
Partition Tree Ensembles for Improving Multi-Class Classification
Miran
¨
Ozdogan
1 a
, Alan Jeffares
2 b
and Sean Holden
3 c
1
Department of Computer Science, Oxford University, U.K.
2
Department of Applied Mathematics and Theoretical Physics, Cambridge University, U.K.
3
Department of Computer Science, Cambridge University, U.K.
Keywords:
Multi-Class Classification, Non-Convex Optimization, AutoML.
Abstract:
We propose the Partition Tree Ensemble (PTE), a novel tree-based ensemble method for classification prob-
lems. This differs from previous approaches in that it combines ideas from reduction methods—that decom-
pose multi-class classification problems into binary classification problems—with the creation of specialised
base learners that are trained on a subset of the input space. By exploiting multi-class reduction, PTEs adapt
concepts from the Trees of Predictors (ToP) method to successfully tackle multi-class classification problems.
Each inner node of a PTE splits either the feature space or the label space into subproblems. For each node
our method then selects the most appropriate base learner from the provided set of learning algorithms. One of
its key advantages is the ability to optimise arbitrary loss functions. Through an extensive experimental eval-
uation, we demonstrate that our approach achieves significant performance gains over the baseline ToP and
AdaBoost methods, across various datasets and loss functions, and outperforms the Random Forest method
when the label space exhibits clusters where some classes are more similar to each other than to others.
1 INTRODUCTION
Ensemble methods have gained popularity in super-
vised machine learning as a strategy for maximising
performance by aggregating the predictions of multi-
ple base learners. It is often advantageous to organ-
ise these base learners using a tree structure. Exam-
ples of such tree-based ensembles include Nested Di-
chotomies (NDs) (Frank and Kramer, 2004), Model
Trees (MTs) (Quinlan et al., 1992) and Trees of Pre-
dictors (ToPs) (Yoon et al., 2018b; Yoon et al.,
2018a).
ToPs employ tree structures to decompose classi-
fication problems. They apply distinct base learners
to various subsets of the feature space, and optimise
for arbitrary loss functions. This can be an advan-
tage, especially in the field of healthcare where it is
often appropriate to evaluate classifier performance
in terms of AUC (Cortes and Mohri, 2003) or other
non-standard measures (Yoon et al., 2018a; Beneke
et al., 2023). Yoon et al. (Yoon et al., 2018b) demon-
strated that ToPs can yield substantial performance
a
https://orcid.org/0009-0008-7776-6107
b
https://orcid.org/0009-0003-8744-0876
c
https://orcid.org/0000-0001-7979-1148
gains when compared to frequently utilised methods
such as Random Forests and AdaBoost (Yoon et al.,
2018b). However, their evaluation of the ToPs ap-
proach was limited to binary classification problems.
Our preliminary experiments showed that their per-
formance compared to baselines diminishes with an
increasing number of classes.
This inspired us to devise an ensemble method
that, similarly to the ToP approach, is able to optimise
for arbitrary loss functions and overcome the short-
comings of ToPs for multi-class problems. We intro-
duce the PTE, a novel tree-based ensemble approach
that combines ideas from reduction techniques (Frank
and Kramer, 2004), which decompose multi-class
classification problems, with the creation of spe-
cialised base learners that are trained on a subset of
the input space, as exemplified by the ToP approach.
We draw inspiration from the Nested Dichotomies
(NDs) (Frank and Kramer, 2004) reduction tech-
nique, which achieves state-of-the-art performance
compared to competitors such as One-VS-One or
One-VS-Rest (Wever et al., 2023; Leathart, 2019). In
PTEs, each inner node of a PTE splits either the fea-
ture space or the label space. For the latter, we divide
the set of possible classes into two subsets that de-
fine three classification problems: “Which of the two
Özdogan, M., Jeffares, A. and Holden, S.
Partition Tree Ensembles for Improving Multi-Class Classification.
DOI: 10.5220/0013163400003905
In Proceedings of the 14th International Conference on Pattern Recognition Applications and Methods (ICPRAM 2025), pages 55-69
ISBN: 978-989-758-730-6; ISSN: 2184-4313
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
55

subsets does a sample belong to?” and “Given that
a sample is part of one of the subsets, which of its
classes does it belong to?”.. By exploiting such Class
Dichotomies, we can more effectively decompose the
original learning problem, and extend the advantages
of ToPs to multi-class problems.
We assess the value of this approach through an
experimental evaluation, comparing its performance
with ToPs, Random Forests and AdaBoost. Our eval-
uation employs nine datasets from the UCI reposi-
tory (Dua and Graff, 2017). In addition, we conduct
experiments on optimising for two different loss func-
tions, specifically the AUC (Cortes and Mohri, 2003)
and the F1-Measure (Chicco and Jurman, 2020).
In summary, our contributions are as follows:
• We introduce the PTE and provide a comprehen-
sive formal description of the method.
• We present a detailed mathematical derivation
of the computational complexity associated with
training PTEs.
• In an extensive experimental evaluation, we com-
pare PTEs against ToPs and additional baselines
of Random Forests (Breiman, 2001) and Ad-
aBoost (Freund et al., 1999). We are able to
show that PTEs can lead to substantial perfor-
mance gains over ToPs and AdaBoost.
2 RELATED WORK
An overview of the respective differences between
PTEs, ToPs, NDs and Model Trees can be found
in Table 1. We acknowledge that other tree-
Table 1: Comparison of PTEs with Existing Tree-Based En-
semble Methods.
Method PTE MT ND ToP
Multiple Base Learners ✔ ✘ ✘ ✔
Partition X ✔ ✔ ✘ ✔
Partition Y ✔ ✘ ✔ ✘
Non-Convex Loss ✔ ✘ ✘ ✔
Classification ✔ ✘ ✔ ✔
Regression ✘ ✔ ✘ ✔
based ensemble methods exist, including Filter
trees (Beygelzimer et al., 2007) and Kernel Regres-
sion Tree (Torgo, 1997b). However, we will focus on
NDs, ToPs and Model Trees as these are the methods
most similar to PTEs.
Figure 1: Illustrative example of a Nested Dichotomy
with Y = {c
1
,c
2
,c
3
,c
4
}. Figure adapted to our notation
from (Wever et al., 2023).
2.1 Nested Dichotomies
Nested Dichotomies (NDs) (Frank and Kramer, 2004)
are ensembles that employ a binary tree structure to
recursively break down a multi-class classification
problem into a set of binary classification problems.
They facilitate the application of classifiers that are
inherently binary to multi-class problems.
We can represent an ND as a binary tree, where
each node corresponds to a subset of classes from the
original multi-class classification problem. The sub-
trees of a node recursively divide its set of classes into
distinct subsets, forming the meta-classes that corre-
spond to its two children until a subset contains only
a single element. At each internal node, the division
constitutes a binary classification problem that is ad-
dressed by a base learner (Frank and Kramer, 2004).
An illustration of this method can be found in Fig-
ure 1.
Since the initial work by Frank and Kramer (Frank
and Kramer, 2004), extensive research has been
conducted on the selection of good ND structures
(Leathart, 2019). We now discuss two contemporary
state-of-the-art methods. Leathart et al. (Leathart,
2019) sought to create trees with easier binary clas-
sification problems by employing dichotomies that
group together classes that are “similar”. This method
is called Random Pair Nested Dichotomies (RPNDs).
In this approach, an initial pair of classes is ran-
domly selected. Then each of the other classes is
grouped with one of those two. The assignment of
other classes is determined by training a binary clas-
sifier on the initial classes. This classifier is applied
to the remaining data and the remaining classes are
assigned based on the classifier’s predictions through
hard voting. Note, that the binary classifier employed
for class grouping is discarded, and a new binary clas-
sifier is trained to differentiate between the two meta-
classes. Leathart et al. (Leathart, 2019) demonstrated
that ensembles constructed using this technique ex-
hibit state-of-the-art predictive performance while be-
ing less runtime-intensive than competing methods.
Wever et al. (Wever et al., 2023) propose a co-
evolutionary algorithm CONDA aimed at discovering
advantageous ensembles of NDs. In contrast with
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
56

Figure 2: Illustrative example of a Tree of Predictors.
RPNDs, this requires a comprehensive optimisation
of the entire tree structure, rather than the successive
growth of the ND tree. It demonstrates superior pre-
dictive performance when compared to RPNDs, al-
beit at the expense of a significant increase in run-
time (Wever et al., 2023).
