
Using LLMs to Extract Adverse Drug Reaction (ADR) from Short Text
Monika Gope and John Wang
School of Information and Communication Technology, Griffith University, Gold Coast, Australia
Keywords:
LLM, ADR Extraction, ADR Detection/Classification, ML.
Abstract:
Adverse drug reactions (ADRs) are unexpected negative effects of a medication despite being used at its nor-
mal dose. Awareness of ADRs can help pharmaceutical companies refine drug formulations or adjust dosing
guidelines to make medications safer and more effective. Twitter (X) can be a handy platform to extract unbi-
ased ADR data from a large and diverse group of people. However, extracting ADRs from short texts such as
tweets presents challenges due to the informal, noisy, and diverse nature of the text, which includes variations
in user language, abbreviations, and misspellings. These factors make it difficult to accurately identify ADRs.
Hence, it is important to identify the most effective strategies for extracting reliable ADR information. In
this paper, we comprehensively evaluate various large language models (LLMs) and ML approaches for ADR
extraction and detection. Using multiple ADR datasets and a range of prompt formulations, we compare the
performance of each model. By systematically testing the effectiveness of these techniques across different
combinations of models, datasets, and prompts, we aim to identify the most effective strategies for extracting
reliable ADR information. Our study shows that LLMs excel in extracting ADRs, for example, with GPT-4
achieving an F1 score of 0.82, surpassing the previous ML methods of 0.64 for the SMM4H dataset. This in-
dicates that LLMs are more effective and simpler alternatives to machine learning models for ADR extraction.
1 INTRODUCTION
Adverse drug reactions refer to unintended and unfa-
vorable responses to taking medications. For an effect
to be considered an ADR, there should be a clear and
reasonable connection between the appropriate dose
of a medication and the harmful effect. ADR extrac-
tion and detection from social media data is an ac-
tive area of research (Luo et al., 2024). Various ML
techniques are used to detect adverse drug reactions
(ADRs). In a sequence labelling-based approach,
(Song et al., 2017) used a conditional random field
(CRF)(Lafferty et al., 2001) model to label ADRs
and indications in tweets. Neural network-based tech-
niques such as self-attention (Vaswani et al., 2017)
capsule networks (Hinton et al., 2011), adversarial
networks (Denton et al., 2015), recurrent neural net-
work (Gupta et al., 2018), convolutional neural net-
work with recurrent neural network (Zhang and Geng,
2019) and deep convolutional neural network (Span-
dana and Prakash, 2024) have also proven effective.
Neural network approaches like Bidirectional long
short-term memory (BLSTM) models have also been
utilized, with (Cocos et al., 2017) and (Ding et al.,
2018) for tweets. (Florez et al., 2018) use BLSTM
for medical notes with extensive feature engineering.
(Tutubalina and Nikolenko, 2017) combined BLSTM
and CRF for ADR detection in user reviews. How-
ever, these methods often struggle to keep up with
this dynamic linguistic landscape (Hughes and Song,
2024).
Transfer learning-based methods like BERT
(Vaswani et al., 2017) and graph neural networks
(Zhao et al., 2020), have shown significant effective-
ness in ADR detection. A hybrid model that com-
bines the BERT and CNN has also proven effective
in ADR detection (Li et al., 2020). Furthermore,
BioBERT (Lee et al., 2020), a variant of BERT pre-
trained on biomedical text in various biomedical NLP
tasks, has also shown effective results on ADR de-
tection (Breden and Moore, 2020). Similarly, models
like RoBERTa (Liu, 2019) have been fine-tuned for
specific applications of transfer learning methods in
handling NLP challenges like ADR detection (Kalyan
and Sangeetha, 2020). Another transformer-based ar-
chitecture for ADR extraction is (Scaboro et al., 2023)
which performs an extensive evaluation of models
like BERT and RoBERTa, showing their superior per-
formance on different datasets.
The rapid progress of Large Language Models
548
Gope, M. and Wang, J.
Using LLMs to Extract Adverse Drug Reaction (ADR) from Short Text.
DOI: 10.5220/0013160700003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineer ing Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 548-555
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

(LLMs) demonstrates the ability to understand and
generate human-like text across multiple domains.
However, despite their impressive performance in dif-
ferent fields, little work has been done to use LLMs
to extract ADRs(Li et al., 2024). In this work, we
use LLMs to extract ADRs from social media text and
structured short texts and compare their performance
with existing state-of-the-art approaches. We make
the following contributions:
• We evaluate the performance of seven different
LLMs over four benchmark data sets and com-
pare them with that of ML methods for extracting
ADR.
• We evaluate the performance of LLMs to classify
ADR and compare them with ML methods.
The rest of the paper is organized as follows: In Sec-
tion 2, we describe our approach. Section 3 reports
the experimental results. Finally, in section 4, we
present the conclusion.
2 OUR APPROACH
2.1 Extraction
In this section, we describe the process of extract-
ing and detecting ADRs from short text using large
language models (LLMs) via APIs using appropriate
prompts. This involves several steps: LLM API in-
tegration, prompting LLMs with different techniques,
etc. After extraction, to evaluate the results, we label
each sentence with the “ADR” term and perform an
approximate match with the labeled sentences.
2.1.1 Accessing Language Models
Accessing Language Models via APIs: The LLMs
used in this experiment are accessed through APIs
that allow users to send text input and receive outputs.
The API call sends a POST request to the model’s
server, where the dataset and the chosen prompt are
included in the request body. The LLM processes the
input text according to the commands provided in the
prompt and returns a structured response. In this case,
the response is a list of identified ADRs.
2.1.2 Prompting for ADR Extraction
To optimize the extraction of ADRs, we apply three
prompting techniques: simple prompting, few-shot
prompting, and chain-of-thought (CoT) prompting
(Ahmed and Devanbu, 2023). Each of these tech-
niques provides the model with a different level of
context and task explanation.
In zero-shot/simple prompting, the instruction
given to the LLM is minimal and direct. The model is
asked to extract ADRs based solely on the current in-
put without any additional examples or explanations.
Few-shot prompting involves giving the model a few
examples of how to extract ADRs from texts. In this
work based on our experiments, we use three exam-
ples in few-shot. Chain-of-Thought prompting en-
courages the model to explain its reasoning step by
step. This is particularly useful for complex or am-
biguous texts where ADRs may not be immediately
obvious. The model is asked to break down the text,
identify relevant phrases, and then filter for ADRs.
