
Impact of Biased Data Injection on Model Integrity in Federated
Learning
Manuel Lengl
1∗ a
, Marc Benesch
1∗ b
, Stefan R
¨
ohrl
1 c
, Simon Schumann
1 d
, Martin Knopp
1,2 e
,
Oliver Hayden
2 f
and Klaus Diepold
1 g
1
Chair of Data Processing, Technical University of Munich, Germany
2
Heinz-Nixdorf Chair of Biomedical Electronics, Technical University of Munich, Germany
Keywords:
Federated Learning, Data Bias, Privacy Violation, Membership Inference Attack, Blood Cell Analysis,
Quantitative Phase Imaging, Microfluidics, Flow Cytometry.
Abstract:
Federated Learning (FL) has emerged as a promising solution in the medical domain to overcome challenges
related to data privacy and learning efficiency. However, its federated nature exposes it to privacy attacks and
model degradation risks posed by individual clients. The primary objective of this work is to analyze how
different data biases (introduced by a single client) influence the overall model’s performance in a Cross-Silo
FL environment and whether these biases can be exploited to extract information about other clients. We
demonstrate, using two datasets, that bias injection can significantly affect model integrity, with the impact
varying considerably across different datasets. Furthermore, we show that minimal effort is sufficient to infer
the number of training samples contributed by other clients. Our findings highlight the critical need for robust
data security mechanisms in FL, as even a single compromised client can pose serious risks to the entire
system.
1 INTRODUCTION
In recent years, Federated Learning (FL) has shown
promising results across various machine learning
fields, offering solutions to some key challenges. FL
successfully addresses critical issues such as insuffi-
cient training data, centralizing sensitive data, and low
training efficiency (Xu et al., 2022). However, every
type of collaborative training also introduces poten-
tial risks. As more parties become involved in the
training process, the likelihood of someone introduc-
ing flawed data increases (Xu et al., 2022). While this
can occur unintentionally, for example, due to differ-
ing measurement setups, it can also be exploited in-
tentionally to disrupt the training process. Addition-
ally, Jegorova et al. (2022) showcase several scenarios
of privacy attacks against FL environments, demon-
strating their effects on data privacy.
a
https://orcid.org/0000-0001-8763-6201
b
https://orcid.org/0009-0005-6004-9644
c
https://orcid.org/0000-0001-6277-3816
d
https://orcid.org/0000-0002-7074-473X
e
https://orcid.org/0000-0002-1136-2950
f
https://orcid.org/0000-0002-2678-8663
g
https://orcid.org/0000-0003-0439-7511
*
These authors contributed equally to this work.
A field where these concerns are particularly criti-
cal is the medical sector. The General Data Protection
Regulation (Voigt and von dem Bussche, 2017) in Eu-
rope imposes stringent requirements on patient data
privacy, making FL an attractive option for collabora-
tion between hospitals (Sohan and Basalamah, 2023).
However, any disturbances in the collaborative pre-
diction models are intolerable, as they could directly
impact patient safety. Therefore, it is crucial to inves-
tigate the potential risks posed by clients introducing
unintended or malicious changes to the training pro-
cess.
Quantitative Phase Imaging (QPI) is an emerging
technology in biology that generates complex image-
based data. It was already successfully applied in
several fields, including oncology (Lam et al., 2019)
and hematology (Fresacher et al., 2023; Klenk et al.,
2023). To harness its potential for solving medical
challenges with machine learning, it requires (like
many image classification tasks) large datasets to de-
velop accurate classification models. This depen-
dence on extensive data makes QPI an ideal use case
for exploring potential threats posed by injected bias,
which can severely affect model accuracy and relia-
bility.
This work aims to address key challenges in FL by
exploring two critical scenarios. First, we analyze the
432
Lengl, M., Benesch, M., Röhrl, S., Schumann, S., Knopp, M., Hayden, O. and Diepold, K.
Impact of Biased Data Injection on Model Integrity in Federated Learning.
DOI: 10.5220/0013153800003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 432-443
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

impact of artificially inducing biases by one client to
intentionally degrade the model’s generalization ca-
pability and overall performance. The primary objec-
tive is to assess the extent to which injected biases
affect a model trained within a FL setup. Second,
we examine a specific white-box attack resembling
a Membership Inference Attack (Salem et al., 2019),
where the adversary seeks to reconstruct partial infor-
mation from other clients’ datasets, raising significant
privacy concerns. In this scenario, we consider a ma-
licious actor with access to a single client’s data pool.
We analyze both scenarios on two distinct datasets
and explore whether systematically designing these
biases could successfully disturb the model and/or
extract insights into other clients’ data. Addition-
ally, we perform a statistical evaluation to determine
whether inherent dataset characteristics provide re-
silience against these attacks, i.e., whether one dataset
shows greater robustness compared to the other.
2 DATA
2.1 Image Acquisition
To show the impact of biased data on real-world ex-
amples, we used a Quantitative Phase Imaging (QPI)
setup to measure human blood samples.
2.1.1 Quantitative Phase Imaging
In general, microscopes based on QPI operate us-
ing the principle of interference between an object
beam and a reference beam to capture the phase shift
of light ∆φ. The shift provides information about
the optical density of the sample interrupting the
object beam. By combining QPI with a microflu-
idic channel and focusing system, this setup allows
for high-throughput sample measurement. Therefore,
this technology is particularly valuable for biomedi-
cal applications (Jo et al., 2019), as it addresses the
key challenge of traditional bright-field microscopy.
In bright-field microscopy, the transparent nature of
biological cells often results in low-contrast images,
making it necessary to apply molecular staining (Bar-
cia, 2007; Klenk et al., 2019). This staining step is
not only time-consuming but can also introduce addi-
tional errors in the processing pipeline. Phase imag-
ing, on the other hand, provides far more detailed in-
sights into cellular structures than intensity images,
without requiring prior labeling.
In this work, we used a customized differential
holographic microscope from Ovizio Imaging Sys-
tems, as illustrated in Figure 1. This system enables
Light Source
Fluidics Chip
Objective
Sheath Flows
Figure 1: Microscope setup.
label-free imaging of untreated blood cells in sus-
pension. Our approach is closely related to off-axis
diffraction phase microscopy (Dubois and Youras-
sowsky, 2008), but it uses a low-coherence light
source and does not require a reference beam. Cells
are precisely focused within a 50×500 µm polymethyl
methacrylate microfluidic channel. Four sheath flows
are used to center the blood cells within the chan-
nel, preventing contact with the channel walls. This
setup allows for measuring 105 frames per second
with an average of 5 cells per frame. The resulting
frames have a size of 384×512 pixels, with an exam-
ple shown in Figure 2. Further details about this mi-
croscope are available in Dubois and Yourassowsky
(2011) and Ugele et al. (2018).
20 µm
0
1
2
3
4
∆φ in rad
Figure 2: Sample frame.
2.1.2 Pre-Processing
To prepare the image frames for further analysis, sev-
eral additional steps are necessary.
Background Cleaning. The output frames of the
microscope setup may contain background artifacts
and noise originating from the microfluidic channel or
camera lens. Due to the fixed orientation of the lens,
camera, light, and microfluidics, these disturbances
tend to remain consistent during individual measure-
ments and, therefore, can be effectively approximated
by calculating the median of all images. By subtract-
ing this background approximation from each frame
individually, the resulting images are much cleaner
and ready for further processing.
