
Federated Learning in Multi-Center, Personalized Healthcare for COPD
and Comorbidities: The RE-SAMPLE Platform
Jakob Lehmann
1 a
, Gesa Wimberg
1 b
, Serge Autexier
1 c
, Alberto Acebes
2 d
,
Christos Kalloniatis
3 e
, Costas Lamprinoudakis
4 f
, Thrasyvoulos Giannakopoulos
4 g
,
Andreas Menegatos
4 h
, Agni Delvinioti
5 i
, Giulio Pagliari
5 j
, Nicoletta di Giorgi
5 k
,
Jarno Raid
6 l
, Danae Lekka
7 m
, Aristodemos Pnevmatikakis
7 n
, Sofoklis Kyriazakos
7 o
,
Konstantina Kostopoulou
7
and Monique Tabak
8 p
1
Deutsches Forschungszentrum f
¨
ur K
¨
unstliche Intelligenz (DFKI), Enrique-Schmidt-Str. 5, 28359 Bremen, Germany
2
Atos IT Solutions and Services Iberia, S.L. Calle de Albarrac
´
ın, 25. 28037 Madrid, Spain
3
Department of Cultural Technology and Communication, University of the Aegean, University Hill, 81100 Mytilene,
Greece
4
Department of Digital Systems, University of Piraeus, 150 Androutsou St., Piraeus 18532, Greece
5
Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Largo Agostino Gemelli, 8, 00168 Rome, Italy
6
Tartu
¨
Ulikooli Kliinikum, Ludvig Puusepa 8, 50406 Tartu, Estonia
7
Innovation Sprint, Clos Chapelle-aux-Champs 30, Bte. 1.30.30 1200 Brussels, Belgium
8
University of Twente, Drienerlolaan 5, 7522 NB Enschede, Netherlands
{jakob.lehmann, gesa.wimberg}@dfki.de
Keywords:
Federated Learning, Interpretable Machine Learning, Chronic Obstructive Pulmonary Disease, Personalized
Care, Real-World Data.
Abstract:
Federated learning is becoming more and more popular, also in healthcare applications. The platform, de-
veloped within a multidisciplinary consortium, is enabling privacy-preserving training of machine learning
models generating predictions for patients with chronic obstructive pulmonary disease and comorbidities.
Moreover, data synchronization and monitoring is made possible using the HL7 FHIR standard. The platform
provides two front ends; a patient facing smartphone app and a healthcare professional facing dashboard that
is used inside three different hospitals in Italy, Estonia and the Netherlands. The overall architecture and im-
plementation into practice is shown in this paper.
a
https://orcid.org/0009-0000-2331-0640
b
https://orcid.org/0000-0002-0694-2942
c
https://orcid.org/0000-0002-0769-0732
d
https://orcid.org/0000-0002-0840-2915
e
https://orcid.org/0000-0002-8844-2596
f
https://orcid.org/0000-0003-3101-5347
g
https://orcid.org/0000-0002-3453-1892
h
https://orcid.org/0000-0002-2469-5535
i
https://orcid.org/0000-0002-2402-9444
j
https://orcid.org/0000-0001-8481-1529
k
https://orcid.org/0000-0002-8033-5411
l
https://orcid.org/0009-0005-7549-3406
m
https://orcid.org/0009-0005-7789-2662
n
https://orcid.org/0000-0002-9623-6354
o
https://orcid.org/0000-0002-8841-6558
p
https://orcid.org/0000-0001-5082-1112
1 INTRODUCTION
Chronic Obstructive Pulmonary Disease (COPD) is
a heterogeneous lung condition characterized by
chronic respiratory symptoms (dyspnoea, cough, ex-
pectoration, exacerbations) due to abnormalities of
the airways (bronchitis, bronchiolitis) and/or alveoli
(emphysema) that cause persistent, often progressive,
airflow obstruction (Agust
´
ı et al., 2023). COPD
is one of the three leading causes of death world-
wide (Patel, 2024) and is one of the high-impact dis-
eases with an increasing prevalence, mortality and
morbidity, with a high burden of disease because of
deterioration of symptoms and highly prevalent acute
exacerbations. Around 65 million people live with
moderate or severe COPD (PRASAD, 2020).
Many patients with COPD have comorbidities like
Lehmann, J., Wimberg, G., Autexier, S., Acebes, A., Kalloniatis, C., Lamprinoudakis, C., Giannakopoulos, T., Menegatos, A., Delvinioti, A., Pagliari, G., di Giorgi, N., Raid, J., Lekka, D.,
Pnevmatikakis, A., Kyriazakos, S., Kostopoulou, K. and Tabak, M.
Federated Learning in Multi-Center, Personalized Healthcare for COPD and Comorbidities: The RE-SAMPLE Platform.
