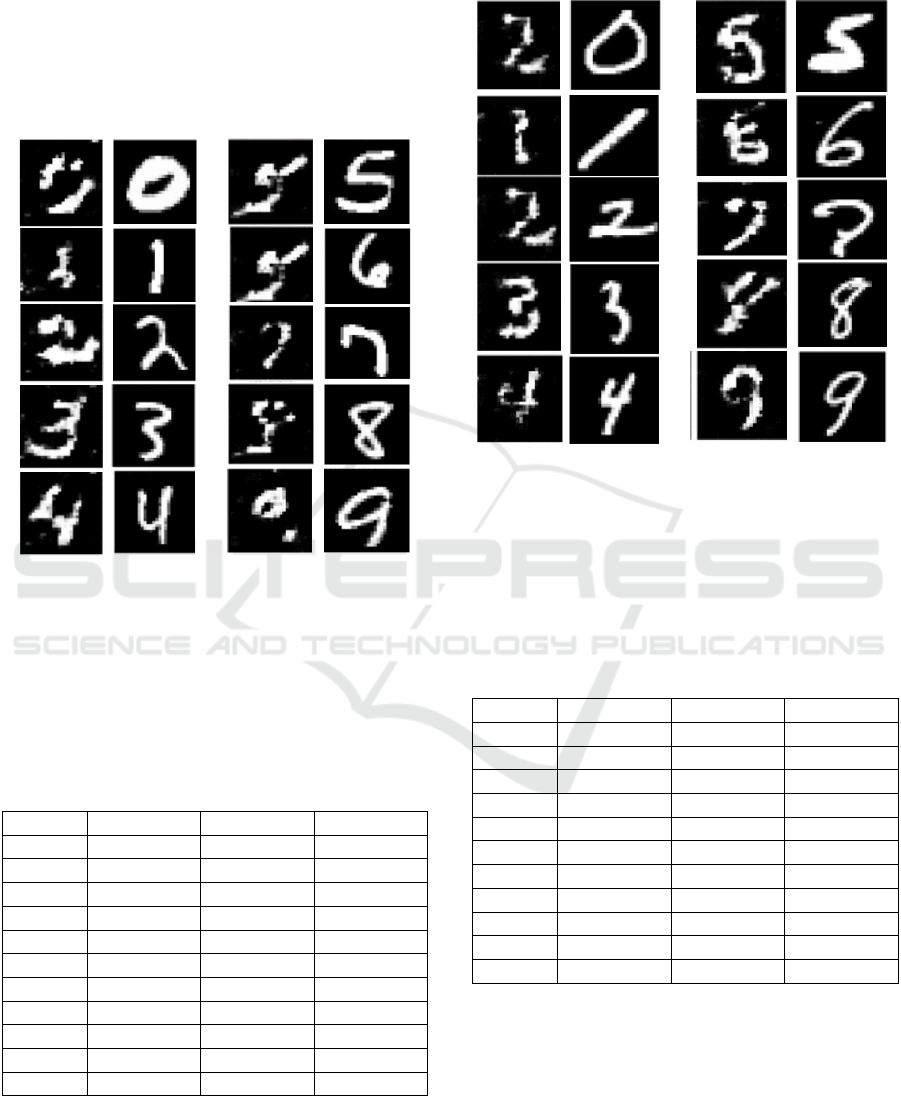
From Figure 10 it is clear that the AC-GAN with
modulation layer and multiplication of the modulated
latent vector with the conditioning class embedding is
able to reconstruct well all digits with the exception
of ‘6’, which confuses it with ‘5’. The SSIM score for
‘6’ is 0.118 (see Table 6). Similarly, the SSIM score
for ‘5’ is 0.115 even though the model is able to
reconstruct the correct digit.
Figure 10: Reconstruction of digit (0-9) images (columns 1
and 3) compared to the MNIST digit (0-9) images (columns
2 and 4) by the AC-GAN with a modulation layer and
multiplication of the modulated latent vector with the
conditional class embedding (see Fig. 4b for GAN
architecture).