Interestingly, Mohr et al. (Mohr et al., 2018)
found that, for multi-class problems, implementing
partial NDs of depth 1 (referred to simply as “Di-
chotomies” in the statistical literature) can sometimes
enhance performance, when compared to using indi-
vidual base learners.
2.2 Trees of Predictors
In this section, we describe the ToPs ensemble
method as introduced in (Yoon et al., 2018a; Yoon
et al., 2018b). We will assume that categorical fea-
tures are one-hot encoded and continuous features are
normalised such that X
j
∈ [0, 1] for every feature j
and X ⊆ [0, 1]
d
. For subsets Z
1
,Z
2
⊆ X of the feature
space, if predictors h
1
: Z
1
7→ Y and h
2
: Z
2
7→ Y
are constructed such that Z
1
∩ Z
2
=
/
0, we define
h
1
∨h
2
: Z
1
∪Z
2
7→ Y as h
1
∨h
2
(z) = h
i
(z) for z ∈ Z
i
.
2.2.1 Formal Definition
A Tree of Predictors can be represented as a binary
tree, denoted T . Each node N within this tree corre-
sponds to a subset X of the feature space and is associ-
ated with a classifier h
N
. The corresponding subset is
used interchangeably to refer to a node and vice versa.
The root node of T must corresponds to the entire fea-
ture space X . For any given node N ∈ T its children,
denoted {N
+
,N
−
}, partition N. For a node N ∈ T , its
corresponding classifier h
N
is generated by training
an algorithm A
N
∈ A on a set N
∗
, where N
∗
= N or
N
∗
corresponds to one of the predecessors of N. For-
mally, this relationship is expressed as h
N
= A
N
(N
∗
)
with N
∗
∈ N
↑
.
The set of leaves, denoted T , forms a partition of
the feature space X . We denote the leaf correspond-
ing to some feature vector x ∈ X as N(x). An illus-
trative example of a Tree of Predictors is provided in
Figure 2.
Note that this approach allows for a node’s classi-
fier to be trained on a set that corresponds to one of
its predecessors. This design choice is driven by the
understanding that partitioning the training set may
result in performance enhancement for one subset,
while degrading performance for another.
2.2.2 Growing a Tree of Predictors
Growing a ToP requires a finite set A = A
1
,. . ., A
m
of
base learner algorithms as input, as well as a training
set D and two validation sets V
1
and V
2
. The pre-
dictor that is obtained by training an algorithm A ∈ A
on some subset E ⊆ D of the training data is denoted
A(E). Let E be a collection of subsets of the training
data D. We then define A(E ) = {A(E) : A ∈ A, E ∈
E} as the set of predictors obtained from training each
algorithm in A on every set in E. Given a subset
N ⊂ X of the feature space and a subset E ⊂ D of the
dataset, we let E(N) = {(x
i
,y
i
) ∈ E : x
i
∈ N}. When
provided with disjoint sets E
1
,. . ., E
t
and correspond-
ing predictors h
l
on E
l
, we denote the overall loss as
L (
W
h
i
,
S
E
i
).
The process is initialised with the root node X and
the corresponding classifier h
X
. The classifier is cho-
sen such that h
X
= A(D(X )), where A is selected from
the set of algorithms A such that it minimises the val-
idation loss on V
1
.
Then the children of each leaf N ∈ T are re-
cursively created. For this, the corresponding fea-
ture space is split along the feature i and threshold
τ
i
∈ [0,1] that result in the best validation loss mea-
sured on V
1
. We write
N
−
τ
i
= {x ∈ N : x
i
< τ
i
},N
+
τ
i
= {x ∈ N : x
i
≥ τ
i
} (1)
In practice, for each continuous feature up to 9 can-
didate thresholds are selected at the 10th, 20th, ...,
90th percentiles. For every candidate threshold τ
i
,
the predictors h
−
∈ A(N
−↑
τ
i
), h
+
∈ A(N
+↑
τ
i
) are cho-
sen such that they minimise the validation loss where
N
↑
to denotes the set consisting of node N and all its
predecessors that correspond to the same label space.
From these predictors we then determine h
N
−
τ
∗
i
, h
N
+
τ
∗
i
that minimise the total validation loss.
A node N is only split if its loss is strictly higher
than the total loss of its children N
−
τ
i
and N
+
τ
i
. The
pseudo-code for this can be found in Algorithm 1.
2.2.3 Weighting Predictors for Optimal
Predictions
Once the locally optimal tree is established, the next
step is to determine how to weight the various pre-
dictors. The prediction for a sample x is determined
by the sequence of nodes
∏
, which spans the path
from the root node to the leaf node N(x) that corre-
Partition Tree Ensembles for Improving Multi-Class Classification
57

Algorithm 1: Growing the Optimal Tree of Predic-
tors. Adapted from (Yoon et al., 2018b).
Require: Feature space X , set of algorithms A,
training set D, first validation set V
1
h
X
← arg min
h∈A(D)
L(h,V
1
)
T ← (X , h
X
),
Recursive step:
Input: Current Tree of Predictors T
for each leaf N ∈ T do
for each feature i and threshold τ
i
do
N
−
τ
i
← {x ∈ N : x
i
< τ
i
}
N
+
τ
i
← {x ∈ N : x
i
≥ τ
i
}
end
let h
−
τ
i
∈ A(N
−
τ
i
), h
+
τ
i
∈ A(N
+
τ
i
)
(i
⋆
,τ
⋆
i
,h
N
−
τ
i
,h
N
+
τ
i
) ←
argminL
h
−
τ
i
∨ h
+
τ
i
;V
1
(N)
end
Stopping criterion:
L(h
N
;V
1
(N)) ≤
min
L
h
−
τ
i
∨ h
+
τ
i
;V
1
(N
−
τ
i
) ∪V
1
(N
+
τ
i
)
sponds to x. Each leaf node is assigned an individ-
ual weight vector ⃗w = (w
N
)
N∈
∏
, which defines the
weights of predictors corresponding to nodes on the
root-leaf path. These weights are used to generate
predictions for samples associated with N(x). The en-
tire weight vector of a leaf is individual; that is, the
common ancestor of two leaves might have different
weights assigned by them.
Weights are required to be non-negative and sum
to one. For each leaf, weight vectors are chosen by
utilising linear regression to minimise the empirical
loss on the corresponding subset of the second vali-
dation set V
2
(N(x)).
2.3 Model Trees
Model Trees (Quinlan et al., 1992), are a tree-based
ensemble method for solving regression tasks. They
combine a conventional Decision Tree structure with
the possibility of linear regression functions at the leaf
nodes (Wang and Witten, 1997).
In the first phase of Model Tree construction, a
Decision Tree induction algorithm (Breiman et al.,
1984) is employed. As a splitting criterion it min-
imises the variation in class values within each sub-
set across the branches. During the second phase, it
prunes the tree, starting from each leaf and working
upwards. This aims to reduce overfitting by removing
or replacing less informative nodes (Breiman et al.,
1984; Quinlan, 1986). A key distinction from conven-
tional pruning methods is that, when pruning back to
an internal node, the method involves replacing that
node with a regression plane rather than a constant
value (Wang and Witten, 1997).
Several works have expanded on the original
Model Tree method. Torgo (Torgo, 1997a) introduced
an approach that allows for different models within
the leafs of the tree. However, Torgo’s method re-
mains constrained to growing the tree based on a sin-
gle base learner. Malerba et al. (Malerba et al., 2001)
proposed a Model Tree induction algorithm that has
some similarities with our PTE method. It employs a
splitting criterion based on how well linear models fit
the data at a node during tree growth.
2.3.1 Differences to Partition Tree Ensembles
Primarily, Model Trees are designed to address re-
gression tasks, whereas PTEs are designed for clas-
sification problems. Additionally, PTEs do not utilise
pruning or smoothing techniques. Instead, they em-
ploy Stacked Generalisation (Wolpert, 1992) to deter-
mine the weights assigned to models along the path
from the root to the leaf node. Furthermore, Model
Trees solely partition the feature space, whereas PTEs
have the versatility to partition both the feature space
and the label space.
Importantly, Model Trees are induced using a sin-
gle base learner algorithm while PTEs provide the
flexibility to select the most suitable base learner at
each tree node.
Lastly, Model Trees are restricted to using convex
loss functions (Yoon et al., 2018b). This can be disad-
vantageous when optimising for F-score or AUC, as is
frequently done in medical applications. In contrast,
PTEs offer the flexibility to optimise non-convex loss
functions.