DOI: 10.5220/0013149800003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 499-506
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
499

diabetes mellitus or chronic heart failure that further
increase patient burden, mortality and costs. Patients
often struggle with the complex handling of the dis-
ease, especially if they have other comorbidities and
therefore suffer from overlapping symptoms. Cur-
rent disease management and monitoring of patients
with COPD and comorbidities relies on information
acquired during time-based scheduled visits when pa-
tients are usually stable, whereas the actual symp-
toms and changes during common daily life triggers
are not quantified. As such disease management is
a big challenge for Healthcare Professionals (HCPs)
due to the complexity and heterogeneity of this multi-
morbidity and since they lack appropriate information
(e.g. Real-World Data (RWD), like patient activity
and symptom data, and environmental data) to pre-
dict exacerbations, tailor the disease management and
treatment, and support self management.
EHealth technologies and smart tools, such as
data platforms and home diagnostics allowing real-
time, objective, and longitudinal monitoring at home,
or virtual coaches offering coaching or personalized
treatment suggestions, have a large potential to im-
prove the health condition and quality of life of the
patient and to relieve burden on HCPs, by for example
preventing exacerbations. To develop eHealth tech-
nologies that offer functionalities based on Machine
Learning (ML) or artificial intelligence, large datasets
are necessary. However, knowledge and parameters
that could be important for predicting exacerbations
of COPD and comorbidities are distributed among
different data sources and representations, includ-
ing evidence from clinical studies, Electronic Health
Records (EHRs) and RWD. To overcome the discrep-
ancy of low resources for conducting a large study
and the necessity for a large dataset, Federated Learn-
ing (FL) offers a promising solution. Smaller datasets
from different hospitals can be used to train a global
model with a higher robustness without transferring
the sensitive medical patient data.
The RE-SAMPLE platform presented in this pa-
per provides support for leveraging data of patients
with COPD and comorbidities distributed in several
places to support the HCP and patient in the manage-
ment of the disease. The key contributions made by
the RE-SAMPLE platform are:
1. RE-SAMPLE offers data storage, synchronisa-
tion and management of patients’ data both from
the hospital information systems, the patient app,
RWD from wearables as well as weather and air
quality RWD in order to improve patient monitor-
ing and coaching.
2. The platform offers privacy-preserving federated
learning of ML models to predict the risk of an
upcoming COPD exacerbation and the quality of
life of the patients.
3. The exacerbation risk and quality of life predic-
tions and the influence of clinical, behavioural
or environmental factors computed from the ML
models are provided in a user interface to the clin-
icians at the collaborating hospitals to be used dur-
ing shared decision-making.
4. The platform has been deployed in highly-secured
hospital IT environments by adopting a privacy by
design approach across all stages of the develop-
ment life-cycle and taking into account all legal
and technical requirements of the General Data
Protection Regulation (GDPR).
This paper presents the technical development
of the platform, describing the different components
within the platform and their interplay. To this end
the paper is organized as follows: Section 2 presents
relevant related work on IT platforms for healthcare
to provide ML-based support. Section 3 describes
the architecture of the platform from a functional per-
spective along the data processing workflows of data
ingestion, ML model training, ML model prediction
and visualization for shared decision-making. Sec-
tion 4 provides an insight on the privacy-by-design
process of the platform development and introduced
technical security and privacy measures. Section 5
provides an overview on how the platform has been
deployed in production in three different hospital en-
vironments in different European countries and the
challenges posed to connect to the very diverse local
hospital information system environments. Section 6
concludes the paper and highlights future steps.
2 RELATED WORK
ML is used more and more in heathcare applications
(Rahman et al., 2023), but there is still potential to im-
prove the support from these systems regarding, e.g.,
collection and storage of data (Habehh and Gohel,
2021). So, the approach of RE-SAMPLE is partic-
ular, since we make use of clinical data from EHRs
one the hand, and of RWD coming from an app, wear-
ables and external environmental services on the other
hand.
In the following, relevant applications and related
projects are listed. The RETENTION project (Abde-
laziz et al., 2018) has a cloud-based approach, they
created a platform to support personalised interven-
tions for patients with heart failure. It also supports
ML model training and continuous monitoring of pa-
tients, but not with a federated infrastructure.
HEALTHINF 2025 - 18th International Conference on Health Informatics
500
CrowdHEALTH is another example for a cloud-
based approach to combine EHRs and other sources
to obtain useful insights into outcomes of prevention
strategies, health policies and efficiency of care (Kyr-
iazis et al., 2019). However, the final platform was a
set of different collections of data to be analysed on
the cloud and had no FL approach. It thus required to
extract the data from the hospitals to a central place,
which hospitals are typically very reluctant to do.