Part of Our Few-shot Prompt
Your task is to carefully read the tweet, and
pinpoint the portion of the tweet that contains
the adverse drug reaction (ADR) For example
Tweet: I don’t know what that has to do with
me. (medicine name) has hurt my connective
tissue, lungs, and thyroid. I guess I should feel
lucky
Output: hurt my connective tissue, lungs, and
thyroid
Part of Our CoT Prompt
Start by carefully reading the tweet. Think
through the following steps:
1. Identify Potential adverse drug reactions
(ADRs): Look for any phrases that might indi-
cate an adverse drug reaction. These are often
symptoms or negative effects.
2. Ignore Irrelevant Content: Disregard any
drug names or unrelated content that doesn’t
describe a reaction.
3. Extract the ADR: Pinpoint the specific part
of the tweet that mentions the ADR and extract
it.
2.2 Evaluation Methods
Our task is to extract ADRs using LLMs and com-
pare the results with previous methods (Kayesh et al.,
2022). To enable us to do a fair comparison with the
ML models, we model the problem similar to (Kayesh
et al., 2022) as follows:
Once we have the extracted ADRs, use preci-
sion, recall, and other metrics for measuring the per-
formance of ADR extraction. To calculate preci-
sion, etc., we utilize approximate match (Cocos et al.,
Using LLMs to Extract Adverse Drug Reaction (ADR) from Short Text
549

2017). Using the ADRs extracted by the LLMs and
the ADRs from the ground truth, we create two la-
beled versions of the tweets/sentences. In approxi-
mate match, these two types of labeled tweets or text
are given as input.
2.2.1 Sequence Labeling
Each word in the tweet is labeled as either “ADR” (if
the word is part of an ADR) or “O” (if the word is
not part of an ADR). This is called sequence label-
ing. The approximate match then compares these two
labeled versions and checks the matching. For label-
ing ADR words in a text, we use the following equa-
tion(Kayesh et al., 2022). Given a sequence of words
W = [w
1
, w
2
, . . . , w
n
], where n is the total number of
words, the corresponding sequence of labels for W is:
L = [l
1
, l
2
, . . . , l
n
] (1)
where for each i ∈ {1, 2, . . . , n}:
l
i
=
(
ADR if w
i
is an ADR word
O otherwise
(2)
For labeling, we have two inputs: ADR pat-
terns(which can be phrases or single words) and the
original text. To find the match for these ADR pat-
terns, we check all possible sub-phrases in a sequence
and measure their similarity with the patterns.
We use the DistilBERT (Sanh, 2019) model to
generate contextual embeddings for both sub-phrases
in the text and the ADR patterns. The process iden-
tifies the closest matching sub-phrases using two key
metrics: Levenshtein distance and cosine similarity
(here we use the embeddings). Levenshtein distance
measures how different two strings are based on char-
acter edits, while cosine similarity compares their se-
mantic closeness using embeddings from DistilBERT.
As Twitter data often contains typos, spelling mis-
takes, and spacing issues, we use these two metrics
instead of simple string matching to achieve more ac-
curate results. If the cosine similarity exceeds 0.8 and
the Levenshtein distance is minimal, we count it as
a match. The matched sub-phrases are then replaced
with the term ”ADR.”
The labeled text is transformed, with each word la-
beled as either ”O” (other) or “ADR,” as shown in Eq
(1) and (2). For example, the sentence “<medicine
name>made me super drowsy but now I feel ex-
tremely high.” would become [O, O, O, ADR,
ADR, O, O, O, O, ADR, ADR]. We make two ver-
sions of the sequence-labeled text for each tweet: one
with the ground truth ADRs and one with the ADRs
extracted by the LLMs.
2.2.2 Approximate Match
In approximate match one of the annotations should
be a substring of the other. Whereas for an ex-
act match (Subramaniam et al., 2003)the annotations
from extracted ADR and ground truth ( human anno-
tated) ADR should be matched exactly.
For the following tweet: “<medicine
name>made me super drowsy but now I feel
extremely high.” Here ground truths are ‘drowsy’ and
‘high’. And the extracted ADRs are ‘super drowsy’
and ‘extremely high’ Using sequence labeling, we
get labeled ADRs from LLM extraction: [O, O, O,
ADR, ADR, O, O, O, O, ADR, ADR] and ground
truth labeled ADRs sequence : [O, O, O, O, ADR,
O, O, O, O, O, ADR]. Then we count the numbers
of ADRs in a tweet where consecutive ADRs are
counted together. After that, all non-ADR words
are replaced with zero and ADR words with the
corresponding ADR count number: extracted ADR
counts:[0, 0, 0, 1, 1, 0, 0, 0, 0, 2, 2]
and ground truth ADR counts: [0, 0, 0, 0, 1,
0, 0, 0, 0, 0, 2]. Then, we find the index of
ADR sequences from the above results: extracted
index of ADR sequences: [[3, 4], [9,10]] and
ground truth ADR sequences: [[4], [10]]. From
the index, an approximate match is calculated by
finding the partial overlaps between ADR sub-
sequence index in the extracted and ground truth. The
subsequence of index [[3, 4], [9, 10]] from the
extraction matches with the ground truth index[[4],
[10]]. Here we see 2 approximate matches.
Part of Our Detection Prompt
Your task is to classify the following texts as
either ADR (Adverse Drug Reaction) or Non-
ADR. An ADR is any negative or unintended
effect experienced by a patient following the
administration of a drug. A Non-ADR text
refers to general mentions of drugs that do not
report adverse effects. For each sentence, de-
termine if the text indicates an ADR or not.
2.3 ADR Detection
We use a text dataset (ADE corpus/PubMed dataset)
to classify the ADR text. We use a similar process to
detect or classify the ADRs as described previously,
e.g., accessing LLMs with API, writing prompts,
evaluating results with ground truths, etc. Here, we
do not use sequence labeling and approximate match.
We use precision and recall metrics from the classi-
fied results. For classification, we use simple prompt-
HEALTHINF 2025 - 18th International Conference on Health Informatics
550
ing. Here we use llma 8b, GPT-4o-mini, and GPT-4
to detect ADR.