3 PARTITION TREE ENSEMBLES
In this section, we introduce PTEs. First, in Sec-
tion 3.1 we explain the motivation for this method.
Then in Section 3.2 we formally introduce Class Di-
chotomies, which constitute the inner nodes of Nested
Dichotomies. (Note that in the statistical literature
these are referred to simply as “Dichotomies”.) In-
spired by (Leathart et al., 2016) we use the Random
Pair method to select Class Dichotomies for PTEs.
We describe this in Section 3.3. In Section 3.4 we
provide a formal definition of PTEs. Then, in Sec-
tion 3.5 we describe our method for inducing them.
Finally, in Section 3.6 we describe how we aggregate
base learner predictions into an overall prediction.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
58

3.1 Motivation
In this section, we succinctly present the motivation
underpinning our PTE algorithm. Previous work by
Yoon et al. (Yoon et al., 2018b) highlighted how a
Tree of Predictors can produce significant improve-
ments over baseline methods such as AdaBoost, and
Random Forest. However, their experimental anal-
ysis was confined to binary classification datasets.
Our preliminary experiments applying ToPs to multi-
class classification problems revealed that their per-
formance advantage over the baselines diminishes
with a higher number of classes.
Achieving high performance in terms of loss func-
tions other than the cross-entropy loss can be piv-
otal, especially in healthcare applications. Conven-
tional performance measures such as cross-entropy
loss or accuracy are often not the most suitable met-
rics in medical settings (Yoon et al., 2018a). In many
cases, employing other loss functions such as AUC-
Loss (Cortes and Mohri, 2003) or the F-Measure (Gu
et al., 2009) may be more appropriate. Hence, our
goal was to identify an ensemble method compara-
ble to Tree of Predictors, which can successfully opti-
mise arbitrary loss functions for multi-class problems
as well.
In response to this challenge, we present PTEs.
Like ToPs, PTEs recursively split the classification
problem into subproblems associated with the nodes
of a binary tree. However, in addition to splitting
the feature space, nodes within the tree can parti-
tion the label space using Class Dichotomies, thereby
giving rise to sub-problems derived from the initial
multi-class classification problem. Since these sub-
problems have a lower number of classes compared
to the original problem, we are able to obtain perfor-
mance gains similar to those we observed for ToPs in
binary classification problems.
3.2 Class Dichotomies
In this section, we provide a formal definition of
Class Dichotomies, which serve as a means to par-
tition a multi-class classification problem into three
distinct sub-problems. Class Dichotomies are utilised
at every inner node of a ND tree. In early ma-
chine learning literature, the individual nodes of
a Nested Dichotomy were themselves referred to
as Nested Dichotomies (Frank and Kramer, 2004).
However, in contemporary usage, the term “Nested
Dichotomies” has evolved to encompass the entire
tree structure (Leathart, 2019).
Given a classification problem with Y =
{c
1
,. . ., c
k
}, a Class Dichotomy partitions the set of
classes into two subsets. Without loss of general-
ity, let these subsets be Y
−
= {c
1
,. . ., c
j
} and Y
+
=
{c
j+1
,. . ., c
k
}. It is important to note that Class Di-
chotomies may define arbitrary non-empty partitions,
and the ordering of classes is not relevant to this. To
construct a predictor that addresses the original multi-
class problem, the Class Dichotomy combines predic-
tions for three sub-problems, which are defined by the
two subsets. First, the Class Dichotomy includes a
predictor h
Y
−
,Y
+
that differentiates between the two
meta-classes determined by Y
−
and Y
+
. This means
that, for a given sample (x,y), h
Y
−
,Y
+
(x) estimates
the probability p(y ∈ Y
−
|x). Also, the Class Di-
chotomy has two predictors, h
Y
−
and h
Y
+
, which es-
timate the class probabilities of a sample, assuming
it belongs to the respective subset; that is, h
Y
r
esti-
mates p(y = c
i
|x,y ∈ Y
r
) for all classes c
i
∈ Y
r
where
Y
r
∈ {Y
−
,Y
+
}. We can simply multiply the condi-
tional probability with the probability of its condition
to obtain an overall class probability estimate:
p(y = c
i
|x) = p(y = c
i
|x,c
i
∈ Y
r
) · p(c
i
∈ Y
r
|x) (2)
Thus, for each class c
i
∈ Y
r
, if Y
r
= Y
−
, we can
estimate p(y = c
i
|x) using h
Y
−
,Y
+
(x) · h
Y
−
(x), and if
Y
r
= Y
+
, using
1 − h
Y
−
,Y
+
(x)
· h
Y
+
(x). We refer
to this overall classifier as h
CD
.
3.3 Random Pair Method
Inspired by (Leathart et al., 2016), we propose the
use of the Random Pair method to generate Class
Dichotomies within our PTEs. Our rationale is as
follows: among methods that rely on iterative split-
ting, RPNDs have demonstrated the best performance
in selecting NDs (Leathart, 2019). Although this
may not directly translate to optimal performance for
choosing individual Class Dichotomies, we believe it
serves as a good proxy. In this section, we present
a detailed explanation of the method along with the
corresponding pseudocode.
The process begins by randomly selecting two
classes, c
−
and c
+
, from the label space Y without
replacement. Following this, we utilise the available
training data for these classes to train a binary clas-
sifier h
c
−
,c
+
to distinguish between the two classes.
This classifier is applied to the remaining training
data and the remaining classes are assigned to one
of two sets Y
−
,Y
+
based on the classifier’s predic-
tions through hard voting. Specifically, if the classi-
fier predicts that the majority of data points belonging
to class c
i
are in class c
−
, class c
i
will be allocated
to the subset Y
−
surrounding c
−
. Otherwise, it will
be assigned to the subset Y
+
. Once the two meta-
classes Y
−
and Y
+
are formed, we discard h
c
−
,c
+
Partition Tree Ensembles for Improving Multi-Class Classification
59

and train a new classifier h
Y
−
,Y
+
using the training
set {(x,0)|(x,y) ∈ D, y ∈ Y
−
}∪ {(x,1)|(x, y) ∈ D, y ∈
Y
+
} to distinguish between the two meta-classes. To
form a complete Class Dichotomy, we also train and
select h
Y
−
= argmin
A∈A
L
A(D
−
),V
−
1
and h
Y
+
=
argmin
A∈A
L
A(D
+
),V
+
1
which constitute the opti-
mal models for the two respective sub-problems.
We choose this method to create Class Di-
chotomies because it has been used to develop state-
of-the-art NDs (Leathart et al., 2016). Although evo-
lutionary algorithms NDEA (Wever et al., 2018) and
CONDA (Wever et al., 2023) have recently outper-
formed RPNDs, these methods optimise the entire
ND tree instead of individual dichotomies. Conse-
quently, they are not suitable for our purposes.
3.4 Formal Definition
In this section, we provide a formal introduction to the
PTE method. We employ the same notation as in Sec-
tion 2.2, and we continue to assume that categorical
features are represented in binary form and continu-
ous features are normalised. For our PTE method, we
require a finite set of m candidate algorithms, denoted
as A = {A
1
,. . ., A
m
}, as well as a set of m
CD
candi-
date algorithms A
CD
that we use to create predictors
which distinguish between the meta-classes in Class
Dichotomies.
Each node N within this tree corresponds to a sub-
set of the feature space X
N
⊆ X and a subset of the
label space Y
N
⊆ Y . A classifier h
N
is associated
with each node, mapping the feature space to the re-
spective subset of the label space. It is generated by
training an algorithm A
N
∈ A on a subset of the fea-
ture space corresponding to node N
∗
, where N
∗
is
either N or one of its predecessors that corresponds
to the same subset of the label space Y
N
. Formally,
this relationship is expressed as h
N
= A
N
(N
∗
) where
(N
∗
∈ N
∗
↑)∧(Y
N
∗
= Y
N
). For any given node N ∈ T ,
its children, denoted as {N
−
,N
+
}, partition the lo-
cal feature space X
N
and correspond to the local label
space Y
N
or vice versa. If and only if a node’s suc-
cessors split the label space, it contains an additional
predictor h
Y
−
N
,Y
+
N
that is generated by training an al-
gorithm A
CD
∈ A
CD
to distinguish between the two
meta-classes defined by the subsets Y
−
N
and Y
+
N
.