Table 6: Summary of reconstruction evaluation results of
AC-GAN with a modulation layer (multiplication of the
modulated latent vector with the conditional class
embedding).
Label Prediction Dice score SSIM score
0 TRUE 0.538 0.128
1 TRUE 0.467 0.474
2 TRUE 0.476 0.184
3 TRUE 0.446 0.218
4 TRUE 0.416 0.193
5 TRUE 0.370 0.115
6 FALSE 0.234 0.118
7 TRUE 0.159 0.164
8 TRUE 0.415 0.281
9 TRUE 0.382 0.199
Average 0.39 0.2074
From Figure 11 it is clear that the AC-GAN with
modulation layer and concatenation of the modulated
latent vector with the conditioning class embedding is
able to reconstruct well all digits with the exception
of ‘0’, which confuses it with ‘2’. The SSIM score for
‘0’ is 0.064 (see Table 7).
Figure 11: Reconstruction of digit (0-9) images (columns 1
and 3) compared to the MNIST digit (0-9) images (columns
2 and 4) by the AC-GAN with a modulation layer and
concatenation of the modulated latent vector with the
conditional class embedding (see Fig. 4c for GAN
architecture).
Table 7: Summary of reconstruction evaluation results of
AC-GAN with a modulation layer (concatenation of the
modulated latent vector with the conditional class
embedding).
Label Prediction Dice score SSIM score
0 FALSE 0.155 0.064
1 TRUE 0.130 0.151
2 TRUE 0.321 0.205
3 TRUE 0.473 0.323
4 TRUE 0.446 0.393
5 TRUE 0.466 0.225
6 TRUE 0.421 0.195
7 TRUE 0.379 0.291
8 TRUE 0.482 0.409
9 TRUE 0.296 0.255
Average 0.3569 0.2511
5 DISCUSSION
Our study has produced interesting results regarding
the reconstruction of mental content (visual stimulus)
from EEG with generative AI by leveraging the latent
vector (not random noise) and a predicted semantic
(class) embedding. The study demonstrated that EEG
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
874

signals can effectively be processed to extract
meaningful features, which, when fed into a trained
classification encoder and a subsequent generative
network, can generate representative images of the
perceived visual stimulus. When comparing the SSIM
scores of the AC-GAN models, we found that the
model with the best reconstruction score was the AC-
GAN without modulation (Average SSIM = 0.291 or
65% average similarity between the generated images
and the ground truth). The model with the worst
reconstruction score was the AC-GAN with
modulation and multiplication (Average SSIM =
0.2074 or 60% average similarity between the
generated images and the ground truth). The average
Dice scores for the three AC-GAN models were
lower than the average SSIM scores (Average Dice
for AC-GAN without modulation = 43.73%; Average
Dice for AC-GAN with modulation and
multiplication =39%; Average Dice for AC-GAN
with modulation and concatenation = 35.69%). Since
the Dice score depends solely on the similarity
between the reconstructed images and the ground
truth on a pixel-by-pixel basis and excludes any
structural information, luminance, and contrast
information from the evaluation, then the SSIM score
is a more accurate reconstruction metric.
In this study, we utilized EEG data collected from
a different group (MindBigData (Vivancos and
Cuesta, 2022)) to which we had no control of their
experimental design, recording and collection
processes. Furthermore, EEG signals by nature are
noisy, full of muscular artifacts and eye blinks
particularly the signals from the frontal electrodes.
Careful preprocessing of the MindBigData EEG
signals reduced the data for training and testing from
51895 samples to 1958 samples, which was only
3.77% of the total data set. Even though the final EEG
dataset used for encoding and classification was
small, our CNN model reached a 92% classification
performance on average, which is close to the current
state-of-the-art (~96% (Mahapatra and Bhuyan,
2023)). Given the single-subject nature of these data,
it is unclear how the results would generalize to a
larger population. We believe this could be a fruitful
area for future investigation.