Impact of Biased Data Injection on Model Integrity in Federated Learning
433

Segmentation. To identify individual cells in the
frames, we apply binary thresholding with a threshold
of 0.3 rad and extract patches with a size of 48 × 48
pixels around each detected cell. From these patches,
we extract binary masks that cover the areas of the in-
dividual cells. To further refine the obtained masks,
we apply a Gaussian filter with a standard deviation
of σ = 0.5 (Gonzalez and Woods, 2002), smoothing
the mask transitions.
Filtering. Although the necessary biological pre-
processing steps are minimal, the high-throughput, to-
gether with the isolation process that is required to
isolate the different subtypes, can still result in the de-
struction of some cells. To filter out these damaged or
fragmented cells, we calculate the 2D area each cell
covers and discard any cell with an area smaller than
357 µm² (equivalent to 30 pixels). Cells or particles
below this threshold are typically remnants from the
isolation process.
2.2 Datasets
In this study, we use both a publicly available bench-
mark dataset and a curated, domain-specific dataset
from the medical field to provide comprehensive in-
sights.
CIFAR-4. The first dataset is derived from CIFAR-
10 (Krizhevsky, 2009), a well-known benchmark in
machine learning research, which we use as a refer-
ence for comparing results obtained from the Leuko-
cyte dataset. CIFAR-10 comprises 60,000 images,
each 32 × 32 pixels, evenly distributed across 10
classes. To facilitate a more realistic comparison, we
reduced this dataset to include only four classes —
airplane, automobile, ship, and truck — resulting in
the CIFAR-4 dataset, which aligns with the four-class
classification structure of the Leukocyte dataset.
Figure 3: CIFAR-4 examples.
Leukocyte. The second dataset, acquired using our
setup described in Section 2.1, contains samples from
various leukocyte subtypes, with the goal of per-
forming a four-part differential
1
. This classification
1
Basophils are excluded due to their low occurrence.
distinguishes between monocytes, lymphocytes, neu-
trophils, and eosinophils. We obtained the sepa-
rated cell types from whole blood samples by apply-
ing the isolation protocol according to Ugele (2019)
and Klenk et al. (2019). The complete dataset in-
cludes 447,541 images of white blood cells from
three healthy donors, each paired with a correspond-
ing segmentation mask. To align with our reference
dataset, CIFAR-4, which contains only 24,000 im-
ages, we reduced the Leukocyte dataset to the same
size. The sample distribution across the four classes
is balanced, and we applied a Min-Max scaling with
min = −1 and max = 7, normalizing the data to the
range [0, 1] (Bishop, 2006). It is important to note that
this dataset contains single-channel images, in con-
trast to the CIFAR-4 dataset with three-channel im-
ages.
Figure 4: Leukocyte examples. (Phase shift of the single-
channel images is color mapped to imitate the appearance
of a Giemsa stain (Barcia, 2007)).
3 METHODOLOGY
3.1 Classification Model
For our experiments, we used a Convolutional Neu-
ral Network based on the AlexNet architecture
(Krizhevsky et al., 2017), which is well-established
in image classification tasks. Our objective in this
work is not to further enhance the already high perfor-
mance of state-of-the-art models but rather to evaluate
the impact of data perturbations. Therefore, AlexNet
is an ideal choice for this purpose, as it offers strong
accuracy while maintaining a relatively low network
complexity, helping to minimize potential confound-
ing factors.
We retained the original AlexNet architecture,
only adapting the first fully connected layer to accom-
modate the different numbers of input channels.
3.2 Federated Learning
As opposed to traditional machine learning, Feder-
ated Learning (FL) enables the cooperation of several
clients to train a common model; without needing to
exchange, share, and store data centrally (McMahan
et al., 2017). Instead of sharing data, the participat-
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
434

ing clients exchange only weight updates from their
locally trained models. In this work, we focus on a
setup with an aggregation server that serves as a cen-
tral orchestrating entity. The server updates its global
model by applying the exchanged weight updates and
returns them to the clients. Each cycle of this process
can be described as one training round.
The overall FL setting can be categorized based
on the topology and data partitioning (Rieke, 2020).
A Cross-Device topology is characterized by a scal-
able and often large number of clients, which may not
always be available. In contrast, a Cross-Silo topol-
ogy involves a smaller number of clients that typi-
cally have identical setups and are always reachable
(Kholod et al., 2020). Data partitioning is typically
classified into two primary types. In horizontal par-
titioning, each client holds a different subset of data
that shares the same features. Converseley, in vertical
data partitioning, the data is split based on features
rather than samples.
In this work, we focused on a horizontal Cross-
Silo setup, which fits well with our clinical context
and imaging data. For this setting, we can describe
the problem of training a machine learning model in
a federated manner as minimizing the objective func-
tion
f(w
1
,..., w
K
) =
1
K
K
∑
k=1
f
k
(w
k
), w
k
∈ R
d
, (1)
where K is the number of clients and f
k
(w
k
) is the
local objective function of one client with model
weights w
k
corresponding to that specific client’s
dataset. We assume a theoretical dataset D =
{(x
1
,y
1
),..., (x
N
,y
N
)} with N being the total num-
ber of samples, partitioned across the K clients.
The dataset is partitioned such that D =
S
k
D
k
and
T
k
D
k
= {}, hence each client holds n
k
= |D
k
| sam-
ples. The local optimization problem can be formu-
lated as
min
w
k
ℓ(x,y;w
k
) = min
w
k
1
n
k
∑
i∈D
k
ℓ(x
i
,y
i
;w
k
), (2)
where ℓ(·) is some loss function.
For the aggregation of the weights on the cen-
tral server, we use the Federated Average approach
designed by McMahan et al. (2017). Each train-
ing session starts with the central server distributing
a random set of initial weights w
(t)
(with t = 0) to
the clients. Note that w
(t)
denotes the global model
weights, while w
(t)
k
represents the local model weights
for client k. A local update is then performed using,
for example, gradient descent
w
(t+1)
k
← w
(t)
− η∇ℓ(·;w
(t)
k
), (3)
where η is the learning rate.
The server aggregates the updated local weights
using a weighted average to compute the global model
for the next time step
w
(t+1)
← w
(t)
− η
K
∑
k=1
n
k
n
∇ℓ(·;w
(t)
k
), (4)
where
∑
K
k=1
n
k
n
∇ℓ(·;w
(t)
k
) = ∇ℓ(·;w
(t)
). Multiple local
updates using Equation 3 can be performed before the
central aggregation continues with the next time step
t + 1 (McMahan et al., 2017).
In our experiments, we used an independent and
identically distributed data split to avoid additional
complexity or noise. Our implementation is based on
the Flower
2
Python framework, designed for simulat-
ing FL procedures.
3.3 Bias Types
To investigate the impact of a single client contribut-
ing biased images, we applied various types of biases.
Each bias has a variable that controls the severity of
the bias, we later refer to this as bias strength. Note
that if the application of a bias results in pixel values
outside the normalized range [0,1], these values are
clipped.
Brightness. The brightness of an image refers to the
overall lightness or luminance of the image. It is a
perceptional term that describes how light or dark an
image appears to a viewer. Brightness is generally
associated with the intensity of light that the viewer
perceives from the image (Gianfrancesco et al., 2018).