To grow a Partition Tree, we randomly partition
the available dataset into the training set D and two
validation sets V
1
and V
2
. Given a subset of the feature
space N ⊂ X and a subset of the dataset Z ⊂ D, we
let Z(N) = {(x
i
,y
i
) ∈ Z : x
i
∈ N}. Our general model
accommodates the optimisation of arbitrary loss func-
tions. An illustrative example of a PTE is provided in
Figure 3.
Figure 3: Illustrative example of a Partition Tree Ensemble
with Y = {c
1
,c
2
,c
3
,c
4
}.
3.5 Growing a Partition Tree
In this subsection, we introduce our algorithm for
growing Partition Trees, building upon the algorithm
for growing ToPs from Section 2.2. This extension
enables splits of the label space, utilising the concept
of Class Dichotomies introduced earlier.
Similarly to the procedure for growing ToPs, we
begin with the trivial Partition Tree containing only
the root node corresponding to (X ,Y ). When creat-
ing children for each node, we first estimate the op-
timal split of the feature space, as for ToPs. Instead
of immediately generating the children based on this
optimal split, we further create a candidate Class Di-
chotomy for each A
CD
∈ A
CD
. For this purpose, we
utilise the Random Pair method, as outlined in Sec-
tion 3.3, to create two meta-classes. Then, the al-
gorithm A
CD
is trained on these meta-classes, yield-
ing h
Y
−
,Y
+
. Using the validation set V
1
, we select
the optimal algorithms A
−
,A
+
∈ A to create h
Y
−
and
h
Y
+
. These three predictors collectively form h
Y ,CD
.
The optimal h
∗
Y ,CD
is chosen from the m
CD
Class Di-
chotomies created in this manner, based on the valida-
tion set V
1
. If h
∗
Y ,CD
demonstrates a lower loss com-
pared to the optimal split of the feature space, then
we create the node’s children based on this Class Di-
chotomy instead of splitting the feature space. In this
case, the children will correspond to partitions of the
local label space Y
−
and Y
+
, while sharing the same
local feature space. Additionally, the current node
will be assigned the meta-classifier h
Y
−
,Y
+
, which is
used to combine the predictions of its children. This
recursive process of creating the children’s offspring
continues until neither the best Class Dichotomy nor
the optimal feature space split can further improve
the local loss. The pseudocode for PTEs is in Algo-
rithm 2.
If a node’s children partition the label space, it is
necessary to assign them a new second validation set
V
2
. To achieve this, we split off a new validation set
from the training set at the current node and pass it to
the children as the V
2
set. This step is crucial because,
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
60

Algorithm 2: Growing the Optimal Partition Tree
Ensemble.
Require: Training set D, the first validation set
V
1
, a set of base estimators A, a set of binary
base estimators for Class Dichotomies A
CD
h
X
← argmin
A∈A
L(A(D);V
1
)
root ← (X ,h
X
),
Recursive step:
Require: node (N,h
N
) for a feature i and a
threshold τ
i
do
N
−
τ
i
← {x ∈ N : x
i
< τ
i
}
N
+
τ
i
← {x ∈ N : x
i
≥ τ
i
}
end
let h
−
τ
i
∈ A(D(N
−
τ
i
)), h
+
τ
i
∈ A(D(N
+
τ
i
)) :
(i
⋆
,τ
⋆
i
,h
N
−
τ
i
,h
N
+
τ
i
) ← arg min L
h
−
τ
i
∪ h
+
τ
i
;V
1
(N)
h
CD
← randomPair(D(N),V
1
(N),A, A
CD
)
L
τ
i
← L(h
−
τ
⋆
i
∪ h
+
τ
⋆
i
,V
1
(N))
L
CD
← L (h
CD
,V
1
(N)))
L
N
← L (h
N
,V
1
(N))
if (L
τ
i
< L
N
) ∧ (L
τ
i
≤ L
CD
) then
node.le f t ← (N
−
τ
⋆
i
;h
+
τ
⋆
i
)
node.right ← (N
+
τ
⋆
i
;h
+
τ
⋆
i
)
recurse(node.le f t)
recurse(node.right)
end
if (L
CD
< min(L
N
,L
τ
i
) then
node.CD ← h
CD
recurse(node.CD.le f t)
recurse(node.CD.right)
end
Stopping criterion:
(L
N
≤ min(L
CD
,L
τ
i
)
without it, stacking would be biased towards assign-
ing higher weights to predictions resulting from Class
Dichotomies. The reason for this will become more
clear in the next section.
3.6 Weighting and Assembling
Predictions
In PTEs, we cannot assemble predictions in the same
manner as in ToPs. This is because base learners be-
low a Class Dichotomy are trained on a sub-problem
with fewer classes. Therefore, integrating their pre-
dictions into the original multi-class problem requires
the use of predictor h
Y
−
,Y
+
, which has been trained
on the meta-classes at the Class Dichotomy node.
To tackle this, we conceptually regard nodes con-
taining a Class Dichotomy as leaf nodes, and their
subtrees as independent PTEs. This strategy enables
us to integrate the two subtrees and the meta-classifier
at a Class Dichotomy into a single predictor h
CD
.
Consequently, our tree now resembles a tree of pre-
dictors, with the exception that we have an added
predictor h
CD
at each leaf node containing a Class
Dichotomy. We can then employ linear regression
to define a weight vector ⃗w, allocating non-negative
weights to each of the predictors on the path from the
root to the leaf node, including the additional h
CD
.
However, to make this feasible, the subtrees of the
Class Dichotomy must be functional PTEs, implying
that weight vectors must have already been assigned
to them. Therefore, we initiate this process at the low-
est Class Dichotomies and work our way up the tree.
Now, the need for creating new V
2
sets for the chil-
dren at nodes with Class Dichotomies becomes appar-
ent. If we use the same validation set V
2
to assign the
weight vectors to the children, the estimated perfor-
mance of h
CD
on V
2
will be biased, and the weight
assigned to it will likely be higher than optimal.
4 IMPLEMENTATION
We implement PTEs in Python 3.11.1 and adhere
to the guidelines for developing scikit-learn estima-
tors (Pedregosa et al., 2011).
In our implementation we introduce an additional
hyperparameter, minLeafSize, which serves as an ad-
ditional stopping criterion. If either the training or
one of the two validation sets at a node contains fewer
samples than minLeafSize, we will not split the node
further. The rationale behind this is that a small vali-
dation set may lead to a higher probability of overfit-
ting.
4.1 Computational Complexity
The computational complexity of PTE can be ex-
pressed as:
O
n
nd
m
∑
i=1
(T
i
(n,d)) +
m
CD
∑
j=1
T
CD
j
(n,d) +
m
∑
i=1
(T
i
(n,d))
!!
. (3)
Here T
i
(n,d) denotes the computational complex-
ity of the i
th
base learner algorithm, m represents the
number of such algorithms, T
CD
j
(n,d) signifies the
computational complexity of the jth base learner al-
gorithm used to create Class Dichotomies, and m
CD
is the number of those algorithms. As previously de-
fined, n refers to the number of samples, and d to the
feature count. We provide a proof for this complexity
bound in Appendix 6.
When using Logistic Regression as the only base
learner our method has a training time complexity
Partition Tree Ensembles for Improving Multi-Class Classification
61

Table 2: Properties of the UCI datasets we use in our exper-
iments.
Dataset Task
ID
Size Classes Features
mfeat-fourier 14 2000 10 76
mfeat-karhunen 16 2000 10 64
mfeat-morph 18 2000 10 6
mfeat-pixel 20 2000 10 240
mfeat-zernike 22 2000 10 47
robot-navigation 9942 5456 4 4
plants-shape 9955 1600 100 64
semeion 9964 1593 10 256
cardiocotography 9979 2126 10 35
of O(n
3
d
2
). This follows from inserting O(nd)—
the computational complexity of Logistic Regres-
sion (Singh, 2023)—into the above formula:
O (n(nd(nd) +(nd + nd))) = O
n
3
d
2
. (4)
5 EVALUATION
In this section, we conduct a thorough experimental
evaluation of our proposed PTEs. In conducting these
experiments, our main objectives are twofold. First,
we aim to evaluate whether PTEs can exploit Class
Dichotomies to more effectively decompose multi-
class problems, compared to ToPs. Second, we inves-
tigate whether the capability of our approach to op-
timise arbitrary loss functions provides a competitive
edge over widely-used ensemble methods for classifi-
cation, such as AdaBoost and Random Forest. A per-
formance comparison to NDs is not included in our
analysis. This is because the primary goal of NDs is
not to enhance the predictive performance for a spec-
ified base learner, but to reduce a multi-class problem
into a set of binary problems.