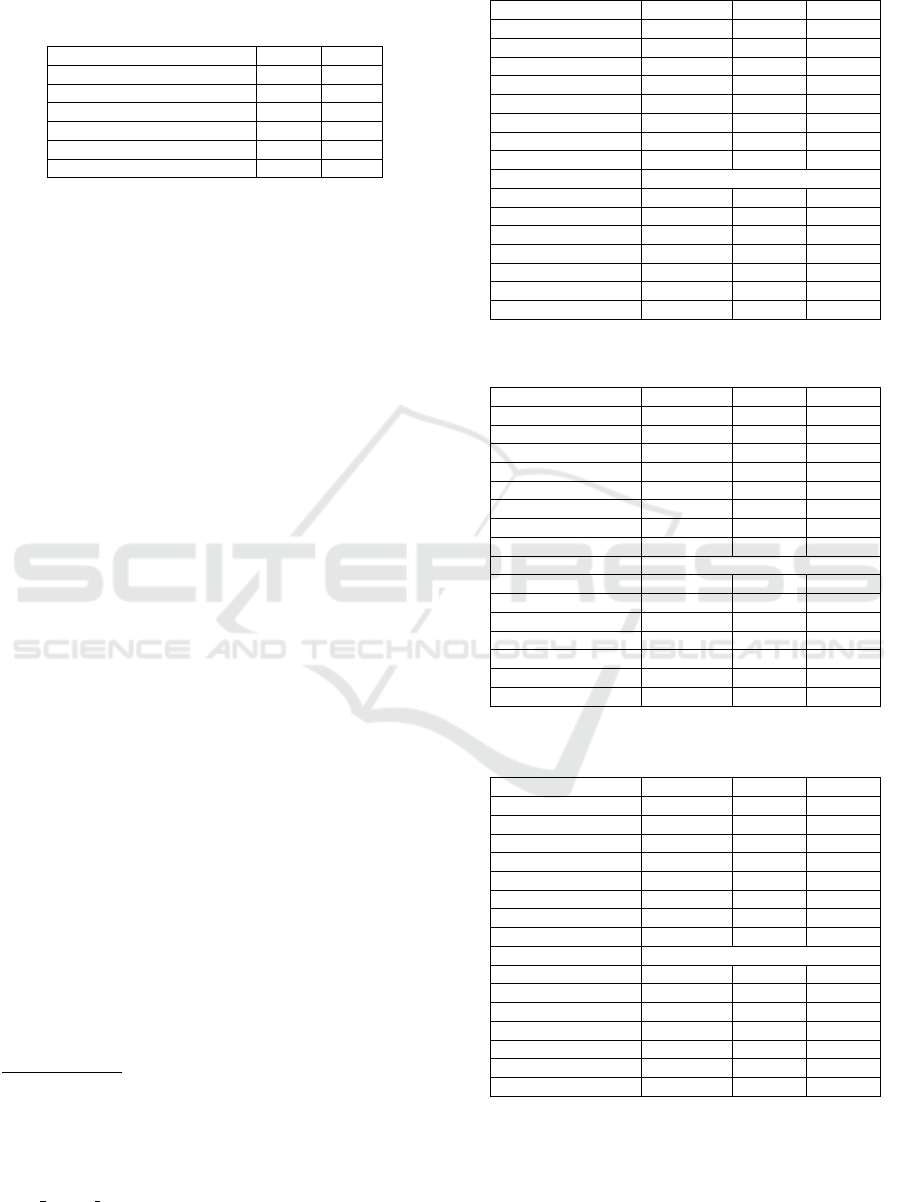
Table 1: Nunber of Tweet/Text in Dataset.
Dataset Total Test
ASU-CHOP 492 172
(SMM4H) 1368 464
WEB-RADR 561 187
Combined 2421 823
ADE Corpus (Extraction) 6.82k 1000
ADE Corpus (Detection) 6.82k 500
3 EVALUATION AND RESULTS
ANALYSIS
For experimental analysis and to ensure consistency
with other methods and datasets, we focused on ex-
tracting ADR from the tweets primarily where each
tweet contains only one drug cause-effect relationship
(Kayesh et al., 2022). In our experiment, the machine
configuration is an Intel i5 processor with 16 GB of
RAM. We run the experiments with Ollma
1
, Grog-
Cloud
2
, and OpenAI
3
.
3.1 Dataset
In this experiment, four publicly available human-
annotated benchmark datasets were used. Three of
these datasets—ASU-CHOP Dataset (Cocos et al.,
2017), Social Media Mining for Health Applications
(SMM4H) Dataset
4
, and WEB-RADR Dataset (Diet-
rich et al., 2020)—are Twitter-based and were com-
bined to create a fifth dataset, referred to as the Twit-
ter dataset. These datasets were provided to us by
the authors of (Kayesh et al., 2022), and we directly
compare our results with the findings presented in
their paper. Besides Twitter data, we used Adverse
Drug Event (ADE) Corpus Version 2
5
which is cre-
ated from medical case reports. Table 1 shows a sum-
mary of the dataset used for the experiments. The
total column indicates the number of total tweets or
texts in the dataset, and the test column shows the
number of texts used in the experiments.
1
https://ollama.com/
2
https://console.groq.com/playground
3
https://openai.com/
4
https://healthlanguageprocessing.org/smm4h19/
5
https://huggingface.co/datasets/ade-benchmark-corpu
s/ade corpus v2
Table 2: Experimental results on the ASU-CHOP Dataset
for LLMs with zero shot with ML methods.
ML Methods Precision Recall F1
CRF 0.8824 0.4688 0.6122
Cocos 0.7189 0.8313 0.7710
CausalBLSTM 0.7770 0.7188 0.7468
CharMHA 0.6748 0.8688 0.7596
CharCausalMHA 0.8235 0.7000 0.7568
MHA 0.7440 0.7813 0.7622
CausalMHA 0.6636 0.8875 0.7594
SCAN 0.7470 0.7750 0.7607
LLM Models
Mistral 7b 0.82 0.69 0.75
Llama 3.1 8b 0.93 0.74 0.82
Gemma2 9b 0.89 0.69 0.78
GPT-4o-mini 0.90 0.74 0.81
GPT-4o 0.96 0.70 0.81
GPT-4-turbo 0.97 0.72 0.83
GPT-4 0.96 0.81 0.88
Table 3: Experimental results on the SMM4H dataset for
LLMs with zero shot with ML methods.