Technically, brightness in an image can be quantified
by the average intensity of the pixels in the image.
Each pixel has a brightness value, which is typically
represented on a scale from 0 to 255 for all three
color channels, where 0 represents black (no bright-
ness) and 255 represents white (full brightness). By
adding a constant value to the pixel values, one can
artificially let the image be perceived as brighter or
darker. In this work we added different values in the
range of [−1,1] to achieve this.
Contrast. Contrast refers to the spread of the pixel
intensities of an image. High contrast images dis-
play a large difference between light and dark areas,
whereas low contrast images might appear flat or dull
due to the closer range of tones. Contrast manipula-
tion is a common technique extensively described by
Gonzalez and Woods (2002). In 8-bit images normal-
ized to pixel values in the range [0, 1], we can define
2
https://flower.ai/
Impact of Biased Data Injection on Model Integrity in Federated Learning
435

a neutral value, called midpoint in our implementa-
tion, around which contrast adjustments are centered.
Given the pixel range of the images, this value is typ-
ically 0.5. We can manipulate the contrast by scaling
the difference from the midpoint by a contrast fac-
tor α, and then adding the midpoint back to the pixel
value.
X
new
= α· (X
old
− 0.5) + 0.5. (5)
The transformation in Equation 5 scales the pixel val-
ues of image X
old
around the midpoint based on the
contrast factor α, where the operations are applied
element-wise.
Gaussian. Adding Gaussian noise to a clear im-
age can be viewed as simulating real-world condi-
tions for testing image processing algorithms since
almost no imaging technology is free of noise (Gon-
zalez and Woods, 2002). Using it as artificial and sys-
tematic bias requires drawing from the same distribu-
tion z ∼ N(µ,σ
2
), but varying a parameter ε such that
a pixel value x
i, j
is transformed as
x
new,i, j
= x
old,i, j
+ ε· z. (6)
We used Equation 6 with z ∼ N(0,1) in our imple-
mentation.
Edge. Convolving an image with special kernel ma-
trices ω
ω
ω, which are well-known from image process-
ing, apply some effect to the image depending on the
kernel. This can be used to amplify or decrease the
extracted features from the original image X. A gen-
eral approach can be mathematically formulated as
X
new
= X
old
+ b· M, (7)
where M is a binary mask to identify the entries of the
matrix that are greater than a threshold T
M = ω
ω
ω∗ X
old
> T =
(
0, x
i j
≤ T
1, x
i j
> T
. (8)
The entries of M are amplified by a scalar bias value
b, which can be negative or positive. For the sake
of simplicity, Equations 7 and 8 take only gray-scale
images into consideration. But the same can be ap-
plied to multi-channel images, too. We mainly used
an edge-detection kernel with
ω
ω
ω =
−1 −1 −1
−1 8 −1
−1 −1 −1
(9)
to emphasize the edges within the image, enhancing
their visibility.
Box Blur. We can introduce a Box Blur bias to sim-
ulate the effect of some imperfections by smoothing
out the image details (Gonzalez and Woods, 2002),
which mimics the loss of fine detail often seen in real-
world data acquisition. One can also use this to build
a more robust model with higher generalization ca-
pability. We will examine this effect to see to what
extent this type of bias leads to more robustness and
when it results in performance degradation.
Similar to the edge detection, we convolve an im-
age with a kernel ω
ω
ω:
ω
ω
ω =
1
9
1 1 1
1 1 1
1 1 1
, (10)
which averages a pixel value based on the neighbor-
ing pixels. The size of the kernel determines the blur
effect. We stick to one blur effect, but instead adjust
the strength of it by using Alpha Blending (Hughes,
2014). Essentially, we interpolate between the origi-
nal image and the blurred image to control how much
of the blurred image is mixed with the original image
based on the parameter ρ. Mathematically, we per-
form the following transforming steps:
X
filtered
= ω
ω
ω∗ X
old
, (11)
X
new
= (1 − ρ) · X
old
+ ρ· X
filtered
. (12)
Adversarial. Adversarial attacks involve intention-
ally adding malicious perturbations to a model’s input
to deceive it. While there are different types of attacks
(Alhajjar et al., 2021), this work focuses on extraction
attacks in a white-box scenario, as they align with our
second goal of gaining knowledge from other clients.
Before diving further into the details of our approach,
we will provide a linear explanation of why adversar-
ial examples are effective.
Due to limited input feature precision and quanti-
zation in digital images, for instance, a pixel intensity
below the threshold of
1
255
(for 8-bit representation,
2
8
= 256) cannot be captured and is effectively dis-
carded. If we add a perturbation z smaller than the
feature precision to the original signal x, we obtain
˜
x = x + z. A model will not be able to distinguish be-
tween x and
˜
x, as long as the perturbation is bounded
by {z :∥ z ∥
∞
= max
i
|z
i
| ≤ ε}. Now, consider a ma-
chine learning model with a weight vector w. Ap-
plying this weight vector to the adversarial example
˜
x
gives:
w
⊤
˜
x = w
⊤
x + w
⊤
z. (13)
This implies two things. First, w
⊤
z influences the
activation. Second, since ∥ z ∥
∞
is independent of
the dimension of z, but w
⊤
z increases (or decreases)
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
436
with the dimension of w, this causes a compound ef-
fect, leading to a significant change in the output for
high dimensional spaces (which is particularly rele-
vant for deep learning models). We can amplify this
effect by selecting z = sign(w) while ensuring the
constraint on z holds. Note that this does not mean
z exceeds the ε-bound; instead, we choose ε such that
∥ ε · sign(w) ∥
∞
= ε (Madry et al., 2017). Also, note
that the sign function is applied element-wise (Good-
fellow et al., 2014).
Goodfellow et al. (2014) introduced a computa-
tionally efficient technique to approximate these per-
turbations: the Fast Gradient Sign Method (FGSM).
This method assumes a loss function ℓ(w,x, y), where
w are the parameters of a neural network. We can then
obtain an optimal max-norm constrained perturbation
(Goodfellow et al., 2014)
z = ε· sign(∇
x
ℓ(w,x, y)). (14)
Varying ε controls the magnitude of the perturba-
tion added to the image, and its optimal value ranges
heavily depend on the dataset and domain.