We assess the performance of PTEs across differ-
ent objectives. In Section 5.1, we examine the perfor-
mance on the AUC-Loss function. This is followed
by an evaluation with the F1-Score in Section 5.2.
For our experiments, we use nine distinct datasets
from the UCI repository (Dua and Graff, 2017) (refer
to Table 2
for more detail). We deliberately chose datasets
featuring a sample size above 1500, and with a max-
imum of 5500. This range was selected to allow for
the potential growth of a tree to a depth where the
impact of our ensemble method could be adequately
demonstrated. Larger datasets, while potentially in-
formative, were not considered due to our computa-
tional constraints and the substantial processing time
required. It is important to highlight that the quan-
tity of features in a dataset also significantly influ-
ences the runtime of our method. The majority of
the datasets chosen for our study comprise 10 classes.
The underlying rationale for selecting datasets with
a variety of classes is to provide sufficient opportuni-
ties for partitioning the label space to enable the PTEs
method to show its effects in comparison to the ToPs
method.
To ensure both comparability and reproducibil-
ity of our results, we adhere to a standardised test-
ing methodology. We apply the same hyperparame-
ter tuning procedure across all methods. Detailed in-
formation on this hyperparameter tuning process can
be found in Appendix 6. To ensure the reliability of
our findings, we utilise the Wilcoxon Signed Rank
Test (Rey and Neuh
¨
auser, 2011) for all performance
results with a significance level, α, of 0.05.
For benchmarking purposes, we compare our
PTEs against ToPs, using the same two sets of base
learners as employed in (Yoon et al., 2018b), but with
a slight modification: we replace Linear Regression
with Logistic Regression. Logistic Regression is typ-
ically more appropriate for classification tasks, and
the reason for the original choice of Linear Regres-
sion in (Yoon et al., 2018b) remains unclear. The two
sets of base learners are LR = {Logistic Regression}
and ALL = {Logistic Regression, Random Forest,
AdaBoost}. Motivated by the successful application
of Logistic Regression in NDs (Wever et al., 2018),
we choose it as the singular base learner for creating
Class Dichotomies in PTEs. From this point forward,
we will denote ToPs and PTEs with the abbreviations
ToP-LR, ToP-ALL, PTE-LR, and PTE-ALL to indi-
cate the specific set of base learners being referred to.
In addition to ToPs, we also evaluate the performance
of our model against the two conventional ensemble
methods Random Forest and AdaBoost, to provide
additional baseline comparisons. We also include the
performance of simple Logistic Regression to provide
additional context for the evaluation. All our experi-
ments were conducted in a high performance comput-
ing cluster using a dual Xeon Gold 6142 16C 2.6GHz
CPU and 384GiB of RAM on nodes that run the Sci-
entific Linux 7 operating system, which is based on
CentOS 7.
5.1 Experiments Using AUC-Loss
In this section, we discuss our experimental re-
sults when using a loss function based on the Area
Under the Receiver Operator Characteristic curve
(AUC) (Cortes and Mohri, 2003). We employ a multi-
class extension of the regular AUC-Measure, as de-
tailed in Section 5.1.2.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
62

Table 3: Loss of PTE-LR and baselines optimised for ROC-
AUC.
Task ID PTE-LR ToP-LR LR
14 0.0191±0.0041 0.0203±0.0048 • 0.0189±0.0041
16 0.003±0.0016 0.0033±0.0021 0.0034±0.0019 •
18 0.0614±0.0262 0.0887±0.014 • 0.0873±0.0138 •
20 0.002±0.0011 0.0022±0.0015 0.0019±0.001
22 0.0209±0.003 0.0207±0.0042 0.0207±0.0029
9942 0.0013±0.0028 0.0005±0.0008 ◦ 0.0011±0.0006
9955 0.0271±0.0055 0.0281±0.0055 0.0283±0.0052
9964 0.0045±0.0028 0.0052±0.0028 • 0.0045±0.0026
9979 0.0434±0.0134 0.1517±0.0247 • 0.1464±0.0242 •
5.1.1 Motivation
In (Cortes and Mohri, 2003) Cortes and Mohri pro-
vide a statistical analysis of the AUC and theoreti-
cally determine its expected value and variance given
a fixed error rate. They point out that algorithms
minimising the error rate do not necessarily lead to
optimal AUC values. It is an important measure in
cases where we are interested in the quality of a clas-
sifier’s ranking, thus directly optimising it can yield
substantial benefits in such cases. Hand and Till ar-
gue in (Hand and Till, 2001) that such a ranking-based
loss can often be useful in multi-class cases as well,
primarily because it is challenging to assign realistic
costs to different types of misclassification in prac-
tice. Moreover, we want to ensure comparebility with
Yoon et al. (Yoon et al., 2018b) who adopted AUC as
a loss function in their introduction of ToPs.
5.1.2 Multi-Class AUC
The multi-class generalisation of AUC we use is com-
puted by assessing the average “separability” between
each pair of classes. In this context, the separability
between two classes c
i
and c
j
is quantified as the av-
erage of two probabilities: 1) the probability that a
randomly drawn instance of class c
j
has a lower esti-
mated probability of belonging to class c
i
than a ran-
domly drawn instance of class c
i
, and 2) the equiv-
alent probability with c
i
and c
j
reversed (Hand and
Till, 2001).
5.1.3 Experimental Results
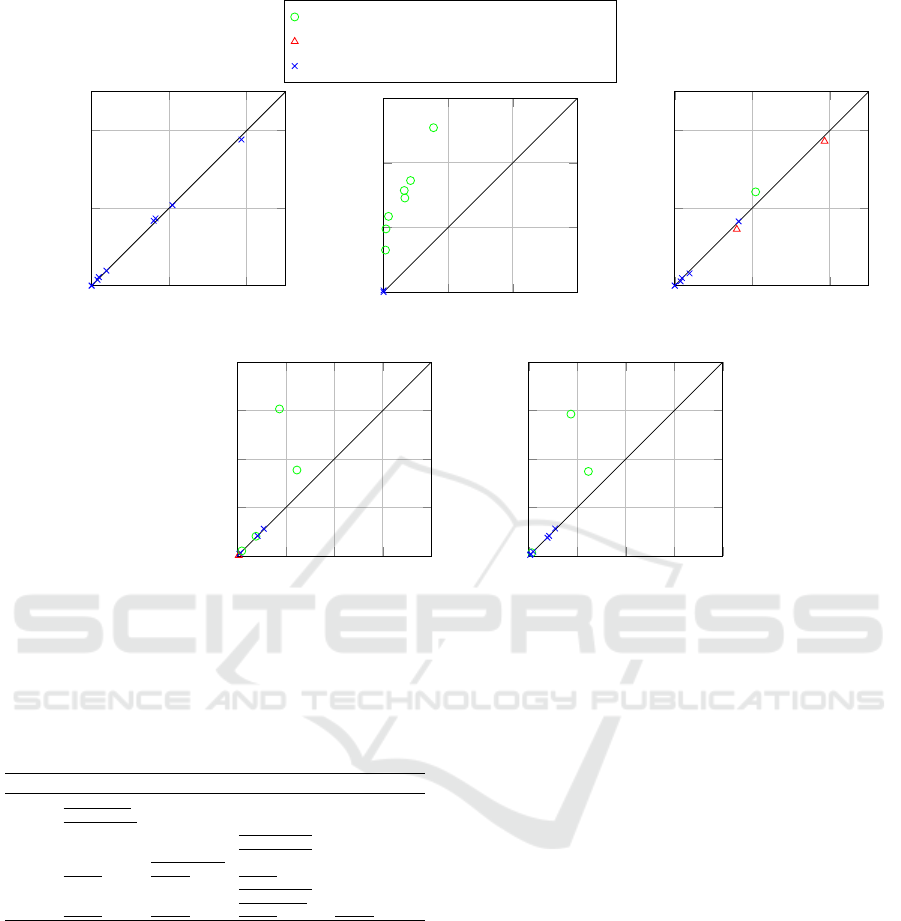
In Table 3 we present the results obtained from
PTE-LR, ToP-LR, and individual Logistic Regres-
sion, each optimised using the multi-class AUC-Loss
function as detailed above. The datasets are identified
by their OpenML Task ID. The table provides the av-
erage loss and the corresponding standard deviation
as obtained from cross-validation. Moreover, it high-
lights instances of statistical significance where ap-
plicable. A significant improvement of PTE-LR over
a baseline is highlighted with symbol •. Conversely,
a significant degradation in performance is indicated
by the symbol ◦. To further elucidate our findings,
Figure 4 provides a visual representation of the re-
sults obtained. We find that when employing Logistic
Regression as the sole base learner, PTEs outperform
ToPs in seven out of nine instances. Interestingly, in
four of these seven cases, the improvement is statis-
tically significant, while only one instance of signifi-
cant degradation is observed.