Despite the AC-GAN model's ability to generate
conditioned images, it is unable to rectify
misclassifications based solely on the encoded EEG
latent vector and conditioning label. As a result, when
the model generates an image that is incorrectly
classified, the resulting image exhibits a low SSIM
score, indicating poor similarity to the ground truth
(e.g. reconstructed ‘2’ digit by the AC-GAN without
modulation (see Fig. 9), or reconstructed ‘0’ digit by
the AC-GAN with modulation and concatenation (see
Fig. 11)). Paradoxically in some cases, this
misclassified image may still receive a relatively high
score, suggesting a potential discrepancy between the
model's perception of the image and its actual fidelity
to the ground truth (e.g. reconstructed ‘6; digit by the
AC-GAN with modulation and multiplication (see
Fig. 10)). This is because in the current model design
the predicted class conditioning label has a higher
influence on the predicted image over the encoded
latent vector.
A future extension to our study would be to
include a reinforcement learning agent applied to the
class label prediction. This agent would penalize the
incorrect class label thus reducing its influence on the
predicted image over the encoded latent vector.
Which in cases where the classification is incorrect
the agent would provide unsupervised learning
adjustments which may help to correct the AC-GAN
prediction and validity to indicate an uncertain score.
ACKNOWLEDGEMENTS
This work was supported by the EU HORIZON 2020
Project ULTRACEPT under Grant 778062.
REFERENCES
Fulford J, Milton F, Salas D, Smith A, Simler A, Winlove
C, Zeman A. (2018). The neural correlates of visual
imagery vividness = An fMRI study and literature
review. Cortex. 105: 26-40
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-
Farley D, Ozair S, Courville A, Bengio Y. (2014)
Generative Adversarial Nets. https://arxiv.org/abs/14
06.2661
Gramfort A, Luessi M, Larson E, Engemann D, Strohmeier
D, Brodbeck C, Goj R, Jas M, Brooks T, Parkkonen L,
Hämäläinen M. (2013) MEG and EEG data analysis
with MNE-Python. Frontiers in Neuroscience.
7(267):1-13
Gurumurthy S, Sarvadevabhatla R, Babu R. (2017)
DeLiGAN: Generative Adversarial Networks for
Diverse and Limited Data. IEEE Conference on
Computer Vision and Pattern Recognition (CVPR).
IEEE. 166-174. https://doi.org/10.1109/cvpr.2017.525
Jiao Z, You H, Yang F, Li X, Zhang H, Shen D. (2019)
Decoding EEG by visual-guided deep neural networks.
IJCAI, pp. 1387–1393
Khare S, Choubey RN, Amar L, Udutalapalli V. (2022).
NeuroVision: perceived image regeneration using
cProGAN. Neural Computing and Applications 34:
5979–5991
Generative Adversarial Network for Image Reconstruction from Human Brain Activity
875
Kavasidis I, Palazzo S, Spampinato C, Giordano D, Shah
M. (2017) Brain2image: Converting brain signals into
images. Proceedings of the 25th ACM international
conference on multimedia, 1809–1817
Mahapatra NC, Bhuyan P. (2023). EEG-based
classification of imagined digits using a recurrent
neural network. J Neural Eng. 20(2)
Mishra R, Sharma K, Bhavsar A. (2021) Visual Brain
Decoding for Short Duration EEG Signals. 29th
European Signal Processing Conference (EUSIPCO).
IEEE. 1226-1230. https://doi.org/10.23919/eusipco54
536.2021.9616192.
Mishra R, Sharma K, Jha RR, Bhavsar A. (2023).
NeuroGAN: image reconstruction from EEG signals
via an attention-based GAN. Neural Computing and
Applications. 35(12): 9181-9192. https://doi.org/10.10
07/s00521-022-08178-1
Odena A, Olah C, Shlens J. (2017) Conditional Image
Synthesis with Auxiliary Classifier GANs. Proceedings
of the 34th International Conference on Machine
Learning. 70: 2642-2651
Sanei S, Chambers JA. (2013). EEG signal processing.
John Wiley & Sons
Stytsenko K, Jablonskis E, Prahm C. (2011). Evaluation of
consumer EEG device emotiv epoc. In MEi: CogSci
Conference 2011, Ljubljana, 2011.