One of the key aspects of the RE-SAMPLE plat-
form is to provide close to real-time results to support
the decision making during patient consultation. For
example, there is an implemented system tested on
multiple historical medical datasets (diabetes, heart
diseases, breast cancer) (Hassan et al., 2020). How-
ever, this system does not support FL.
There are also projects with very similar objec-
tives introducing a FL platform (Lampropoulos et al.,
2021). This project focuses on patients with cancer
and their healthcare providers, aiming to enhance the
quality of life for patients by offering personalized
intervention recommendations and support. Even if
this also used different data sources, a difference to
RE-SAMPLE is that the HCPs only had access to the
data stored in the edge node, while in RE-SAMPLE
the technical challenge was to allow both access to
the data in the edge node as well as to data on the
cloud which belongs to the disease self-management
app used by the patients in the different hospital sites.
There are other works on predicting COPD ex-
acerbations that do not use ML (Adibi et al., 2020).
To our knowledge, there is no FL platform to predict
COPD exacerbations.
However, there are deep learning approaches
regarding COPD. One approach for prediction of
COPD exacerbation is based on a dataset with 94 pa-
tients and frequent data (Nunavath et al., 2018). A
Long Short-Term Memory (LSTM) is trained for clas-
sification with three classes (stable, significant deteri-
oration, urgent need for follow up) achieving a good
accuracy, but the prediction is only for a couple of
days in advance. The model is more accurate for pre-
dicting the stable state. Moreover, LSTMs are used
to study COPD disease progression based on the four
GOLD standard levels (Tang et al., 2018). While ex-
tracting data from clinical notes, the irregular time
visits in the data are handled.
3 ARCHITECTURE
A key idea of RE-SAMPLE is to enrich the patients’
EHRs with RWD like weather and air quality data, an-
swers to questionnaires and activity and sensor data.
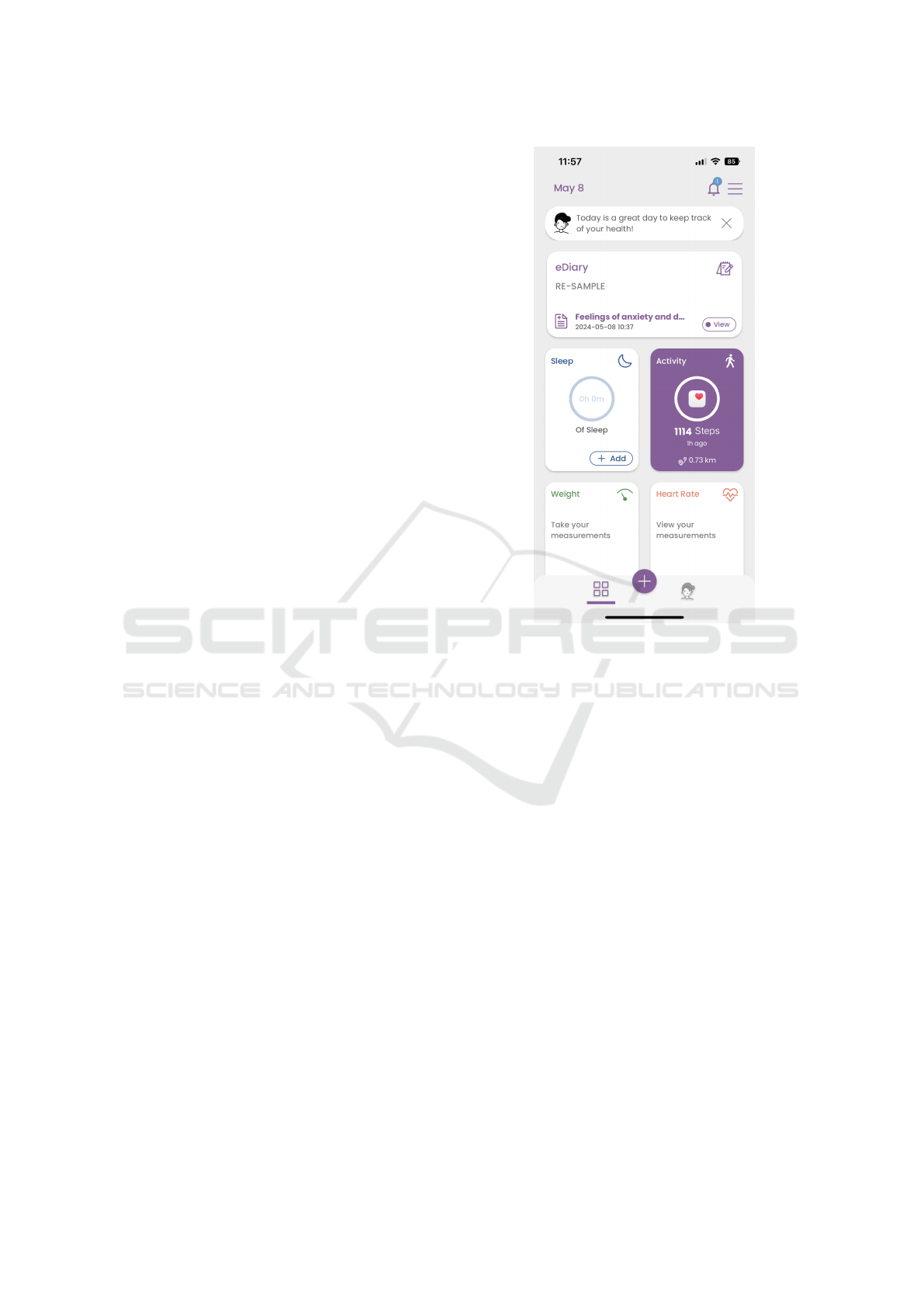
Figure 1: Patient smartphone app.