This divergence in performance, where we ob-
serve slight performance degradation for some
datasets and significant improvement for others, could
potentially be attributed to the inherent nature of the
classes within these datasets. In the field of statis-
tics, the recommendation to employ NDs is contin-
gent upon having a substantial rationale for selecting
specific dichotomies. For instance, if there is some
particular ordering of classes, it might be advanta-
geous to cluster together classes that fall within the
same order spectrum, either lower or higher (Fox,
2016). Hence, we hypothesise that datasets demon-
strating significant improvements may possess such
intrinsic characteristics, making them more apt for the
application of Class Dichotomies.
We substantiate this hypothesis by examining the
dataset where PTEs exhibit the most significant per-
formance enhancement over ToPs. In the case of the
cardiotocography dataset, we note a reduction in loss
of over two-thirds, which is more than twice the rate
of improvement observed in any other case. This
dataset categorises each instance based on the class
(1-10) of the fetal heart rate pattern, with classes 1-8
corresponding to normal baselines, class 9 to mod-
erate bradycardia, and class 10 to severe bradycar-
dia (Maso et al., 2012). Our conjecture is that, for
this dataset, the similarity among certain classes (1-
8, 9-10) makes the application of a Class Dichotomy
especially advantageous.
Conversely, for datasets where such properties are
absent, PTEs might experience minor degradations
due to factors such as noise, or increased overfit-
ting owing to their higher flexibility. Moreover, it is
important to highlight that, in certain instances, the
random-pair method might more readily discover a
beneficial partitioning of classes compared to other
scenarios.
We hypothesise that the substantial performance
degradation observed on the wall-robot-navigation
dataset is likely attributable to overfitting. Interest-
ingly, this dataset has only four classes, while all oth-
ers contain at least ten classes. This observation sug-
gests that Class Dichotomies might be more advanta-
geous for datasets with a greater number of classes.
While PTE-LR statistically outperforms simple
Partition Tree Ensembles for Improving Multi-Class Classification
63
Dataset with Significant Improvement
Dataset with Significant Deterioration
Dataset with Neither
0 0.02 0.04
0
0.02
0.04
PTE-ALL
ToP-ALL
0
0.05
0.1
0.15
0
0.05
0.1
0.15
PTE-ALL
AdaBoost
0 0.02 0.04
0
0.02
0.04
PTE-ALL
RF
0
0.05
0.1
0.15
0.2
0
0.05
0.1
0.15
0.2
PTE-LR
ToP-LR
0
0.05
0.1
0.15
0.2
0
0.05
0.1
0.15
0.2
PTE-LR
LR
Figure 4: Scatter plots contrasting the AUC-Loss of PTE-ALL with baselines ToP-ALL, Random Forest, and AdaBoost, as
well as the AUC-Loss of PTE-LR compared to ToP-LR and Logistic Regression. Each data point in these plots represents the
mean loss obtained over the ten cross-validation folds for a specific OpenML task. Data points are color-coded to highlight
the statistical significance of the performance difference between PTE and the respective baselines.
Table 4: Loss of PTE-ALL and baselines optimised for
ROC-AUC.
Task ID PTE-ALL ToP-ALL RF AdaBoost
14 0.0165±0.003 0.0174±0.0039 0.0166±0.0032 0.0728±0.0184 •
16 0.0019±0.0009 0.0022±0.0012 0.0019±0.0012 0.0489±0.0118 •
18 0.0387±0.0073 0.0377±0.0082 0.0372±0.0072 ◦ 0.1272±0.02 •
20 0.0015±0.0011 0.0015±0.001 0.0011±0.0008 0.0325±0.0105 •
22 0.0209±0.003 0.0207±0.0042 0.0242±0.0036 • 0.0863±0.0397 •
9942 0.0±0.0 0.0±0.0 0.0±0.0 0.0013±0.0042
9955 0.016±0.0033 0.0167±0.0042 0.0145±0.0025 ◦ 0.0787±0.0112 •
9964 0.0038±0.0015 0.0039±0.0017 0.0032±0.001 0.0586±0.0166 •
9979 0.0±0.0 0.0±0.0 0.0±0.0 0.0±0.0
Logistic Regression in three instances, we note that
Logistic Regression slightly surpasses PTE-LR on
five datasets. A similar pattern of degraded perfor-
mance is observed when comparing ToP-LR to Logis-
tic Regression. These observations suggest that both
tree-based ensemble methods might be overfitting the
training data, thereby offering an avenue for further
investigation and optimisation of these methods.
In Table 4, we present the outcomes derived from
PTE-ALL, ToP-ALL, Random Forest, and AdaBoost.
Exploiting a broader set of base learners, we note
outcomes akin to those observed when using solely
Logistic Regression. Specifically, PTE-ALL out-
performs ToP-ALL in five scenarios, while the re-
verse occurs in two instances, with no statistically
significant differences in these comparisons. How-
ever, PTE-ALL exceeds the performance of Random
Forest in three cases and falls behind in four, with
one instance of statistically significant improvement
and two cases of statistically significant degradation.
Meanwhile PTE-ALL consistantly outperforms Ad-
aBoost. Taking into account these results, we can
assert that for the AUC-Loss function, incorporating
Class Dichotomies into tree-based ensembling is in-
deed advantageous. Nevertheless, the comparison to
Random Forest raises doubts about the merits of at-
tempting to enhance Random Forest by employing it
as a base learner in PTEs or ToPs.
Evaluating the performance of the three distinct
base learners, we find that Random Forest is the
most proficient in capturing the underlying distribu-
tion of the datasets we explored. Given that its base
learners—Decision Trees—have an intrinsic ability
to partition the feature space, we hypothesise that
they might not obtain significant advantages from the
tree-based partitioning characteristic of our ensemble
methods.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
64

Table 5: Runtime of PTE-ALL and baselines optimised for ROC-AUC.
Task ID PTE-ALL ToP-ALL PTE-LR ToP-LR RF AdaBoost LR
14 171.6819±58.8443 77.7228±19.3721 ◦ 1.0283±0.0362 ◦ 0.5091±0.0336 ◦ 31.272±6.8324 ◦ 3.5745±0.2582 ◦ 0.1625±0.0078 ◦
16 176.6548±72.4138 59.2395±24.3352 ◦ 1.1891±0.6958 ◦ 0.4631±0.0257 ◦ 27.6619±11.8803 ◦ 2.951±0.2188 ◦ 0.1332±0.0067
◦
18 55.1±44.2909 31.1692±20.3078 2.351±0.6888 ◦ 0.3143±0.0415 ◦ 2.0931±1.7565 ◦ 0.1606±0.1092 ◦ 0.1049±0.0019 ◦
20 147.4708±61.4661 60.185±15.4995 ◦ 19.8405±10.0204 ◦ 12.1076±1.5598 ◦ 3.2289±1.1929 ◦ 9.7781±1.0633 ◦ 0.7686±0.4613 ◦
22 89.3503±18.7013 42.5977±6.5014 ◦ 0.9597±0.2382 ◦ 0.4143±0.0595 ◦ 25.8293±2.9524 ◦ 2.3192±0.1968 ◦ 0.1253±0.0027 ◦
9942 771.8217±519.4681 688.7455±305.1365 15.7081±3.163 ◦ 14.4876±2.9639 ◦ 0.7687±1.1615 ◦ 0.6253±0.04 ◦ 0.119±0.0068 ◦
9955 171.4221±47.3616 69.4383±9.6315 ◦ 31.0588±11.3884 ◦ 10.0724±0.5299 ◦ 22.2991±25.0351 ◦ 3.4312±0.7699 ◦ 0.5485±0.0541 ◦
9964 98.5397±45.7543 38.8915±7.974 ◦ 2.0098±0.628 ◦ 0.6526±0.0767 ◦ 2.4775±0.5695 ◦ 1.2225±0.0923 ◦ 0.2283±0.0333 ◦
9979 31.1584±2.2996 15.5143±0.26 ◦ 4.0713±0.6113 ◦ 0.4265±0.0843 ◦ 0.5081±0.7243 ◦ 0.5961±0.0134 ◦ 0.126±0.005 ◦
Table 5 displays the average runtime of each
method and dataset. We observe that PTEs exhibit
significantly longer runtimes compared to ToPs for
both families of base learners. The increased runtime
can range from two-fold to almost ten-fold in some
cases.