Tirupattur P, Rawat YS, Spampinato C, Shah M. (2018)
Thoughtviz: visualizing human thoughts using
generative adversarial network. Proceedings of the 26th
ACM international conference on multimedia, 950–958
Vivancos D, Cuesta F. (2022). MindBigData 2022: A dataset
of brain signals. https://arxiv.org/pdf/2212.14746
APPENDIX
Supplementary Table 1: EEG encoder/classifier CNN
design.
Layer Output
Shape
Attributes Parameters
Input (None, 9, 32, 1)
BatchNormalization (None, 9, 32, 1) 4
2D Convolution (None, 9, 32,
128)
128 filters,
(9,1), pad 1,
same
640
2D Convolution (None, 9, 32,
64)
64 filters,
(9,1), pad 1,
same
73792
MaxPooling2D (None, 9, 16,
64)
(1, 2) 0
2D Convolution (None, 64, 13,
40)
64 filters, (4,
25), pad 1,
valid
57644
MaxPooling2D (None, 64, 6,
40)
(1, 2) 0
2D Convolution
(None, 15, 5,
128)
128 filters,
(50, 2), pad
1, valid
512128
Flatten (None, 9600) 0
BatchNormalization (None, 9600) 38400
Dense (None, 512) 512 nodes,
Relu
4915712
Dropout (None, 512) 0.1 0
Dense (None, 256) 256 nodes,
Relu
131328
Dropout (None, 256) 0.1 0
Dense (None, 128) 128 nodes,
Relu
32896
Dropout (None, 128) 0.1 0
BatchNormalization (None, 128) 512
Dense (None, 10) Softmas, L2
regularizatio
n
1290
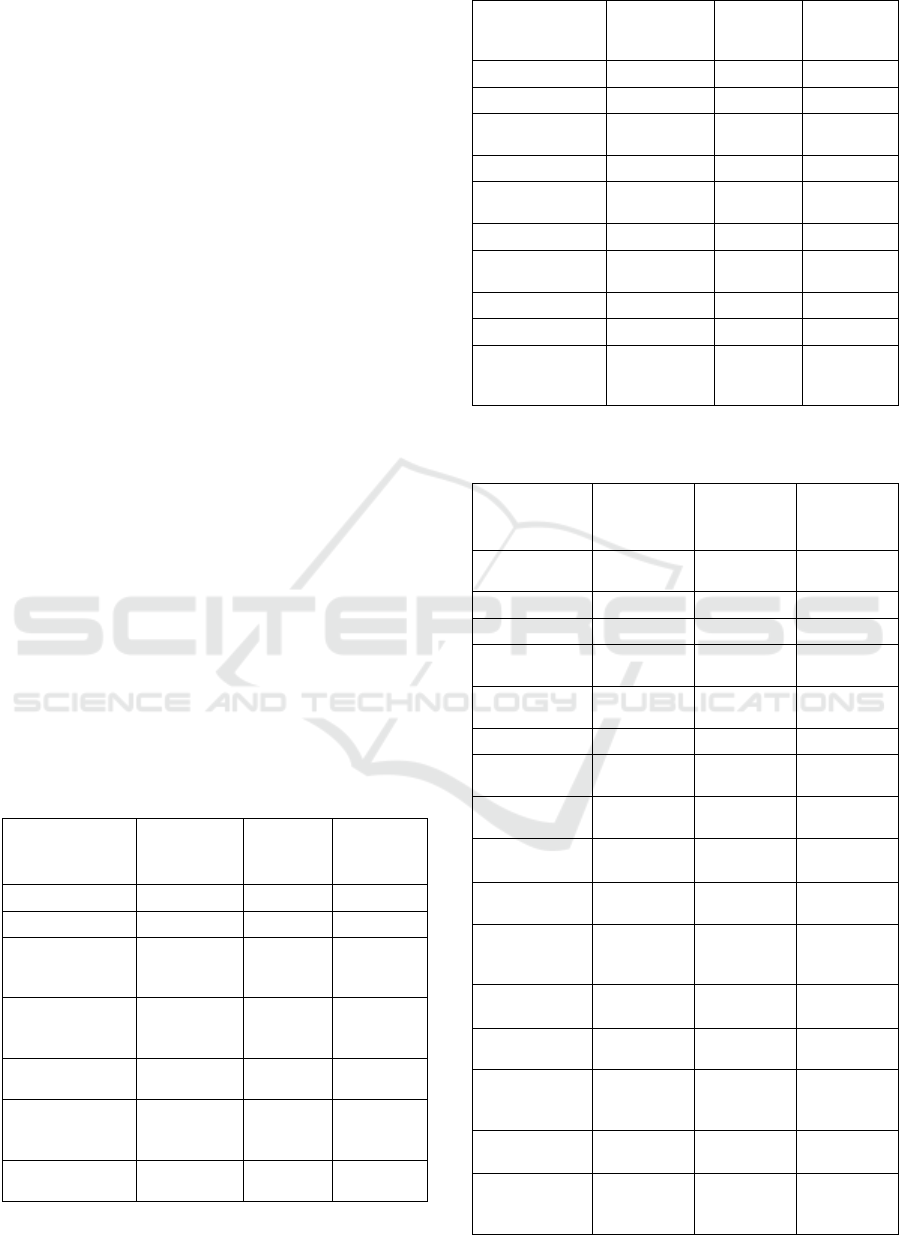
Supplementary Table 2: AC-GAN generator network
model design.
Layer
Output
Shape
Attributes Parameters
Input (class
label)
(None, 1)
0
Embedding (None, 1, 128)
1280
Flatten (None, 128)
0
I
nput (EEG latent
space)
(None, 128)
0
Modulation of
EEG Layer
(None, 128)
256
Multiply (None, 128)
0
Dense (None, 6272)
Nodes 6272,
Relu
809088
Reshape