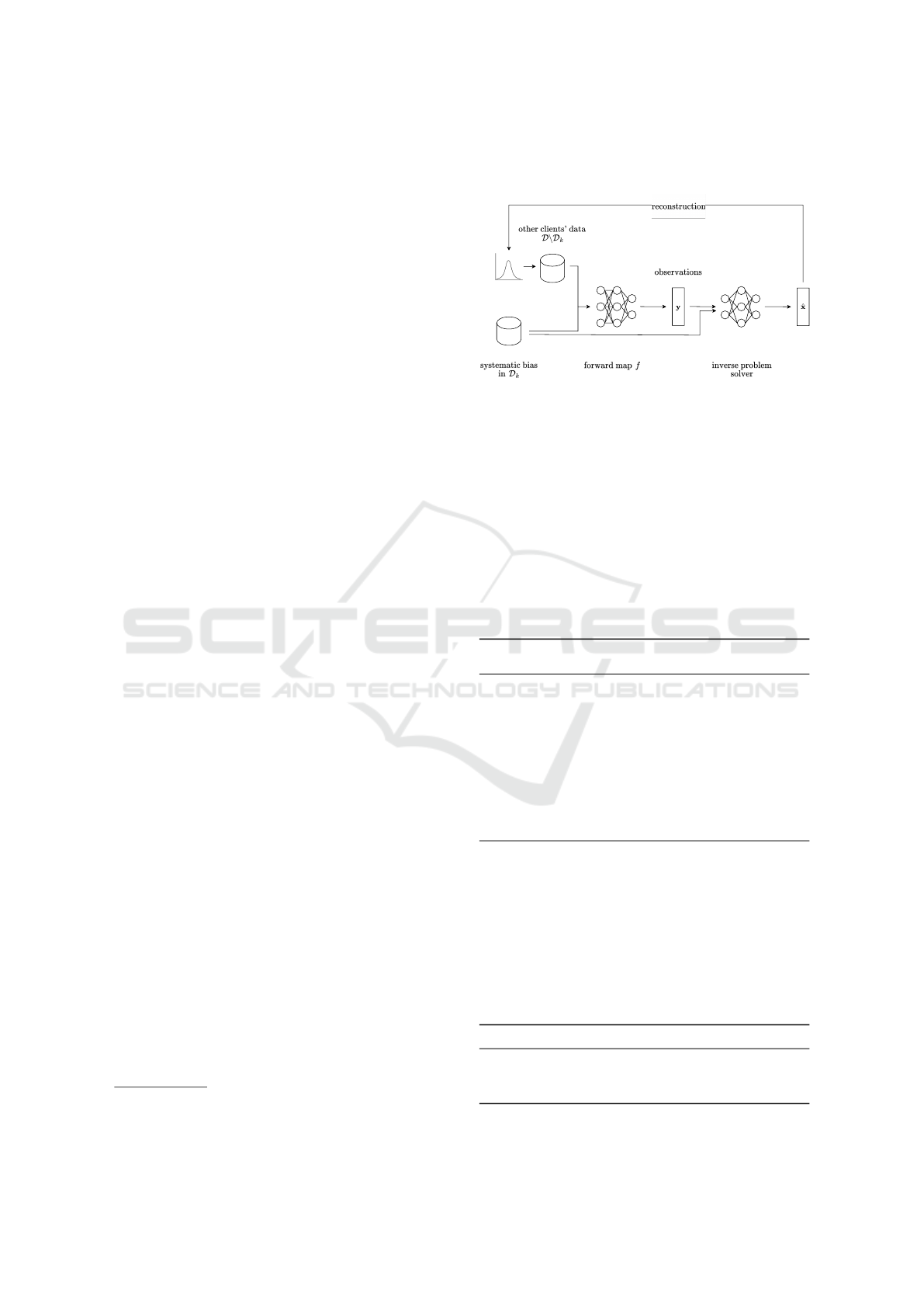
3.4 Inverse Problem Solving
For the goal of inferring information about the
datasets of other clients, we formulate the challenge
as an inverse problem since the malicious client does
not have direct access to the sample counts of other
clients. Mathematically, we assume a fixed dataset
D, where subsets of this dataset represent the data
shares of different clients in a FL framework. D
k
de-
notes the dataset of a compromised client, whereas
D
¯
k
represent the dataset of all other clients, such that
D = D
k
∪ D
¯
k
and D
¯
k
= D \D
k
. Our goal is to esti-
mate |D
¯
k
|, when only D
k
is known. We have access
to the results of a forward map f (D
k
,D
¯
k
), which we
observe as
y = f (D
k
,D
¯
k
) + z, (15)
where y are vectors of metrics from FL training pro-
cesses for all biases and z represents uncertainty in the
forward model, accounting for fluctuations observed
during training. These fluctuations arise from the ran-
domness introduced during bias injection and train-
ing
3
, even with a fixed seed.
Consequently, we employed three different super-
vised machine learning models - Linear Regression
(LR) (James et al., 2013), Support Vector Regression
(SVR) (Vapnik et al., 1996), and Random Forest Re-
gression (RFR) (Breiman, 2001) - as inverse problem
solvers to estimate the number of traning samples the
3
for example due to mini-batch sampling differences and stochastic op-
timization techniques
other clients have contributed D
¯
k
. Figure 5 provides
a visualization of this approach.
Figure 5: Overview of the pipeline setup for solving the
inverse problem.
For simplicity, we assume that the total number of
training examples across all clients remains constant.
A more complex version of this problem could be for-
mulated without this assumption.
We conduct multiple simulations with varying ra-
tios between D
k
and D
¯
k
. In each simulation, we apply
the biases described in Section 3.3 and perform sev-
eral FL trainings. The resulting metric vector is saved
as one observation for each ratio, respectively. This
process is summarized in the following algorithm:
Algorithm 1: Dataset Size Variation Analysis in Federated
Learning with Bias Injection.
1: Fix dataset D
2: for each ratio in set of ratios do
3: Split dataset D into D
k
and D
¯
k
4: for each bias in grid do
5: Apply bias to D
k
6: Perform FL training with D
k
and D
¯
k
7: Calculate metrics as observation y
8: end for
9: end for
After analyzing the loadings of a Principal Com-
ponent Analysis (Bishop, 2006) performed on the ob-
servations, we discard features with redundant infor-
mation. The input to the regression models then in-
clude Bias Type, Strength, Kullback-Leibler Diver-
gence, and Accuracy. Exemplary samples are shown
in Table 1.
Table 1: Exemplary inputs for the regression models.
Bias Type Strength KL-D Accuracy
Gaussian 0.05 0.02 0.95
Edge 0.4 0.07 0.88
...
Impact of Biased Data Injection on Model Integrity in Federated Learning
437

3.5 Evaluation Metrics
For evaluating the performance of the classification,
we calculate Accuracy as a general performance met-
ric and the F1 score to have a balanced metric be-
tween false positives and false negatives (Bishop,
2006). Additionally, we calculate the Kullback-
Leibler Divergence (KL-D) to quantify how the distri-
bution of the predictions of the biased models deviates
from that of the unbiased model (Bishop, 2006). KL-
D is particularly sensitive to small changes, making it
useful for capturing subtle effects of bias variations.
However, it assumes a probability distribution, so it is
necessary to normalize the prediction frequencies by
the total number of predictions. This process is illus-
trated in Figure 6.
Figure 6: Using KL-D in a four-class classification task.
The evaluation of the inverse problem solver
is performed using the Root Mean Squared Error
(RMSE) between the predicted and the actual dataset
size.
3.6 Training Setup
For training AlexNet, we use the Adam optimizer
with a fixed learning rate of 0.001. The model is
trained with a batch size of 32, and we use Cross En-
tropy as the loss function (Goodfellow et al., 2016).
Training is conducted over 4 FL rounds with 3 epochs
each, and 4 participating clients. For the RFR we use
100 trees with a maximum depth of 10. The minimum
number of samples per leaf is set to 2, and the mini-
mum number of samples required to split an internal
node is 5. The SVR is implemented using the radial
basis function kernel, a regularization parameter of 1
and an ε value of 0.1. These parameters were experi-
mentally determined after performing a grid search.
All experiments are run for 5 different random
seeds. For the AlexNet, we always use 4,000 sam-
ples as test set, and the remaining ones are split into
training and validation with a ratio of 75:25.