To this end, the patients are equipped with a wear-
able device capable of measuring activity data like
the number of steps or the heart rate. Additionally, a
smartphone app (see Figure 1) is used to fill-in ques-
tionnaires and to inform the patient. In contrast to
clinical data, the RWD is collected continuously in-
between the patients visits to the hospital. This allows
the ML systems to take into account a more holis-
tic picture of the patients’ state. The data from both
the wearable device and the patient smartphone app
is synchronized to the centralized Healthentia cloud,
while the clinical data remains at the hospital. Since
different hospitals are involved, this means that the
data is distributed. The platform architecture needs to
facilitate data homogenization between these sources
and combine it into a standardized data model — en-
suring that the features as well as their encoding are
the same between the hospitals, which may be using
completely different software solutions and data rep-
resentations internally. Further, datasets that are uni-
form across hospitals need to be created and prepro-
cessed to allow for FL. RE-SAMPLE uses the Flower
framework for the implementation of the FL (Beu-
tel et al., 2020). Since sensitive patient data is in-
volved, highest privacy standards need to be met re-
sulting in a GDPR compliant privacy-by-design sys-
Federated Learning in Multi-Center, Personalized Healthcare for COPD and Comorbidities: The RE-SAMPLE Platform
501

tem. To achieve this, the platform uses an edge com-
puting architecture, with every pilot site running an
edge node. These edge nodes run the components,
which are docker containers, allowing for high porta-
bility and security. Each edge node is physically
hosted in the hospitals’ premises and is under the con-
trol of the pilot sites’ ICT Departments. Due to the
type of data available in each single node, only autho-
rized personnel can have access to the VMs and per-
form installations or work on the datasets. In addition
to the edge nodes, a single orchestrator node manages
the FL and authorization process. The components of
the RE-SAMPLE platform belong to one of the three
parts:
• the Health Data Hub (HDH), consisting itself of
four components which together allow for data in-
gestion and storage,
• the ML components, consisting of five edge node
and one orchestrator node components, managing
ML training and the production of ML results,
• the Local Data Connector (LDC), enabling the
dashboard to display sensitive hospital data if ac-
cessed from within a secure hospital network or
connection.
The architecture is visualized in Figure 4 on p..
3.1 Workflows
3.1.1 Data Ingestion
In RE-SAMPLE, patient data from the EHRs and
RWD from Healthentia is ingested into a central stor-
age component called the Clinical Data Repository
(CDR). It contains the entire patient data, including
any ML results which are produced for the patient.
The only data not in the CDR is the air quality and
weather data, which is collected in a separate compo-
nent and added on-demand to the patient data. The
CDR is a standardized, FHIR-based (Bender and Sar-
tipi, 2013) database storing data as interoperable re-
sources. Built with Java and MariaDB, it runs in a
separate docker container and provides a REST API
for advanced searches, although external access is re-
stricted in production. The HL7 FHIR Implemen-
tation Guide (IG) customizes the standard FHIR re-
sources to meet project needs, using semantics to en-
sure data is correctly formatted. IGs are formal def-
initions for data exchange and, in this project, doc-
ument and validate the use of the HL7 FHIR stan-
dard internally. The Clinical Data Repository API
(CDR-API) is a RESTful API that follows the Ope-
nAPI specification and handles data ingestion and ex-
port for the CDR, following predefined rules in the
Implementation Guide. Built in Java, it validates and
models the data using the RE-SAMPLE-model code
library. The CDR-API is the entry-point for patient
data into the CDR. Hospitals provide data stored in
their EHRs directly to the CDR-API via scheduled
periodic POST requests. The OpenAPI specifica-
tion
1
was used to allow hospitals to easily produce
clients which export their data to the CDR-API. Ac-
tivity, sensor and questionnaire data from Healthen-
tia is requested from Healthentias API with a ded-
icated component called the Clinical Data Repos-
itory Synchronizer (CDR-SYNC). During patient
creation in the CDR, it links the patient to their ex-
ternal Healthentia ID and syncs activity and question-
naire data via scheduled tasks. The CDR, IG, CDR-
API and CDR-SYNC together form the HDH.