ML Methods Precision Recall F1
CRF 0.5452 0.4342 0.4834
Cocos 0.4748 0.8660 0.6134
CausalBLSTM 0.4689 0.9156 0.6202
CharMHA 0.5261 0.8238 0.6422
CharCausalMHA 0.4759 0.9330 0.6303
MHA 0.4957 0.8610 0.6292
CausalMHA 0.5053 0.8313 0.6285
SCAN 0.4950 0.8561 0.6273
LLM Models
Mistral 7b 0.65 0.75 0.70
Llama 3.1 8b 0.68 0.81 0.74
Gemma2 9b 0.72 0.76 0.74
GPT-4o-mini 0.66 0.83 0.74
GPT-4o 0.71 0.74 0.72
GPT-4-turbo 0.70 0.80 0.75
GPT-4 0.71 0.89 0.79
Table 4: Experimental results on the WEB-RADR dataset
for LLMs with zero shot with ML methods.
ML Methods Precision Recall F1
CRF 0.7833 0.2597 0.3900
Cocos 0.5511 0.6851 0.6108
CausalBLSTM 0.5378 0.6685 0.5961
CharMHA 0.4696 0.8122 0.5951
CharCausalMHA 0.4735 0.7403 0.5776
MHA 0.5468 0.8066 0.6518
CausalMHA 0.4940 0.9116 0.6408
SCAN 0.5094 0.9006 0.6507
LLM Models
Mistral 7b 0.66 0.73 0.69
Llama 3.1 8b 0.66 0.73 0.69
Gemma2 9b 0.66 0.62 0.64
GPT-4o-mini 0.65 0.73 0.69
GPT-4o 0.68 0.59 0.63
GPT-4-turbo 0.63 0.65 0.64
GPT-4 0.71 0.86 0.78
Using LLMs to Extract Adverse Drug Reaction (ADR) from Short Text
551
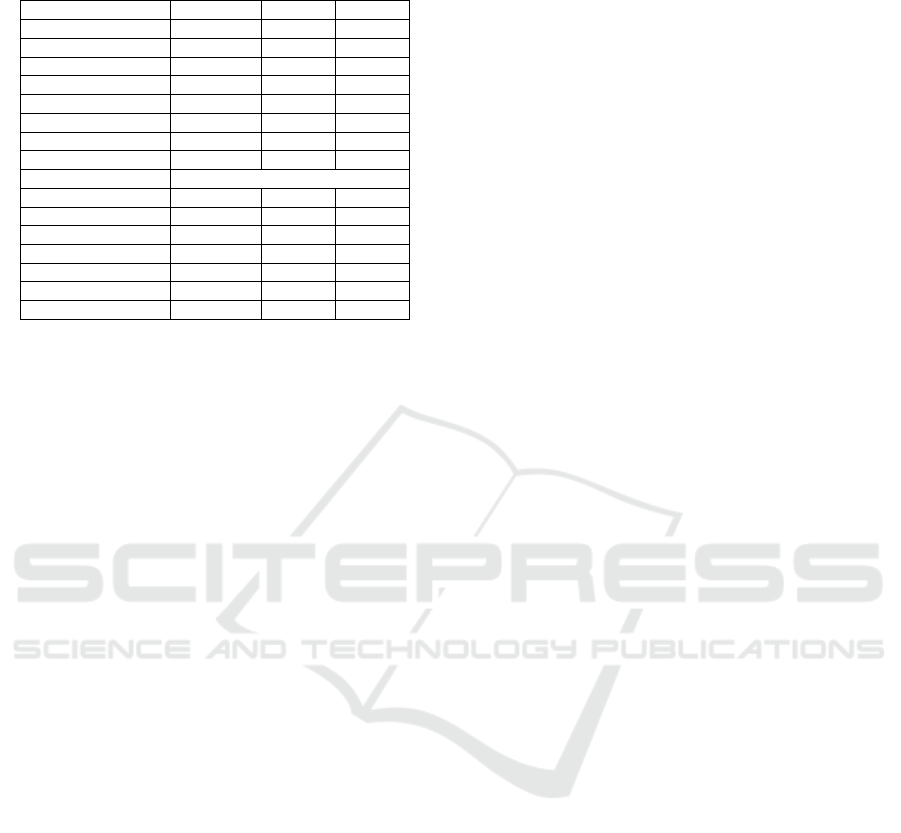
Table 5: Experimental results on the combined Twitter
Dataset for LLMs with zero shot with ML methods.
ML Methods Precision Recall F1
CRF 0.6196 0.4600 0.5280
Cocos 0.5725 0.7993 0.6672
CausalBLSTM 0.5812 0.7371 0.6500
CharMHA 0.5877 0.7798 0.6702
CharCausalMHA 0.5915 0.7869 0.6753
MHA 0.5814 0.7993 0.6731
CausalMHA 0.5772 0.8099 0.6741
SCAN 0.5995 0.7922 0.6825
LLM Models
Mistral 7b 0.68 0.73 0.70
Llama 3.1 8b 0.72 0.78 0.75
Gemma2 9b 0.74 0.71 0.72
GPT-4o-mini 0.70 0.79 0.74
GPT-4o 0.75 0.70 0.72
GPT-4-turbo 0.73 0.75 0.74
GPT-4 0.75 0.87 0.81
3.2 LLMs and ML Methods Used in the
Experiment
Several ML-based ADR extraction approaches with
seven LLMs are used for our experiments. Seven
LLMs are Mistral (7b), Llama 3.1 (8b), Gemma2
(9b), GPT-4, GPT-4o, GGPT-4-turbo, and GPT-4o-
mini. And the ML methods are: Conditional Ran-
dom Field (CRF) model (Lafferty et al., 2001), Co-
cos (Cocos et al., 2017), CausalBLSTM, CharMHA,
CharCausalMHA, MHA, CausalMHA (Kayesh et al.,
2019), shared causal attention network (SCAN)
(Kayesh et al., 2022), GLoVe (Haq et al., 2022),
BERT (Haq et al., 2022), SOTA (Yan et al., 2021),
attentive sequence model (Ramamoorthy and Mu-
rugan, 2018), LSTM (Ding et al., 2018), FARM-
BERT (Hussain et al., 2021), DCNN (Spandana and
Prakash, 2024). (The last four model’s results are
shown in Table 13 for classification experiments.)