4 RESULTS
4.1 Bias Injection Impact on Federated
Learning Performance
In the first experiment, we analyze the impact of a sin-
gle client applying various bias types and strengths to
their share of data on the performance of our two dis-
tinct datasets. Even with identical training procedures
and bias strengths, the impact is expected to vary sig-
nificantly depending on the dataset.
4.1.1 Results on Single Dataset
Initially, we visually assess how different strengths of
biases affect the model performances. For easier vi-
sual comparability, we normalized the bias strengths
to either [−1,1] or [0, 1], depending on the possible
signs of their strength variable. For the analysis, we
focus on Accuracy as the primary performance met-
ric, but the tendencies remain the same across all met-
rics. The resulting curves are expected to show a re-
versed U-shape or V-shape for biases that can have
both positive and negative strengths, indicating that
with stronger absolute bias, performance decreases.
For biases with only positive strengths, the curve will
show a one-sided shape.
To then statistically evaluate the significance of
these effects, we apply a One-Sample t-Test
4
(James
et al., 2013), which compares the mean performance
under a certain bias with a hypothetical mean (i.e., the
performance without bias). Since we ran our simula-
tions with different random seeds, we can calculate
the mean and variance of a specific metric (e.g., Ac-
curacy) for each bias and strength. The hypotheses
for the t-Test are formulated as follows:
• Null Hypotheses (H
0
): There is no significant dif-
ference in performance when a specific bias and
strength is applied.
4
We assume normal distribution due to various sources
of randomness during training (e.g., weight initialization,
optimization, data shuffling). Each model prediction can be
considered a random variable, and according to the Central
Limit Theorem (Moore et al., 2017), the sum or average of
many such random variables tends to follow a normal dis-
tribution. A Shapiro-Wilk test (Moore et al., 2017) further
supports this assumption, although it has lower power for
small sample sizes.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
438
• Alternative Hypotheses (H
1
): The change in per-
formance due to the specific bias and strength is
significant.
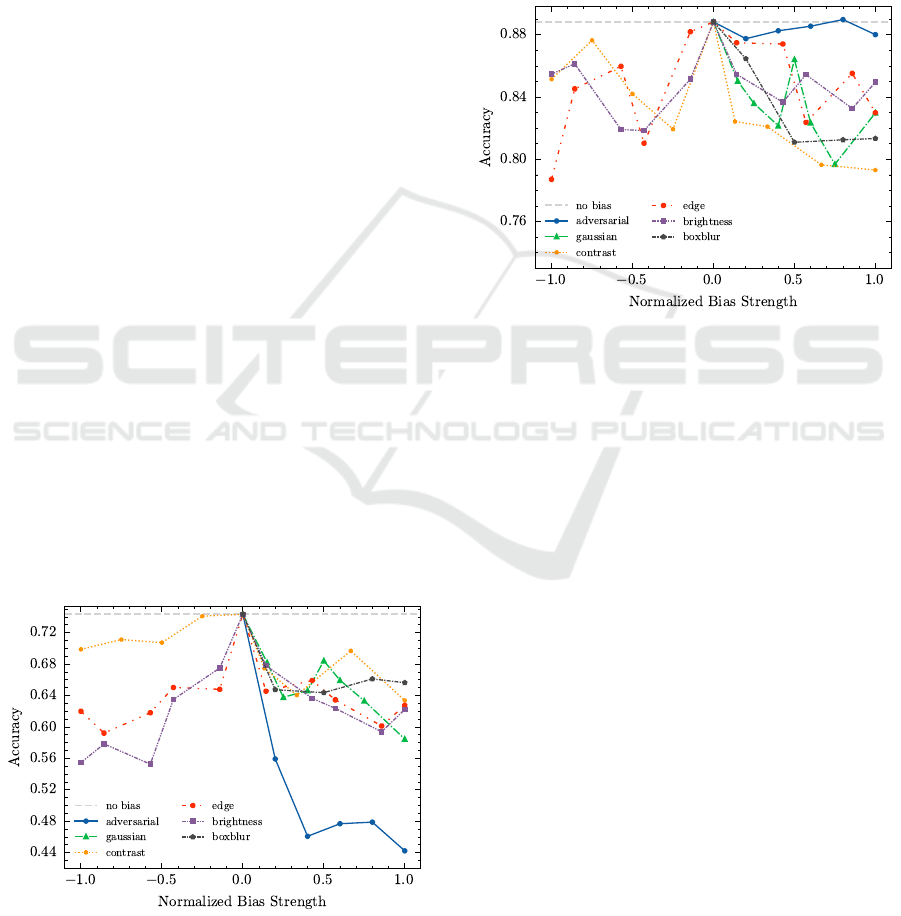
CIFAR-4. For the CIFAR-4 dataset, Figure 7 shows
the Accuracy across different bias types as a function
of bias strength. The corresponding p-values are pre-
sented in Table 5.
Starting with Brightness and Edge biases, the
curves exhibit the expected inverse U-shape. Low
bias has minimal effect on the model’s performances
across all metrics. However, as the absolute bias
strength increases, performance drops significantly,
which is supported by the statistical test showing sig-
nificant impact for almost all strength levels. Inter-
estingly, for Brightness, there is a slight skew notice-
able: negative strength values cause a larger decline
than equivalent positive values, a result that is further
confirmed by the t-Test.
The effect of Contrast bias diverges from expec-
tations. While positive strength results in the ex-
pected performance decreases, negative values do not
show this tendency. They appear to have little in-
fluence, with performance decreasing only slightly
with stronger negative values. Consequently, negative
strengths do not show a significant impact.
Gaussian noise almost consistently degrades per-
formance as strength increases, with a counter-
intuitive outlier for a normalized strength of 0.5. This
anomaly is likely due to experimental errors.
The Box Blur bias shows a steep initial decrease
in performance, which then quickly saturates. Even
at maximum blur (normalized bias strength = 1.0), the
performance drop remains mostly unchanged. Addi-
tionally, this bias has high standard deviations, mak-
ing it difficult to draw definitive conclusions about its
impact.
Figure 7: Accuracy across different bias types and strengths
for the CIFAR-4 dataset. All data points show the mean of
5 runs, standard deviation is not shown for clarity.
Finally, following the theoretical explanation in
Section 3.3, we successfully create Adversarial at-
tacks that highly impact the model’s performance.
Even with small perturbations (e.g., normalized
strength = 0.2), imperceptible to the human eye, the
metrics show a substantial drop with high confidence
(i.e., low standard deviation). Max-norm perturba-
tions above 0.4 induced by the FGSM algorithm dras-
tically reduce Accuracy to between 0.4 and 0.5, which
is quite poor for a four-class classification task. These
findings are further supported by very low p-values.
Figure 8: Accuracy across different bias types and strengths
for the Leukocyte dataset. All data points show the mean of
5 runs, standard deviation is not shown for clarity.
Leukocyte. In the Leukocyte dataset, as shown in
Figure 8, the effects of biases are generally less pro-
nounced, which is supported by the higher p-values in
Table 5 in the appendix.
For Brightness and Edge bias, the decline in per-
formance is minimal. However, a subtle U-shape is
apparent, suggesting that higher bias values have a
slightly greater impact on the model than lower ones.
Brightness shows again a slight skewness, both visu-
ally and statistically, but overall, the impact remains
surprisingly low (especially given that this bias could
alter the apparent size of the cells).