(None, 7, 7,
128)
0
Batch
Normalization
(None, 7, 7,
128)
512
UpSample2D
(None, 14, 14,
128)
nearest 0
2D Convolution
(None, 14, 14,
128)
128, kernel 3,
stride 1, pad
same, Relu
147584
Batch
Normalization
(None, 14, 14,
128)
512
UpSample2D
(None, 28, 28,
128)
Nearest 0
2D Convolution
(None, 28, 28,
64)
64, kernel 3
stride 1, pad
same, TanH
73792
Batch
Normalization
(None, 28, 28,
64)
256
2D Convolution
(None, 28, 28,
1)
1, kernel 3,
stride 1, pad
same, Tanh
577
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
876

Supplementary Table 3: AC-GAN discriminator network
model design.
Layer Output Shape Attributes Parameters
Input (generated
image)
(None, 28, 28, 1)
0
2D Convolution
(None, 14, 14,
16)
16, kernel 3,
stride 2, pad
same
160
LeakyRelu
activation
(None, 14, 14,
16)
0.2 0
Dropout
(None, 14, 14,
16)
0.25 0
2D Convolution (None, 7, 7, 32)
32, kernel 3,
stride 2, pad
same
4640
ZeroPad (None, 8, 8, 64)
0
LeakyRelu
activation
(None, 8, 8, 64) 0.2 0
Dropout (None, 8, 8, 64) 0.25 0
Batch
Normalization
(None, 8, 8, 64) 0.8 128
2D Convolution (None, 4, 4, 128)
64, kernel 3,
stride 2, pad
same
18496
LeakyRelu
activation
(None, 4, 4, 128) 0.2 0
Dropout (None, 4, 4, 128) 0.25 0
Batch
Normalization
(None, 4, 4, 64) 0.8 256
2D Convolution (None, 4, 4, 128)
128, kernel 3,
stride 1, pad
same
73856
LeakyRelu
activation
(None, 4, 4, 128) 0.2 0
Dropout (None, 4, 4, 128) 0.25 0
Flatten (None, 2048 0
Dense (validity
confidence)
(None, 1) 2049
Dense (class
prediction)
(None, 10) 20490
Generative Adversarial Network for Image Reconstruction from Human Brain Activity
877