5.2 Experiments Using F1-Loss
This section outlines the results from our experiments
using the F1-Loss. F1 is a metric that reflects a clas-
sifier’s precision and recall, thereby providing a bal-
anced evaluation of the model’s performance.
5.2.1 Motivation
The F1-Measure is a widely-used evaluation metric,
due to its ability to better manage class imbalances
compared to accuracy. In situations where the dataset
is unbalanced accuracy tends to become an unreliable
measure because it often gives an overly optimistic es-
timation based on the classifier’s performance on the
majority class, thereby obscuring its potential short-
comings in predicting minority classes (Chicco and
Jurman, 2020). Conversely, the F1-Measure repre-
sents the harmonic mean of Precision and Recall.
Given that the harmonic mean of two numbers tends
to be closer to the smaller one, a high F1-Measure
signifies a strong Recall and Precision, thus providing
a more appropriate measure of the classifier’s perfor-
mance in the presence of class imbalances (Gu et al.,
2009).
5.2.2 Multi-Class F1
In our multi-class classification experiments we use
the Macro F1-Score described in (Grandini et al.,
2020). The Macro F1-Score is calculated based on the
Macro Average Precision (MVP) and Recall (MVR).
We calculate the macro average of a measure by ap-
plying it to all classes individually and taking the
arithmetic mean of the results.
Table 6: Loss of PTE-LR and baselines optimised for F1-
Score.
Task ID PTE-LR ToP-LR LR
14 0.1687±0.0302 0.1788±0.0301 • 0.1739±0.0232
16 0.0457±0.0176 0.0425±0.0145 0.0421±0.0162 ◦
18 0.3851±0.1047 0.5287±0.0391 • 0.533±0.032 •
20 0.0285±0.0111 0.0325±0.012 0.029±0.0113
22 0.2003±0.0148 0.1837±0.0366 0.1928±0.0183
9942 0.0174±0.0143 0.0158±0.007 0.0312±0.009 •
9955 0.5356±0.0337 0.5344±0.0314 0.5433±0.0332
9964 0.083±0.0192 0.079±0.0167 0.085±0.0244
9979 0.3761±0.0839 0.645±0.038 • 0.6192±0.0414 •
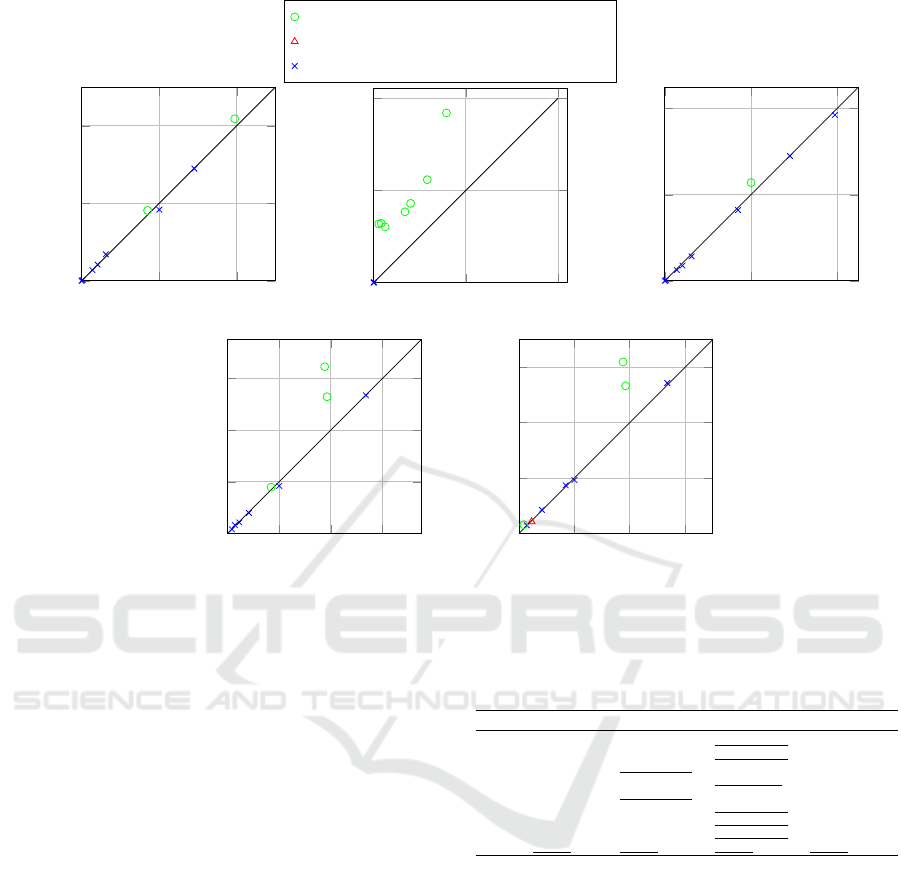
5.2.3 Experimental Results
In Table 6, we present the mean loss obtained for each
dataset using PTE-LR, ToP-LR, and individual Lo-
gistic Regression when optimising for multi-class F1-
Loss.
Figure 5 provides a visual representation of the re-
sults obtained.
We observe that PTE-LR outperforms ToP-LR in
four cases, with three of them showing statistically
significant differences. Conversely, ToP-LR achieves
better results in five instances, although none of these
differences are statistically significant. This pattern,
characterised by slight performance degradation on
some datasets and significant gains on others, mirrors
what we previously observed in our experiments with
AUC-Loss.
In Section 5.1.3 we hypothesised that these vari-
ations in performance can be attributed to certain
datasets possessing intrinsic characteristics that make
them more amenable to the application of Class Di-
chotomies. Interestingly, all three datasets that exhib-
ited significant improvements in F1-Loss also showed
significant improvements in AUC-Loss, reinforcing
our conjecture that, regardless of the loss function em-
ployed, there exists a property in the underlying dis-
tribution that can be more effectively captured when
utilising Class Dichotomies.
On the other hand, the minor performance degra-
dations observed on datasets where this property is
not apparent may again be attributed to factors such
as noise and overfitting, which can pose challenges
for the PTEs.
Partition Tree Ensembles for Improving Multi-Class Classification
65
Dataset with Significant Improvement
Dataset with Significant Deterioration
Dataset with Neither
0 0.2 0.4
0
0.2
0.4
PTE-ALL
ToP-ALL
0
0.5
1
0
0.5
1
PTE-ALL
AdaBoost
0 0.2 0.4
0
0.2
0.4
PTE-ALL
RF
0 0.2 0.4
0.6
0
0.2
0.4
0.6
PTE-LR
ToP-LR
0 0.2 0.4
0.6
0
0.2
0.4
0.6
PTE-LR
LR
Figure 5: Scatter plots contrasting the F1-Loss of PTE-ALL with baselines ToP-ALL, Random Forest, and AdaBoost, as well
as the F1-Loss of PTE-LR compared to ToP-LR and Logistic Regression.
When comparing PTE-LR with simple Logistic
Regression, we continue to observe a similar pattern
as in the previous experiments. PTE-LR outperforms
Logistic Regression in seven cases, with three of them
showing statistically significant improvements. Con-
versely, Logistic Regression outperforms PTE-LR in
two cases, with one of them being statistically signif-
icant. Based on the observed results, it is evident that
overfitting is a significant challenge for both ToP-LR
and PTE-LR, with the more flexible PTE-LR model
being particularly susceptible. This conclusion is sup-
ported by the fact that, in datasets where individual
Logistic Regression outperforms PTE-LR, ToP-LR
also demonstrates performance superior to PTE-LR.
Therefore, in future research it will be essential to ad-
dress the issue of overfitting in order to enhance the
performance of our ensemble method further.
The outcomes of PTE-ALL, ToP-ALL, Random
Forest, and AdaBoost optimised for F1-Loss are pre-
sented in Table 7.
Consistent with previous findings, PTE-ALL out-
performs ToP-ALL in the majority of cases, with two
instances demonstrating statistical significance. Fur-
thermore, there is one case where PTE-ALL exhibits a
significant improvement over Random Forest. We ob-
serve that the runtime for optimising F1-Loss is com-
Table 7: Loss of PTE-ALL and baselines optimised for F1-
Score.