The behavior of Contrast shows again a skewed
U-shape, with a tendency for less impact when Con-
trast is decreased. However, the effect of negative
strengths is more noticeable than in the CIFAR-4
dataset. Overall, no bias strength yields a significant
impact.
For Gaussian noise, we observe a gradually in-
creasing decline in performance. While the impact is
not drastic, each noise level still results in a measur-
able performance drop. The curve looks quite similar
to the pattern seen for CIFAR-4, with a similar peak at
0.5, making it worthwile to investigate further.
With Box Blur, added to the cell images, the
Impact of Biased Data Injection on Model Integrity in Federated Learning
439

model’s performance degrades, but the impact satu-
rates once the normalized strength exceeds 0.5. Fur-
ther increases do not cause any noticeable changes in
performance drop. All impacts are again statistically
significant.
Adversarial attacks show a much smaller effect
than anticipated. While we would expect stronger
adversarial perturbations to severely degrade the
model’s performance, the actual impact is minimal.
The model’s Accuracy remains largely unaffected
across the range of adversarial strengths, indicating
that the dataset is relatively robust to these attacks.
4.1.2 Comparison
The p-values in Table 5 together with the visual in-
spection in the previous sections already suggest that
the CIFAR-4 dataset is generally more sensitive to the
introduced biases compared to the Leukocyte dataset.
To statistically evaluate whether one dataset is inher-
ently more robust against different types of biases, we
conducted a two-way ANOVA (Moore et al., 2017),
assessing the effect of the dataset. This allows us
to test whether the differences in robustness between
the two datasets are statistically significant. While
ANOVA traditionally compares group means, we for-
mulate hypotheses to interpret whether one dataset is
significantly more robust to the biases than the other:
• Null Hypothesis (H
0
): There is no significant
difference in robustness between Leukocyte and
CIFAR-4 when a specific bias is applied, imply-
ing similar performance changes.
• Alternative Hypothesis 1 (H
1
): There is a signif-
icant difference in robustness between Leukocyte
and CIFAR-4 when a specific bias is applied, im-
plying that performance for one dataset is signifi-
cantly more impacted.
The null hypotheses H
0
can be rejected for a given
bias type if the p-value is below 0.05.
Table 2 summarizes the outcome of the analysis.
The results show, that for several bias types, there is
a significant difference in robustness. Four out of six
types have a p-value below 0.05, with extremely low
values in some cases. Confirming with Table 5 to see
which dataset is impacted more heavily shows that the
Leukocyte dataset is more robust in all four cases of
Adversarial, Brightness, Edge, and Gaussian Bias, al-
though the significance for the latter is not as strong
as for the others.
For the Box Blur, both datasets are impacted sig-
nificantly, with no inherent difference, even though
the p-value is relatively small at p = 0.076. Interest-
ingly, in the case of the Contrast bias, there is no dif-
ference at all, suggesting that both datasets are simi-
larly affected (or not affected) by changes in strength.
Table 2: Results of a two-way ANOVA to assess whether
one dataset is inherently more robust against different bias
types. p-values below 0.05 are marked in bold.
Bias Type df(D,R) F-value p-value
Adversarial (1,48) 218.53 1.71 × 10
−19
Box Blur (1,38) 3.33 0.07601
Brightness (1,97) 63.95 2.72 × 10
−12
Contrast (1,78) 0.13 0.72458
Edge (1,97) 31.73 1.73 × 10
−7
Gaussian (1,67) 10.49 0.00187
4.2 Systematic Knowledge Extraction
from Other Clients
In the second experiment, we aim to infer informa-
tion about the datasets of other clients. As an initial
approach, we attempt to estimate the ratio of data con-
tributed by each client - specifically, how many sam-
ples other clients contribute to the training process.
We vary the ratio by adjusting the number of samples
contributed by the compromised client. As input for
the prediction, we use the observations of the metrics
y derived from various FL training runs under differ-
ent ratios and induced biases.
4.2.1 Results on Single Dataset
We first evaluate the success of the attack on individ-
ual datasets. For this purpose, we train regression
models to predict the amount of training data con-
tributed by the other client. Additionally, to support
the general assumption of a correlation between the
malicious clients’ share of data and the resulting drop
in performance, we visually examine whether a trend
is present.
CIFAR-4. Table 3 presents the outcomes for the
three regression models. When considering all bias
types and strengths, RFR performs best with a RMSE
of 3,812. Given that the maximum number of train-
ing samples is 20,000 images, a RMSE of 3,812 is
rather underwhelming. However, further fine-tuning
the training by using only biases that showed to have
a high impact drastically improves the results.
By applying only Brightness bias with strengths
greater than 0, the RMSE for RFR drops to 2,204,
even though we use only a fraction of the original
training samples. For the LR model, the RMSE ac-
tually drops to 1,948. Filtering the input further by
using samples with strengths greater than or equal to
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
440
0.4 improves the results even more. This notable im-
provement suggests that carefully crafting and induc-
ing bias can provide insights into the other clients’
dataset sizes. Knowing which bias type and strength
significantly impacts model performance appears to
aid in this process.
Table 3: RMSEs on the CIFAR-4 dataset. # is the amount of
training data, RMSE is on the test data with 120 samples.
Applied Bias # RMSE
LR SVR RFR
All Bias Types 481 4094 4153 3812
Only Brightness (> 0) 70 1948 3534 2204
Only Brightness (≥ 0.4) 34 1302 2191 1636
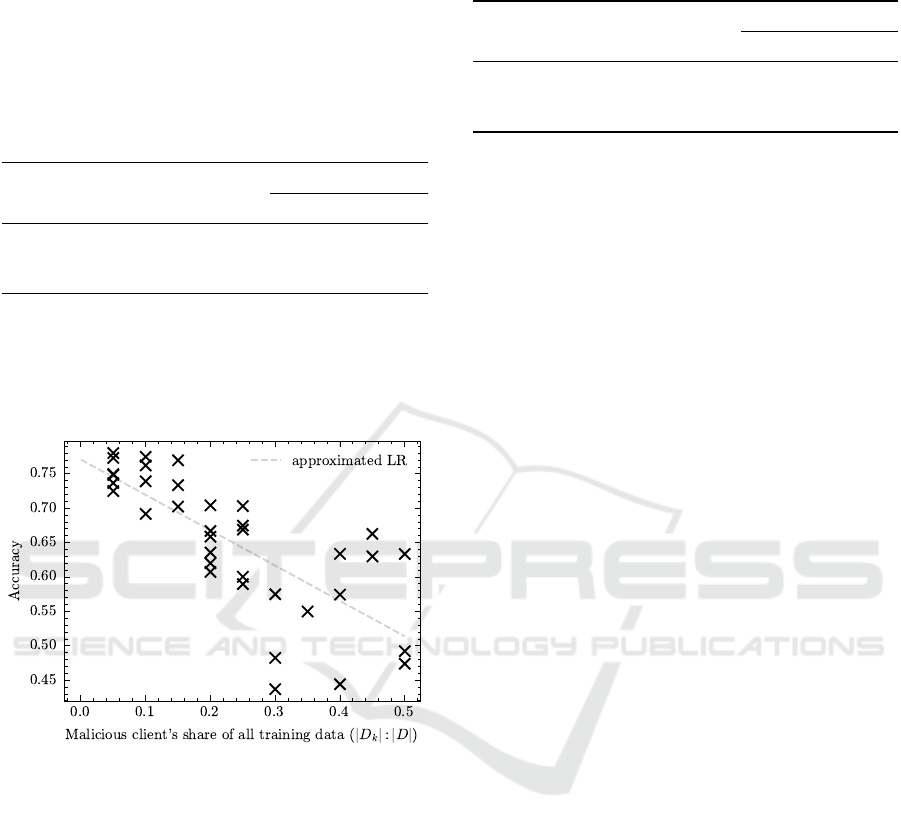
The visual examination of the correlation between
the malicious clients’ share and the overall perfor-
mance, as shown in Figure 9, aligns perfectly with
our expectations; a higher contribution of biased data
results in decreased performance.