Air quality and weather data for the locations of
all patients in the CDR are continuously collected by
the ML Environmental Data Manager. In order to
use the collected data for ML purposes, the ML Data
Manager retrieves it periodically from the CDR-API
and ML Environmental Data Manager and splits it
into different training and inference datasets. The
datasets have classification and regression targets and
contain a varying number of features as input — de-
pending on how many patient hospital visits are avail-
able in the hospitals EHRs.
3.1.2 ML Training
When the ML Data Manager refreshes the patient data
from the CDR-API, it creates the new datasets for ML
training as well as inference requests datasets, adding
aggregated environmental data in the process. If it
finds that new data has been added since the last syn-
chronization, it triggers ML training. When ML train-
ing is triggered, any datasets that are not suitable for
training (for example because of too few data-points
or missing examples for a class in classification) are
filtered out. The ML Data Manager informs the ML
Training Manager of the suitable datasets with re-
cent changes. The ML Training Manager then initi-
ates both local and federated ML training for a pre-
defined set of ML models for each training target.
The process for local training is that the ML Train-
ing Manager requests the dataset from the ML Data
Manager. It then runs missing value imputation, using
a mean-matching scheme (Morris et al., 2014) wher-
ever possible. The dataset is then normalized, ML
models are iteratively fitted on it, performance metrics
are calculated and if the metrics are above a threshold,
the model is sent to the ML Model Manager.
1
https://edge1-db.test.re-sample.eu/clinical-data-
repository-api/swagger-ui/index.html#/
HEALTHINF 2025 - 18th International Conference on Health Informatics
502

Figure 2: Prediction explanation.
3.1.3 ML Results
For federated training, the process is similar but
also involves the Federated Learning Coordina-
tion, which is located in the orchestrator node at
the secured coordinating central server outside of the
hopistals. Instead of simply starting training like in
the local case, the ML Training Manager submits a re-
quest to the Federated Learning Coordination to start
training of a dataset. If other edge nodes also have
this dataset available, the Federated Learning Coor-
dination initiates federated training and starts the FL
server. The ML Training Managers on all edge nodes
periodically request what dataset and model should
be trained next from the Federated Learning Coordi-
nation. If there is any to start, the ML Training Man-
agers will each start a FL client and request and im-
pute the dataset from their respective ML Data Man-
agers like in the local case. What is different, how-
ever, is that the ML Data Manager will apply differen-
tial privacy (Dwork et al., 2014) to the dataset for ad-
ditional privacy protection of the patients’ data. The
FL clients all fit their model on the dataset and send
the parameters to the server who averages them and
sends them back. This process continues for a speci-
fied number of training rounds. Finally, the FL clients
calculate performance metrics with their own test data
and send the jointly trained model to the ML Model
Manager.
The ML Results Manager periodically retrieves
inference requests for the latest patient data from the
ML Data Manager to calculate updated ML results.
ML results include predictions for every data target as
well as accompanying SHAP values (Molnar, 2020)
to explain it. In addition, counterfactual predictions
are produced for classification targets that show what
changes in the patient data could produce a different
prediction outcome. For this, only the data values that
are actually changeable are considered, such as, e.g.,
activity data or the body mass index. The ML Re-
sults Manager also calculates different simulated pre-
dictions, for which it changes a set of patient data in
a predefined way in order to fully convey what ef-
fect these changes are predicted to have on the target
variable value, visualized in Figure 3. All ML results
are submitted to the CDR-API and stored in the HL7
FHIR CDR, where it can be retrieved for the user in-
terface used by HCPs. Figure 5 shows how the pre-
dicted target values change over time within the dash-
board.
In addition, Figure 2 shows the plot of SHAP val-
ues for the latest target value prediction, which indi-
cates for all input variables with their current values
how much these increased (blue values) or decreased
(red values) the computation of the predicted score.
This provides a mean for the HCPs to assess which
and how values of that specific feature contributed to
the prediction.
3.1.4 Use for Shared Decision-Making
The ML results contain a lot of information about the
patients. As such, they are sensitive data from a pri-
vacy perspective that cannot be disclosed to external
systems. To still be able to display them alongside
the less sensitive activity and questionnaire data, the
LDC was introduced. It is a software component used
in hospital edge nodes to give the Healthentia web
portal access to patient clinical data. This access is
only available within the hospital’s secure network,
as data cannot leave the hospital due to strict pri-
Figure 3: ML simulations.