3.3 Comparison of ML Models with
LLMs for ADR Extraction and
Detection
In this work, the focus is on extracting and detect-
ing ADR words from the Twitter and ADE corpus
datasets. To maintain consistency with (Kayesh et al.,
2022) we evaluated the results for ADR extraction for
the Twitter dataset. We compare the results for ADR
extraction for short text (ADE corpus - extraction
dataset/ Pubmed dataset) with (Haq et al., 2022). For
ADR detection we compare the results with (Span-
dana and Prakash, 2024) for non-Twitter short text
(ADE corpus/Pubmed dataset ).
3.3.1 Comparison of ML Models with LLMs for
Twitter Dataset
The results of the different datasets with different
models are shown in Tables 2, 3, 4, and 5 for ASU-
CHOP, SMM4H, WEB-RADR, and the combined
dataset, respectively, for zero-shot prompts. From
these tables, we see that the overall results of LLM
models are better than the ML methods. For all four
datasets, the LLM model GPT-4 achieves the highest
F1 scores for zero-shot, which are 0.88, 0.79, 0.78,
and 0.81 for ASU-CHOP, SMM4H, WEB-RADR,
and combined datasets respectively. GPT-4-turbo
(0.97), Gemma2 (0.72), and GPT-4 (0.75) achieved
the highest precision for ASU-CHOP, SMM4H, and
combined datasets respectively.
For ASU-CHOP and WEB-RADR datasets, the
ML model CausalMHA achieves the highest re-
call. For the SMM4H dataset, the ML model Char-
CausalMHA achieves the highest 0.75 recall. Here
we see CRF has the lowest F1 score and Recall in
all four datasets. Nevertheless, MHA, CausalMHA,
and SCAN demonstrate consistent F1 scores across
all four datasets.
To achieve better performance, we experiment
with few-shot and CoT prompting shown in Tables
6, 7, 8, and 9. From these tables, we can see that for
all three datasets(ASU-CHOP, SMM4H, and WEB-
RADR), GPT-4 achieves the highest F1 score for
the few-shot. Gemma2 reaches the highest precision
score for three (SMM4H, WEB-RADR, and the com-
bined) datasets out of four datasets for few-shot. For
the ASU-CHOP dataset, GPT-4o-mini achieves the
highest precision. In the ASU-CHOP dataset, the
zero-shot, few-shot, and cot-promoting achieve the
highest precision for GPT-4-turbo. For the WEB-
RADR dataset, Mistral achieves the highest recall.
For all datasets, GPT-4 achieves the best recall (0.90)
and the highest F1 score (0.85). Our experiments
focus on few-shot examples, which is why the few-
shot results showed consistently good performance.
In our results, we find that our recall and precision
are consistent in LLMs. Our experiments show that
LLMs can capture over 60 percent of ADRs while
still achieving good precision, which is vital for ADR
extraction. This balance ensures that more ADRs
are identified without compromising the quality of
the results. A summary of the improvement rates of
F1 score achieved by the LLMs models against Scan
(Kayesh et al., 2022) is shown in Table 10 for few-
shot prompting. Table 10 summarizes the results,
showing that most LLM models outperform the ML
models in three datasets. Notably, the top F1-score
improvement is 19.27 percent on the SMM4H dataset.
GPT-4 shows the best results in F1 score improve-
HEALTHINF 2025 - 18th International Conference on Health Informatics
552
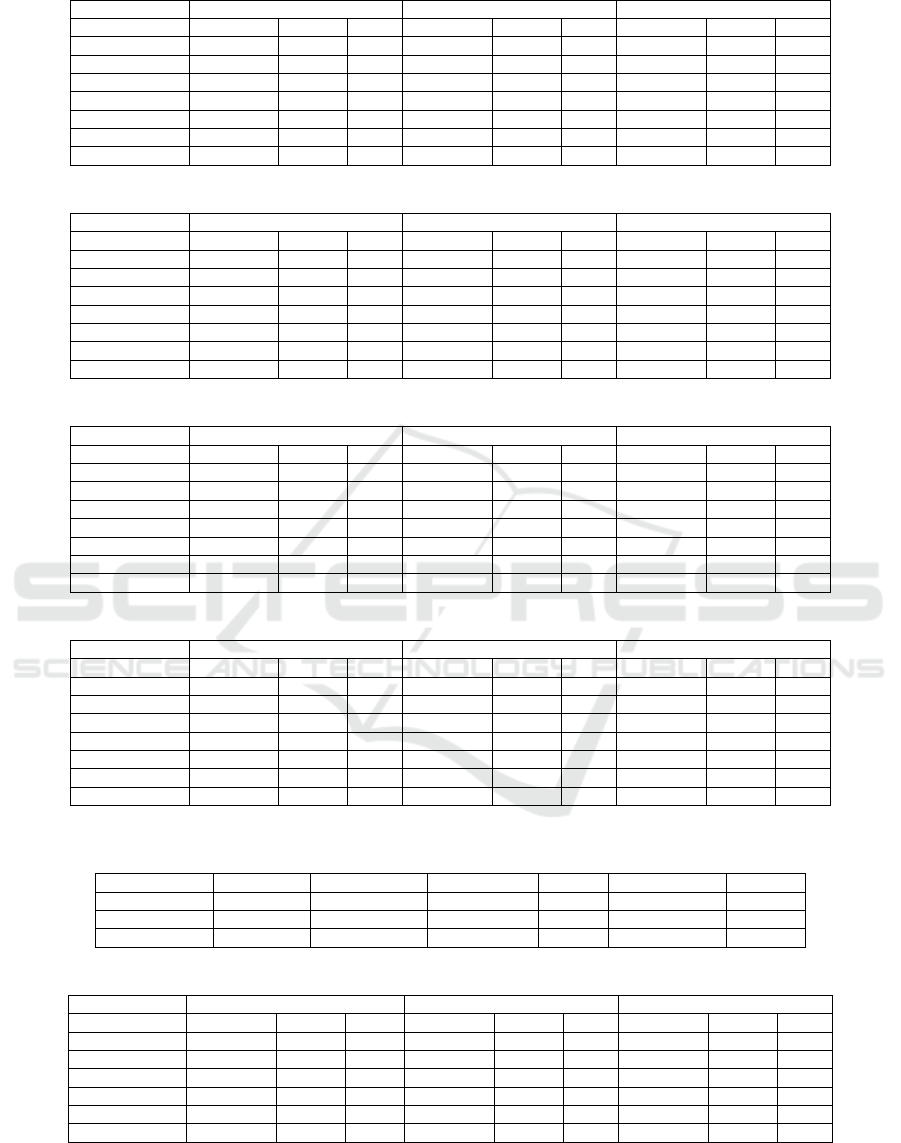
Table 6: Experimental results on the ASU-CHOP Dataset with three prompts.