Figure 9: Model Accuracy for strong Brightness biases
(strength ≥ 0.4) as function of the proportion of data the
malicious client contributed. Results shown for CIFAR-4.
Leukocyte. Using the same approach as in the pre-
vious section, we perform training on the Leukocyte
dataset with all biases, as well as with selected Bright-
ness biases. The results for all models are shown in
Table 4. This time, the RFR model outperforms the
others in all runs. With a RMSE of 1,430 when us-
ing all biases and a RMSE of 688 when only using
strong Brightness biases, the RFR demonstrates ex-
cellent performance. An error of 688 samples when
predicting a dataset size of 20,000 corresponds to a
relative error of 3.44%.
Table 4: RMSEs on the Leukocyte dataset. # is the amount
of training data, RMSE is on the test data with 120 samples.
Applied Bias # RMSE
LR SVR RFR
All Bias Types 481 1729 2481 1430
Only Brightness (> 0) 70 1621 2749 1314
Only Brightness (≥ 0.4) 34 1181 1688 688
4.2.2 Comparison
Comparing Tables 3 and 4, this experiment shows a
clear trend: the Leukocyte dataset is more vulnera-
ble to this attack. Across all runs and for each model
tested, the RMSE values are consistently lower than
for the CIFAR-4 dataset. Given the clear pattern of
these results, we decided not to pursue further statis-
tical analyses, as the observed trend is both strong and
conclusive for the scope of this experiment.
5 CONCLUSION
Summarizing the findings from the experiments, this
work provides valuable insights into how bias types,
strengths, and dataset characteristics influence the
performance of models in Federated Learning (FL)
environments. Additionally, we demonstrated how
these factors can be exploited by a malicious adver-
sary in a white-box scenario, revealing vulnerabilities
in FL systems under adversarial conditions.
In the first experiment, we systematically ana-
lyzed the effect of six bias types across two datasets,
revealing that the performance drop due to bias varies
significantly depending on the dataset and the bias
type. The CIFAR-4 dataset was generally more sensi-
tive to most biases compared to the Leukocyte dataset,
with Adversarial, Brightness, Edge, and Gaussian bi-
ases showing particularly strong effects. This sug-
gests that the Leukocyte dataset is inherently more ro-
bust to these types of perturbations, possibly due to
the nature of the images.
The statistical analyses, including the One-
Sample t-Tests and two-way ANOVA, supported
these findings by confirming significant differences
between the datasets, particularly for Adversarial and
Brightness biases. However, contrast bias showed
minimal impact on both datasets, indicating that the
model is relatively unaffected by this type of bias.
In the second experiment, we explored the pos-
sibility of estimating the number of samples other
clients contributed by inducing bias and evaluating
the regression models’ performance. Here, the Leuko-
cyte dataset demonstrated more vulnerability, with
Impact of Biased Data Injection on Model Integrity in Federated Learning
441

consistently lower RMSE values across all models,
particularly when Brightness bias was selectively ap-
plied. This suggests that biases can be exploited to
infer client data contributions, though the effective-
ness varies between datasets. Although the strategy
employed in this work is from a more theoretical na-
ture, we empirically proved that the Leukocyte dataset
is highly vulnerable to such threats. Only a few
collected data points were sufficient for a successful
knowledge retrieval.
In conclusion, the results highlight the importance
of understanding how bias type and dataset character-
istics interact to affect FL model performance. These
insights can help designing more robust and secure
FL systems, particularly in settings where data hetero-
geneity and malicious clients may pose risks. Overall,
one cannot draw general conclusions across different
datasets. Experiments must be carefully planned and
executed when it comes to data manipulation, such as
the injection of biases. Given the highly sensitive na-
ture of human health data, we recommend conducting
even more nuanced research regarding these datasets.
Especially in FL, where each client constitutes a vul-
nerability, one compromised client can cause serious
trouble, making it essential to pursue state-of-the-art
data security mechanisms.
For future work, it would be interesting to exam-
ine additional bias types to strategically extract dif-
ferent information from honest clients. Additionally,
none of the models presented in this work were op-
timized, and we used the same architectures to en-
sure a fair comparison. However, given that different
datasets can yield completely different conclusions
even with the same architecture and circumstances,
optimizing models for specific datasets and rerunning
the same attacks could be beneficial. Considering the
promising results, we believe this approach could lead
to a significant performance boost and would be worth
further investigation.
ACKNOWLEDGEMENTS
The authors would like to especially thank their colleagues
from the Heinz-Nixdorf Chair of Biomedical Electronics -
D. Heim and C. Klenk - for performing sample preparation
and measurements.
REFERENCES
Alhajjar, E., Maxwell, P., and Bastian, N. (2021). Ad-
versarial machine learning in Network Intrusion De-
tection Systems. Expert Systems with Applications,
186:115782.
Barcia, J. J. (2007). The Giemsa Stain: Its History and Ap-
plications. International Journal of Surgical Pathol-
ogy, 15(3):292–296.
Bishop, C. M. (2006). Pattern Recognition and Ma-
chine Learning. Information Science and Statistics.
Springer, New York.
Breiman, L. (2001). Random Forests. Machine Learning,
45(1):5–32.
Dubois, F. and Yourassowsky, C. (2008). Digital holo-
graphic microscope for 3D imaging and process using
it.
Dubois, F. and Yourassowsky, C. (2011). Off-axis interfer-
ometer.
Fresacher, D., R
¨
ohrl, S., Klenk, C., Erber, J., Irl, H.,
Heim, D., Lengl, M., Schumann, S., Knopp, M.,
Schlegel, M., Rasch, S., Hayden, O., and Diepold,
K. (2023). Composition counts: A machine learning
view on immunothrombosis using quantitative phase
imaging. Proceedings of Machine Learning Research,
219:208–229.
Gianfrancesco, M. A., Tamang, S., Yazdany, J., and Schma-
juk, G. (2018). Potential Biases in Machine Learn-
ing Algorithms Using Electronic Health Record Data.
JAMA Internal Medicine, 178(11):1544.
Gonzalez, R. C. and Woods, R. E. (2002). Digital Image
Processing. Prentice Hall, Upper Saddle River, N.J,
2nd ed edition.
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep
Learning. Adaptive Computation and Machine Learn-
ing. The MIT Press, Cambridge.
Goodfellow, I. J., Shlens, J., and Szegedy, C. (2014). Ex-
plaining and harnessing adversarial examples. CoRR,
abs/1412.6572.
Hughes, J. F. (2014). Computer Graphics: Principles and
Practice. Addison-Wesley, Upper Saddle River, New
Jersey, third edition edition.
James, G., Witten, D., Hastie, T., and Tibshirani, R. (2013).