Federated Learning in Multi-Center, Personalized Healthcare for COPD and Comorbidities: The RE-SAMPLE Platform
503

vacy policies. The LDC allows the clinical dashboard
to display hospital clinical data alongside Healthen-
tia’s data, formatting it for easy visualization in the
Healthentia portal. It also retrieves ML results to be
displayed in the dashboard.
4 RE-SAMPLE PLATFORM
SECURITY AND PRIVACY
In order to ensure patient data security and privacy,
the platform was designed using a privacy by design
approach (Hes and Borking, 1995), meaning that pri-
vacy was taken into account across all stages of the
development life-cycle to ensure that the sensitive
medical data is protected. Furthermore, during the de-
sign phase all legal and technical requirements of the
GDPR were considered. Indicatively, it was ensured
that GDPR principles like the data protection (pur-
pose limitation, data minimisation, accuracy, account-
ability), the lawfulness of processing, and the user
consent, were fully satisfied. In order to accomplish
this a gap analysis was performed on the platform
to identify potential non compliance issues with the
GDPR, and following that a data protection impact
assessment was carried out to identify potential im-
pact of a privacy violation incidents on the data sub-
jects. Finally, a thorough risk analysis was conducted,
in order to estimate the probability of occurrence and
possible consequences of a security incident for the
platform. Through the combination of the aforemen-
tioned procedures a list of both organisational and
technical measures, such as patient anonymity via dis-
tinct IDs per component, authentication, application
hardening, access control, and logging, were imple-
mented to minimize security or privacy incidents.
One of the key aspects of this methodology was to
identify the security domains, in order to ensure ad-
equate measures for their communication. Three se-
curity domains were identified: the hospital network,
the orchestrator node and external domains. The hos-
pital network houses the majority of the components
and is isolated in it’s own docker network with only
the CDR-API having a connection to the hospital in-
formation system, which supplies its data using a se-
cured API call. The LDC exposes the data to the clin-
ician dashboard only when a secure connection is es-
tablished. The orchestrator node houses the federated
learning coordination component for the joint train-
ing of the ML models and is secured by a keycloak
2
based authentication/authorization server. Before ini-
tiating a request a component must first authenticate
2
https://www.keycloak.org/
using the OpenID Connect (OIDC) specification and
then get an authorization token using OAuth 2.0. The
token is then sent to an NGINX reverse proxy, that
forwards the token to the keycloak instance that then
allows access to the endpoint.
The platform’s security scope ensures that the
edge node operates within a secure hospital network,
limiting physical access to aggregated data in the
HDH. Within the edge node, components have re-
stricted access to relevant data, with identifier map-
pings stored in a local database.
5 DEPLOYMENTS IN
PRODUCTION
The platform is currently up and running in three col-
laborating European hospitals, where it is used dur-
ing an interventional clinical study. The data used
to train the machine learning models comprise retro-
spective data from approximately 1,000 patients and
prospective data collected during the observational
study from almost 200 patients. Hospitals typically
store historical data in EHRs or study databases. In
the RE-SAMPLE project, some hospitals have cre-
ated data pipelines that automatically retrieve clini-
cal data from internal hospital information systems.
In one hospital, clinical data such as blood measure-
ments and inpatient access information can be di-
rectly extracted from structured data sources, while
spirometry measurements, six-minute walking test re-
sults, and other relevant details can be extracted from
pseudonymized clinical reports via text mining. One
hospital updated their internal system using forms
to manually ingest the data by implementing new
forms for the study data to send them to the edge
node. The third hospital is working with HiX Digi-
tal Health Services
3
and has built a pipeline to auto-
matically transform the data from the hospital infor-
mation system into the correct uniform format to send
the clinical data to the CDR-API. In all three collab-
orating hospitals, prospective patient data are dynam-
ically collected following the common RE-SAMPLE
data model, with scheduled daily updates that make
it available to the data ingestion components of the
RE-SAMPLE platform. The HCP facing dashboard
is visualized in Figure 5, and the patient-facing smart-
phone app is shown in Figure 1.