Zero Shot Few Shot CoT
LLM Models Precision Recall F1 Precision Recall F1 Precision Recall F1
Mistral 7b 0.82 0.69 0.75 0.84 0.84 0.73 0.87 0.69 0.77
Llama 3.1 8b 0.93 0.74 0.82 0.87 0.71 0.78 0.93 0.70 0.80
Gemma2 9b 0.89 0.69 0.78 0.92 0.60 0.73 0.94 0.71 0.81
GPT-4o-mini 0.90 0.74 0.81 0.95 0.74 0.83 0.90 0.74 0.81
GPT-4o 0.96 0.70 0.81 0.92 0.68 0.78 0.97 0.71 0.82
GPT-4-turbo 0.97 0.72 0.83 0.97 0.71 0.82 0.97 0.66 0.79
GPT-4 0.96 0.81 0.88 0.94 0.78 0.85 0.95 0.73 0.83
Table 7: Experimental results on the SMM4H dataset with three prompts.
Zero Shot Few Shot CoT
LLM Models Precision Recall F1 Precision Recall F1 Precision Recall F1
Mistral 7b 0.65 0.75 0.7 0.68 0.71 0.69 0.66 0.74 0.7
Llama 3.1 8b 0.68 0.81 0.74 0.70 0.82 0.76 0.72 0.81 0.76
Gemma2 9b 0.72 0.76 0.74 0.79 0.69 0.74 0.78 0.8 0.79
GPT-4o-mini 0.66 0.83 0.74 0.74 0.81 0.77 0.69 0.84 0.76
GPT-4o 0.71 0.74 0.72 0.73 0.78 0.75 0.7 0.79 0.74
GPT-4-turbo 0.70 0.80 0.75 0.79 0.79 0.79 0.78 0.77 0.77
GPT-4 0.71 0.89 0.79 0.75 0.90 0.82 0.74 0.84 0.78
Table 8: Experimental results on the WEB-RADR dataset with three prompts.
Zero Shot Few Shot CoT
LLM Models Precision Recall F1 Precision Recall F1 Precision Recall F1
Mistral 7b 0.66 0.73 0.69 0.68 0.64 0.66 0.69 0.76 0.72
Llama 3.1 8b 0.66 0.73 0.69 0.67 0.75 0.71 0.69 0.69 0.69
Gemma2 9b 0.66 0.62 0.64 0.79 0.57 0.66 0.72 0.67 0.69
GPT-4o-mini 0.65 0.73 0.69 0.67 0.68 0.67 0.62 0.71 0.66
GPT-4o 0.68 0.59 0.63 0.71 0.64 0.67 0.67 0.67 0.67
GPT-4-turbo 0.63 0.65 0.64 0.72 0.65 0.68 0.72 0.62 0.67
GPT-4 0.71 0.86 0.78 0.74 0.84 0.79 0.69 0.72 0.70
Table 9: Experimental results on the combined Twitter Dataset with three prompts.
Zero Shot Few Shot CoT
LLM Models Precision Recall F1 Precision Recall F1 Precision Recall F1
Mistral 7b 0.68 0.73 0.70 0.70 0.70 0.70 0.70 0.73 0.71
Llama 3.1 8b 0.72 0.78 0.75 0.72 0.77 0.74 0.75 0.76 0.75
Gemma2 9b 0.74 0.71 0.72 0.82 0.65 0.73 0.8 0.75 0.77
GPT-4o-mini 0.70 0.79 0.74 0.76 0.77 0.76 0.71 0.79 0.75
GPT-4o 0.75 0.70 0.72 0.76 0.73 0.74 0.74 0.74 0.74
GPT-4-turbo 0.73 0.75 0.74 0.81 0.74 0.77 0.80 0.71 0.75
GPT-4 0.75 0.87 0.81 0.74 0.84 0.79 0.77 0.79 0.78
Table 10: Average Increment in F1 (percentage) achieved by different language models against the SCAN model for few
shots.
Dataset Mistral 7b Llama 3.1 8b Gemma2 9b GPT-4 GPT-4o-mini GPT-4o
ASU-CHUP -3.07 1.93 -3.07 8.93 6.93 1.93
SMM4H 6.27 13.27 11.27 19.27 14.27 12.27
WEB-RADR 0.93 5.93 0.93 13.93 1.93 1.93
Table 11: Experimental Extraction Results on the ADE Corpus with three prompts.
Zero Shot Few Shot CoT
LLM Models Precision Recall F1 Precision Recall F1 Precision Recall F1
Mistral 7b 0.92 0.85 0.88 0.93 0.75 0.83 0.94 0.77 0.85
Llama 3.1 8b 0.93 0.87 0.90 0.93 0.50 0.65 0.97 0.65 0.78
Gemma2 9b 0.95 0.82 0.88 0.96 0.38 0.54 0.97 0.62 0.76
GPT-4o-mini 0.92 0.88 0.90 0.94 0.76 0.84 0.94 0.82 0.88
GPT-4-turbo 0.95 0.85 0.90 0.97 0.74 0.84 0.97 0.75 0.85
GPT-4 0.94 0.79 0.86
ment. In a small dataset, Mistral does not demon-
strate strong performance as ASU-CHOP has 172 test
tweets. The scan also achieves better performance
than Mistral and Gemma2 in the small dataset.