An Introduction to Statistical Learning, volume 103
of Springer Texts in Statistics. Springer, New York.
Jegorova, M., Kaul, C., Mayor, C., O’Neil, A. Q., Weir, A.,
Murray-Smith, R., and Tsaftaris, S. A. (2022). Sur-
vey: Leakage and Privacy at Inference Time. IEEE
Transactions on Pattern Analysis and Machine Intel-
ligence, pages 1–20.
Jo, Y., Cho, H., Lee, S. Y., Choi, G., Kim, G., Min, H.-s.,
and Park, Y. (2019). Quantitative Phase Imaging and
Artificial Intelligence: A Review. IEEE Journal of
Selected Topics in Quantum Electronics, 25(1):1–14.
Kholod, I., Yanaki, E., Fomichev, D., Shalugin, E.,
Novikova, E., Filippov, E., and Nordlund, M. (2020).
Open-Source Federated Learning Frameworks for
IoT: A Comparative Review and Analysis. Sensors,
21(1):167.
Klenk, C., Erber, J., Fresacher, D., R
¨
ohrl, S., Lengl, M.,
Heim, D., Irl, H., Schlegel, M., Haller, B., Lahmer,
T., Diepold, K., Rasch, S., and Hayden, O. (2023).
Platelet aggregates detected using quantitative phase
imaging associate with COVID-19 severity. Commu-
nications Medicine, 3(1):161.
BIOINFORMATICS 2025 - 16th International Conference on Bioinformatics Models, Methods and Algorithms
442

Klenk, C., Heim, D., Ugele, M., and Hayden, O. (2019).
Impact of sample preparation on holographic imaging
of leukocytes. Optical Engineering, 59(10):1.
Krizhevsky, A. (2009). Learning multiple layers of features
from tiny images.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2017). Im-
ageNet classification with deep convolutional neural
networks. Communications of the ACM, 60(6):84–90.
Lam, V. K., Nguyen, T., Phan, T., Chung, B.-M., Nehmetal-
lah, G., and Raub, C. B. (2019). Machine Learning
with Optical Phase Signatures for Phenotypic Profil-
ing of Cell Lines. Cytometry Part A, 95(7):757–768.
Madry, A., Makelov, A., Schmidt, L., Tsipras, D., and
Vladu, A. (2017). Towards Deep Learning Models
Resistant to Adversarial Attacks.
McMahan, H. B., Moore, E., Ramage, D., Hampson, S.,
and y Arcas, B. A. (2017). Communication-Efficient
Learning of Deep Networks from Decentralized Data.
Moore, D. S., McCabe, G. P., and Craig, B. A. (2017). In-
troduction to the Practice of Statistics. W.H. Freeman,
Macmillan Learning, New York, ninth edition edition.
Rieke, N. (2020). The future of digital health with federated
learning. page 7.
Salem, A., Zhang, Y., Humbert, M., Berrang, P., Fritz, M.,
and Backes, M. (2019). ML-Leaks: Model and Data
Independent Membership Inference Attacks and De-
fenses on Machine Learning Models. In Proceedings
2019 Network and Distributed System Security Sym-
posium, San Diego. Internet Society.
Sohan, M. F. and Basalamah, A. (2023). A Systematic Re-
view on Federated Learning in Medical Image Analy-
sis. IEEE Access, 11:28628–28644.
Ugele, M. (2019). High-Throughput Hematology Analy-
sis with Digital Holographic Microscopy. PhD thesis,
Friedrich-Alexander-Universit
¨
at, Erlangen-N
¨
urnberg.
Ugele, M., Weniger, M., Stanzel, M., Bassler, M., Krause,
S. W., Friedrich, O., Hayden, O., and Richter, L.
(2018). Label-Free High-Throughput Leukemia De-
tection by Holographic Microscopy. Advanced Sci-
ence, 5(12):1800761.
Vapnik, V., Golowich, S., and Smola, A. (1996). Support
vector method for function approximation, regression
estimation and signal processing. In Mozer, M., Jor-
dan, M., and Petsche, T., editors, Advances in Neu-
ral Information Processing Systems, volume 9. MIT
Press.
Voigt, P. and von dem Bussche, A. (2017). The EU Gen-
eral Data Protection Regulation (GDPR): A Practical
Guide. Springer, Cham, Switzerland.
Xu, A., Li, W., Guo, P., Yang, D., Roth, H., Hatamizadeh,
A., Zhao, C., Xu, D., Huang, H., and Xu, Z.
(2022). Closing the Generalization Gap of Cross-
silo Federated Medical Image Segmentation. In 2022
IEEE/CVF Conference on Computer Vision and Pat-
tern Recognition (CVPR), pages 20834–20843, New
Orleans. IEEE.
APPENDIX
Table 5: p-values of One-Sample t-Test for different bias
types and strengths. Values below 0.05 are marked in bold.
Bias Type Strength Normalized p-value
Strength Leukocyte CIFAR-4
adversarial 0.05 0.2 0.10049 0.00013
adversarial 0.1 0.4 0.26677 0.00005
adversarial 0.15 0.6 0.76482 0.00033
adversarial 0.2 0.8 0.59898 0.01023
adversarial 0.25 1 0.18156 0.00366
boxblur 0.2 0.2 0.04639 0.00657
boxblur 0.5 0.5 0.04319 0.01250
boxblur 0.8 0.8 0.04700 0.03888
boxblur 1.0 1 0.03407 0.01082
brightness -0.7 -1 0.00390 0.00025
brightness -0.6 -0.86 0.07892 0.00003
brightness -0.4 -0.57 0.07115 0.00068
brightness -0.3 -0.43 0.09971 0.01009
brightness -0.1 -0.14 0.08177 0.05862
brightness 0.1 0.14 0.24948 0.01797
brightness 0.3 0.43 0.01723 0.01976
brightness 0.4 0.57 0.02486 0.01203
brightness 0.6 0.86 0.01637 0.00450
brightness 0.7 1 0.06022 0.00134
contrast 0.2 -1 0.04463 0.00147
contrast 0.4 -0.75 0.17572 0.17743
contrast 0.6 -0.5 0.12090 0.08683
contrast 0.8 -0.25 0.09401 0.74708
contrast 1.2 0.13 0.06418 0.05162
contrast 1.5 0.33 0.06123 0.02293
contrast 2.0 0.67 0.11998 0.04025
contrast 2.5 1 0.11422 0.03791
edge -0.7 -1 0.20720 0.00096
edge -0.6 -0.86 0.22619 0.02740
edge -0.4 -0.57 0.10591 0.00995
edge -0.3 -0.43 0.16661 0.00417
edge -0.1 -0.14 0.65725 0.01054
edge 0.1 0.14 0.04556 0.00284
edge 0.3 0.43 0.29284 0.03785
edge 0.4 0.57 0.00126 0.02180
edge 0.6 0.86 0.08363 0.00108
edge 0.7 1 0.06179 0.00508
gaussian 0.03 0.15 0.07305 0.01742
gaussian 0.05 0.25 0.15497 0.00826
gaussian 0.08 0.4 0.03487 0.00389
gaussian 0.1 0.5 0.00217 0.00698
gaussian 0.12 0.6 0.01362 0.00556
gaussian 0.15 0.75 0.05628 0.00454
gaussian 0.2 1 0.16167 0.00021
Impact of Biased Data Injection on Model Integrity in Federated Learning
443