3
https://www.chipsoft.com/en
HEALTHINF 2025 - 18th International Conference on Health Informatics
504
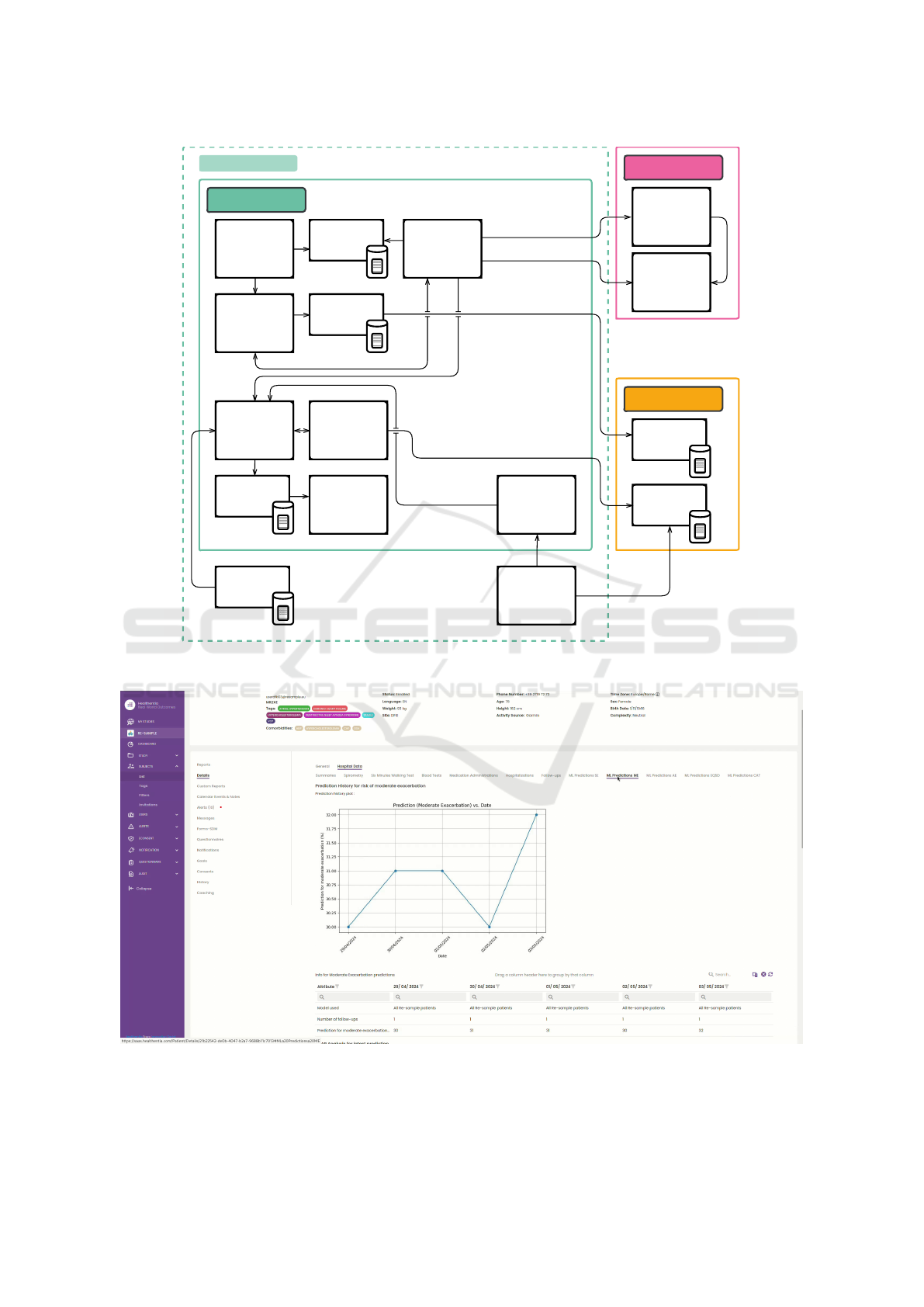
Training
Manager
Results
Manager
Data
Manager
Edge Node
Environmental
Data Manager
Model
Manager
HCP/GP/Admin
Webbrowser
Hospital
Information
System
Orchestrator Node
Federated
Learning
Coordination
Authentication
Server
External
Healthentia
Environmental
Data Services
Local Data
Connector
Clinical Data
Repository
Synchronizer
Clinical Data
Repository API
Clinical Data
Repository
Hospital Network
FHIR
Implementation
Guide
Figure 4: Architecture diagram.
Figure 5: Screenshot of the clinical dashboard.
Federated Learning in Multi-Center, Personalized Healthcare for COPD and Comorbidities: The RE-SAMPLE Platform
505

6 CONCLUSION
In this paper, we described the RE-SAMPLE platform
that can be setup in multiple hospitals for federated
ML model training and to generate personalized treat-
ment suggestions for patients with COPD and comor-
bidities. It enables data storage, synchronisation and
management for patient monitoring for use in shared-
decision making for patients with COPD and comor-
bidities. We described the implemented architecture
of the up-and-running system and the workflows.
To protect patient privacy, we implemented robust
security measures and compliance with healthcare
data protection regulations. Our federated learning
approach ensures patient data remains secure within
each hospital’s environment. All components are
open source.
Future work will include the analysis of the per-
formance of the ML models – in particular comparing
locally trained models to models trained by federated
learning – and the importance of the predictors espe-
cially for COPD exacerbations.