Using LLMs to Extract Adverse Drug Reaction (ADR) from Short Text
553
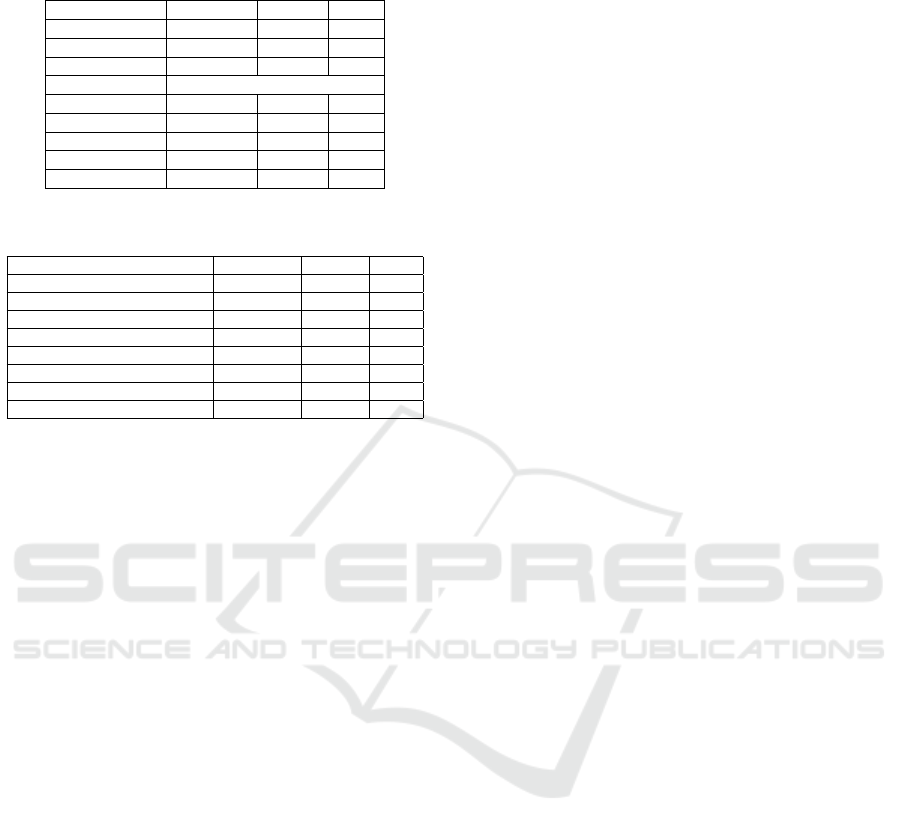
Table 12: Experimental Extraction Results on the ADE Cor-
pus for zero shot.
ML Methods Precision Recall F1
GLoVe 0.93 0.94 0.94
BERT 0.94 0.98 0.96
SOTA 0.91
LLM Models
Mistral 7b 0.92 0.85 0.88
Llama 3.1 8b 0.93 0.87 0.90
Gemma2 9b 0.95 0.82 0.88
GPT-4o-mini 0.92 0.88 0.90
GPT-4-turbo 0.95 0.85 0.90
Table 13: Experimental Results on the ADE Corpus for
classification.
ML Methods Precision Recall F1
Attentive Sequence Model 0.88 0.82 0.85
LSTM 0.86 0.94 0.90
FARM-BERT 0.98 0.96 0.97
DCNN 0.92 0.95 0.93
LLM Models
GPT-4o-mini 1.0 0.91 0.95
GPT-4 1.0 0.97 0.98
Llama 1.0 1.0 1.0
Overall, GPT-4 achieves the best result in all
datasets. GPT-4o-mini and Llama show similar per-
formance across small to moderately large datasets
for ADR extraction in Tweets. However, these two
models have much fewer parameters than GPT-4.
GPT-4o-mini and Llama show very promising perfor-
mance in extracting ADR.
3.3.2 Comparison of ML Models with LLMs for
ADE Corpus Dataset
The results in Table 12 show the comparison of LLMs
with ML methods for the ADE corpus dataset in a
zero-shot setting. We can see from Table 11 that
Gemma and GPT-4-turbo achieved higher precision
for zero-shot than the ML methods. Here, the ML
model BERT (Haq et al., 2022) achieved the highest
F1 score. However, the F1 scores of LLM models
(GPT-4o-mini, GPT-4o-turbo, Llama) are very close
to the ML F1 score.
Results with different prompting techniques
achieving better precision are shown in Table 11.
Here, we see that zero-shot recall performs better than
other techniques, though few-shot and CoT achieve
better precision. This is due to the selection of shots
or examples in the prompts. We can get better re-
sults if we have better shots/examples. The overall
precision of LLM models is better than that of ML
methods. Also, we can see that all the LLMs are
showing better results for the ADE corpus dataset
than the Twitter dataset. This is because Twitter data
is unstructured and informal, whereas the ADE cor-
pus dataset is more structured and formal. In both
datasets, LLMs achieved better performance than ML
methods.
Table 13 shows the detection or classification re-
sults for the ADE corpus dataset with LLMs and ML
methods. Here, we can see that the LLM models show
better results in detection than the previous ML meth-
ods. We have compared three LLM models here, and
all of them showed 100 percent precision in detect-
ing ADR sentences. GPT-4 and Llama showed better
performance.
4 CONCLUSION
ADR extraction from social media and text data
is crucial for patient safety. This paper employs
Large Language Models (LLMs), including GPT-4o-
mini, Llama, and GPT-4-turbo, to extract ADRs us-
ing few-shot and other prompting techniques. Re-
sults show LLMs consistently outperform ML mod-
els, with GPT-4 achieving an F1 score of 0.82 on the
SMM4H dataset, surpassing the previous state-of-the-
art score of 0.64, highlighting their ability to capture
nuanced linguistic patterns.
Our experiments suggest that complex machine-
learning models are unnecessary for effective ADR
extraction. Limitations include using identical
prompts, lack of standardized datasets, and unad-
dressed data privacy concerns. Future work could re-
fine prompts, optimize models like GPT-4 and Llama,
and address ethical, resource, and security challenges
while enhancing performance through model-specific
tuning and error analysis.
REFERENCES
Ahmed, T. and Devanbu, P. (2023). Better patching us-
ing llm prompting, via self-consistency. In 2023
38th IEEE/ACM International Conference on Au-
tomated Software Engineering (ASE), pages 1742–
1746. IEEE.
Breden, A. and Moore, L. (2020). Detecting adverse
drug reactions from twitter through domain-specific
preprocessing and bert ensembling. arXiv preprint
arXiv:2005.06634.
Cocos, A., Fiks, A. G., and Masino, A. J. (2017). Deep
learning for pharmacovigilance: recurrent neural net-
work architectures for labeling adverse drug reactions
in twitter posts. Journal of the American Medical In-
formatics Association, 24(4):813–821.
Denton, E. L., Chintala, S., Fergus, R., et al. (2015). Deep
generative image models using a laplacian pyramid of
adversarial networks. Advances in neural information
processing systems, 28.
Dietrich, J., Gattepaille, L. M., Grum, B. A., Jiri, L., Lerch,
M., Sartori, D., and Wisniewski, A. (2020). Adverse
HEALTHINF 2025 - 18th International Conference on Health Informatics
554

events in twitter-development of a benchmark refer-
ence dataset: results from imi web-radr. Drug safety,
43:467–478.
Ding, P., Zhou, X., Zhang, X., Wang, J., and Lei, Z. (2018).
An attentive neural sequence labeling model for ad-
verse drug reactions mentions extraction. Ieee Access,
6:73305–73315.
Florez, E., Precioso, F., Riveill, M., and Pighetti, R. (2018).
Named entity recognition using neural networks for
clinical notes. In International Workshop on Medica-
tion and Adverse Drug Event Detection, pages 7–15.
PMLR.
Gupta, S., Pawar, S., Ramrakhiyani, N., Palshikar, G. K.,
and Varma, V. (2018). Semi-supervised recurrent neu-
ral network for adverse drug reaction mention extrac-
tion. BMC bioinformatics, 19:1–7.
Haq, H. U., Kocaman, V., and Talby, D. (2022). Mining
adverse drug reactions from unstructured mediums at
scale. In Multimodal AI in healthcare: A paradigm
shift in health intelligence, pages 361–375. Springer.
Hinton, G. E., Krizhevsky, A., and Wang, S. D. (2011).
Transforming auto-encoders. In Artificial Neural Net-
works and Machine Learning–ICANN 2011: 21st In-
ternational Conference on Artificial Neural Networks,
Espoo, Finland, June 14-17, 2011, Proceedings, Part
I 21, pages 44–51. Springer.
Hughes, A. J. and Song, X. (2024). Identifying and align-
ing medical claims made on social media with med-
ical evidence. In Proceedings of the 2024 Joint In-
ternational Conference on Computational Linguistics,
Language Resources and Evaluation (LREC-COLING
2024), pages 8580–8593.
Hussain, S., Afzal, H., Saeed, R., Iltaf, N., and Umair,
M. Y. (2021). Pharmacovigilance with transformers:
A framework to detect adverse drug reactions using
bert fine-tuned with farm. Computational and Mathe-
matical Methods in Medicine, 2021(1):5589829.
Kalyan, K. S. and Sangeetha, S. (2020). Want to iden-
tify, extract and normalize adverse drug reactions in
tweets? use roberta. In Proceedings of the Fifth So-
cial Media Mining for Health Applications Workshop
& Shared Task, pages 121–124.
Kayesh, H., Islam, M. S., and Wang, J. (2019). A causal-
ity driven approach to adverse drug reactions detec-
tion in tweets. In International Conference on Ad-
vanced Data Mining and Applications, pages 316–
330. Springer.
Kayesh, H., Islam, M. S., Wang, J., Ohira, R., and Wang,
Z. (2022). Scan: A shared causal attention network
for adverse drug reactions detection in tweets. Neuro-
computing, 479:60–74.
Lafferty, J., McCallum, A., Pereira, F., et al. (2001). Con-
ditional random fields: Probabilistic models for seg-
menting and labeling sequence data. In Icml, vol-
ume 1, page 3. Williamstown, MA.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). Biobert: a pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Li, Y., Li, J., He, J., and Tao, C. (2024). Ae-gpt: using
large language models to extract adverse events from
surveillance reports-a use case with influenza vaccine
adverse events. Plos one, 19(3):e0300919.
Li, Z., Lin, H., and Zheng, W. (2020). An effective emo-
tional expression and knowledge-enhanced method
for detecting adverse drug reactions. IEEE Access,
8:87083–87093.
Liu, Y. (2019). Roberta: A robustly optimized bert pretrain-
ing approach. arXiv preprint arXiv:1907.11692.
Luo, H., Yin, W., Wang, J., Zhang, G., Liang, W., Luo, J.,
and Yan, C. (2024). Drug-drug interactions predic-
tion based on deep learning and knowledge graph: A
review. Iscience.
Ramamoorthy, S. and Murugan, S. (2018). An attentive se-
quence model for adverse drug event extraction from
biomedical text. arXiv preprint arXiv:1801.00625.
Sanh, V. (2019). Distilbert, a distilled version of bert:
Smaller, faster, cheaper and lighter. arXiv preprint
arXiv:1910.01108.
Scaboro, S., Portelli, B., Chersoni, E., Santus, E., and
Serra, G. (2023). Extensive evaluation of transformer-
based architectures for adverse drug events extraction.
Knowledge-Based Systems, 275:110675.
Song, Q., Li, B., and Xu, Y. (2017). Research on adverse
drug reaction recognitions based on conditional ran-
dom field. In Proceedings of the International Confer-
ence on Business and Information Management, pages
97–101.
Spandana, S. and Prakash, R. V. (2024). Multiple features-
based adverse drug reaction detection from social me-
dia using deep convolutional neural networks (dcnn).
Multimedia Tools and Applications, pages 1–15.
Subramaniam, L. V., Mukherjea, S., Kankar, P., Srivastava,
B., Batra, V. S., Kamesam, P. V., and Kothari, R.
(2003). Information extraction from biomedical liter-
ature: methodology, evaluation and an application. In
Proceedings of the twelfth international conference on
Information and knowledge management, pages 410–
417.
Tutubalina, E. and Nikolenko, S. (2017). Combination
of deep recurrent neural networks and conditional
random fields for extracting adverse drug reactions
from user reviews. Journal of healthcare engineering,
2017(1):9451342.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in neural
information processing systems, 30.
Yan, Z., Zhang, C., Fu, J., Zhang, Q., and Wei, Z. (2021).
A partition filter network for joint entity and relation
extraction. In Proceedings of the 2021 Conference on
Empirical Methods in Natural Language Processing,
pages 185–197.
Zhang, M. and Geng, G. (2019). Adverse drug event detec-
tion using a weakly supervised convolutional neural
network and recurrent neural network model. Infor-
mation, 10(9):276.
Zhao, X., Xiong, Y., and Tang, B. (2020). Hitsz-icrc: A
report for smm4h shared task 2020-automatic classi-
fication of medications and adverse effect in tweets.
In Proceedings of the Fifth Social Media Mining for
Health Applications Workshop & Shared Task, pages
146–149.
Using LLMs to Extract Adverse Drug Reaction (ADR) from Short Text
555