ACKNOWLEDGEMENTS
This paper is part of a project that has received fund-
ing from the European Union’s Horizon 2020 re-
search and innovation programme under grant agree-
ment No. 965315.
REFERENCES
Abdelaziz, A., Elhoseny, M., Salama, A. S., and Riad, A.
(2018). A machine learning model for improving
healthcare services on cloud computing environment.
Measurement, 119:117–128.
Adibi, A., Sin, D. D., Safari, A., Johnson, K. M., Aaron,
S. D., FitzGerald, J. M., and Sadatsafavi, M. (2020).
The acute copd exacerbation prediction tool (accept):
a modelling study. The Lancet Respiratory Medicine,
8(10):1013–1021.
Agust
´
ı, A., Celli, B. R., Criner, G. J., Halpin, D., Anzueto,
A., Barnes, P., Bourbeau, J., Han, M. K., Martinez,
F. J., Montes de Oca, M., et al. (2023). Global initia-
tive for chronic obstructive lung disease 2023 report:
Gold executive summary. American journal of respi-
ratory and critical care medicine, 207(7):819–837.
Bender, D. and Sartipi, K. (2013). HL7 FHIR: An agile and
restful approach to healthcare information exchange.
In Proceedings 26th IEEE international symposium
on computer-based medical systems, pages 326–331.
Beutel, D. J., Topal, T., Mathur, A., Qiu, X., Fernandez-
Marques, J., Gao, Y., Sani, L., Kwing, H. L., Par-
collet, T., Gusm
˜
ao, P. P. d., and Lane, N. D. (2020).
Flower: A friendly federated learning research frame-
work. arXiv preprint arXiv:2007.14390.
Dwork, C., Roth, A., et al. (2014). The algorithmic founda-
tions of differential privacy. Foundations and Trends®
in Theoretical Computer Science, 9(3–4):211–407.
Habehh, H. and Gohel, S. (2021). Machine learning in
healthcare. Current genomics, 22(4):291.
Hassan, F., Shaheen, M. E., and Sahal, R. (2020). Real-time
healthcare monitoring system using online machine
learning and spark streaming. International Journal of
Advanced Computer Science and Applications, 11(9).
Hes, R. and Borking, J. (1995). Privacy-enhancing tech-
nologies: The path to anonymity.
Kyriazis, D., Autexier, S., Boniface, M., Engen, V.,
Jimenez-Peris, R., Jordan, B., Jurak, G., Kiourtis, A.,
Kosmidis, T., Lustrek, M., et al. (2019). The crowd-
health project and the hollistic health records: Col-
lective wisdom driving public health policies. Acta
Informatica Medica, 27(5):369.
Lampropoulos, K., Kosmidis, T., Autexier, S., Savi
´
c, M.,
Athanatos, M., Kokkonidis, M., Koutsouri, T., Vizitiu,
A., Valachis, A., and Padron, M. Q. (2021). ASCAPE:
An open AI ecosystem to support the quality of life of
cancer patients. In 2021 IEEE 9th Int. Conference on
Healthcare Informatics (ICHI), pages 301–310.
Molnar, C. (2020). Interpretable machine learning. Lulu.
com.
Morris, T. P., White, I. R., and Royston, P. (2014). Tuning
multiple imputation by predictive mean matching and
local residual draws. BMC medical research method-
ology, 14:1–13.
Nunavath, V., Goodwin, M., Fidje, J. T., and Moe, C. E.
(2018). Deep neural networks for prediction of ex-
acerbations of patients with chronic obstructive pul-
monary disease. In Engineering Applications of Neu-
ral Networks: 19th International Conference, EANN
2018, Bristol, UK, September 3-5, 2018, Proceedings
19, pages 217–228. Springer.
Patel, N. (2024). An update on copd prevention, diagnosis,
and management: The 2024 gold report. The Nurse
Practitioner, 49(6):29–36.
PRASAD, B. (2020). Chronic obstructive pulmonary dis-
ease (copd). International Journal of Pharmacy Re-
search & Technology (IJPRT), 10(1):67–71.
Rahman, A., Hossain, M. S., Muhammad, G., Kundu,
D., Debnath, T., Rahman, M., Khan, M. S. I., Ti-
wari, P., and Band, S. S. (2023). Federated learning-
based ai approaches in smart healthcare: concepts,
taxonomies, challenges and open issues. Cluster com-
puting, 26(4):2271–2311.
Tang, C., Plasek, J. M., Zhang, H., Xiong, Y., Bates, D. W.,
and Zhou, L. (2018). A deep learning approach to han-
dling temporal variation in chronic obstructive pul-
monary disease progression. In 2018 IEEE Interna-
tional Conference on Bioinformatics and Biomedicine
(BIBM), pages 502–509.
HEALTHINF 2025 - 18th International Conference on Health Informatics
506