Task ID PTE-ALL ToP-ALL RF AdaBoost
14 0.1701±0.0209 0.1818±0.0246 • 0.1644±0.0235 0.3836±0.0634 •
16 0.0411±0.0152 0.0421±0.0114 0.0355±0.0137 0.321±0.1003 •
18 0.2909±0.0233 0.2896±0.0283 0.2899±0.0291 0.5574±0.0789 •
20 0.0279±0.0101 0.028±0.0088 0.0255±0.005 0.318±0.2167 •
22 0.2003±0.0148 0.1837±0.0366 0.2281±0.0291 • 0.4291±0.0626 •
9942 0.0023±0.0074 0.0023±0.0072 0.0014±0.0043 0.0023±0.0074
9955 0.3949±0.049 0.4181±0.0438 • 0.3852±0.0412 0.9187±0.0181 •
9964 0.0624±0.0132 0.0685±0.0114 0.0569±0.0097 0.3017±0.0583 •
9979 0.0±0.0 0.0±0.0 0.0±0.0 0.0±0.0
parable to that of AUC-Loss.
6 CONCLUSION
This paper introduced Partition Tree Ensembles, a
tree-based ensemble method that incorporates ideas
from Nested Dichotomies and Trees of Predictors
to improve multi-class classification performance.
Through an extensive experimental evaluation, we
demonstrated that our approach achieves significant
performance gains over its predecessor Tree of Pre-
dictors across various datasets and loss functions.
Furthermore, we observed significant improvements
over Random Forest on several datasets. In compari-
son with AdaBoost, our method demonstrated signif-
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
66

icantly higher performance in nearly all cases. How-
ever, this comes at the cost of a significantly increased
runtime.
There are several avenues for future research that
stand to be explored. Firstly, we intend to apply PTEs
to other families of base learners. Recent studies
have shown that well-tuned neural networks exhibit
state-of-the-art performance on tabular data (Kadra
et al., 2021), and it would be interesting to investigate
whether ensembling using our Partition Tree Method
can further improve their performance.
Secondly, we suspect that there is potential for en-
hancing performance by employing alternative strate-
gies for selecting and weighting the base learners.
In the current algorithm, the subtrees of each class
dichotomy allocate a portion of the training data
to replace the second validation set. Future work
could explore using greedy forward selection, pro-
posed in (Caruana et al., 2004), to determine the opti-
mal weight for each base learner and the best possible
split using only a single validation set. Furthermore,
it might be valuable to investigate the benefits of re-
taining base learners in the final ensemble that were
trained but not selected as optimal.
Finally, an interesting avenue for future research
would be to explore the strategy of sampling a subset
of attributes to evaluate for each split in the tree con-
struction process. This approach has the potential to
mitigate overfitting and improve runtime efficiency.
In conclusion, the development of Partition Tree
Ensembles presents a promising approach for enhanc-
ing multi-class classification performance. By lever-
aging ideas from Nested Dichotomies and Trees of
Predictors, our method offers significant performance
gains over the Trees of Predictors method.
ACKNOWLEDGMENTS
This work was performed using resources provided
by the Cambridge Service for Data Driven Discov-
ery (CSD3) operated by the University of Cambridge
Research Computing Service (www.csd3.cam.ac.uk),
provided by Dell EMC and Intel using Tier-2 funding
from the Engineering and Physical Sciences Research
Council (capital grant EP/T022159/1), and DiRAC
funding from the Science and Technology Facilities
Council (www.dirac.ac.uk). For the purpose of open
access, the author has applied a Creative Commons
Attribution (CC BY) licence to any Author Accepted
Manuscript version arising from this submission.
REFERENCES
Beneke, M., K
¨
onig, M., and Link, M. (2023). The inverted
pendulum as a classical analog of the eft paradigm.
Beygelzimer, A., Langford, J., and Ravikumar, P.
(2007). Multiclass classification with filter
trees, 2007. Preprint, available at http://hunch.
net/jl/projects/reductions/mc to b/invertedTree. pdf.
Breiman, L. (2001). Random forests. Machine Learning,
45(1):5–32.
Breiman, L., Friedman, J., Stone, C., and Olshen, R. (1984).
Classification and Regression Trees. Taylor & Francis.
Caruana, R., Niculescu-Mizil, A., Crew, G., and Ksikes, A.
(2004). Ensemble selection from libraries of models.
In Proceedings of the Twenty-First International Con-
ference on Machine Learning, ICML ’04, page 18,
New York, NY, USA. Association for Computing Ma-
chinery.
Chicco, D. and Jurman, G. (2020). The advantages of the
matthews correlation coefficient (MCC) over F1 score
and accuracy in binary classification evaluation. BMC
Genomics, 21(1):6.
Cortes, C. and Mohri, M. (2003). Auc optimization vs.
error rate minimization. In Thrun, S., Saul, L., and
Sch
¨
olkopf, B., editors, Advances in Neural Informa-
tion Processing Systems, volume 16. MIT Press.
Dua, D. and Graff, C. (2017). UCI machine learning repos-
itory.
Fox, J. (2016). Applied Regression Analysis and General-
ized Linear Models. SAGE, London.
Frank, E. and Kramer, S. (2004). Ensembles of nested di-
chotomies for multi-class problems. In Brodley, C. E.,
editor, Machine Learning, Proceedings of the Twenty-
first International Conference (ICML 2004), Banff, Al-
berta, Canada, July 4-8, 2004, volume 69 of ACM In-
ternational Conference Proceeding Series. ACM.
Freund, Y., Schapire, R., and Abe, N. (1999). A short in-
troduction to boosting. Journal-Japanese Society For
Artificial Intelligence, 14(771-780):1612.
Grandini, M., Bagli, E., and Visani, G. (2020). Metrics for
multi-class classification: an overview. arXiv preprint
arXiv:2008.05756.
Gu, Q., Zhu, L., and Cai, Z. (2009). Evaluation measures
of the classification performance of imbalanced data
sets. In Cai, Z., Li, Z., Kang, Z., and Liu, Y., editors,
Computational Intelligence and Intelligent Systems,
pages 461–471, Berlin, Heidelberg. Springer Berlin
Heidelberg.
Hand, D. J. and Till, R. J. (2001). A simple generalisation of
the area under the roc curve for multiple class classifi-
cation problems. Machine Learning, 45(2):171–186.
Kadra, A., Lindauer, M., Hutter, F., and Grabocka, J.
(2021). Well-tuned simple nets excel on tabular
datasets. Advances in neural information processing
systems, 34:23928–23941.
Leathart, T., Pfahringer, B., and Frank, E. (2016). Build-
ing ensembles of adaptive nested dichotomies with
random-pair selection. In Frasconi, P., Landwehr,
N., Manco, G., and Vreeken, J., editors, Machine
Learning and Knowledge Discovery in Databases -
Partition Tree Ensembles for Improving Multi-Class Classification
67

European Conference, ECML PKDD 2016, Riva del
Garda, Italy, September 19-23, 2016, Proceedings,
Part II, volume 9852 of Lecture Notes in Computer
Science, pages 179–194. Springer.
Leathart, T. M. (2019). Tree-structured multiclass probabil-
ity estimators. PhD thesis, The University of Waikato,
Hamilton, New Zealand. Doctoral.
Lindauer, M., Eggensperger, K., Feurer, M., Biedenkapp,
A., Deng, D., Benjamins, C., Ruhkopf, T., Sass, R.,
and Hutter, F. (2022). Smac3: A versatile bayesian op-
timization package for hyperparameter optimization.
Journal of Machine Learning Research, 23(54):1–9.
Malerba, D., Appice, A., Bellino, A., Ceci, M., and Pal-
lotta, D. (2001). Stepwise induction of model trees.
In Esposito, F., editor, AI*IA 2001: Advances in Ar-
tificial Intelligence, pages 20–32, Berlin, Heidelberg.
Springer Berlin Heidelberg.
Maso, G., Businelli, C., Piccoli, M., Montico, M., De Seta,
F., Sartore, A., and Alberico, S. (2012). The clini-
cal interpretation and significance of electronic fetal
heart rate patterns 2 h before delivery: an institutional
observational study. Archives of Gynecology and Ob-
stetrics, 286(5):1153–1159.
Mohr, F., Wever, M., and H
¨
ullermeier, E. (2018). Reduction
stumps for multi-class classification. In Duivesteijn,
W., Siebes, A., and Ukkonen, A., editors, Advances in
Intelligent Data Analysis XVII, pages 225–237, Cham.
Springer International Publishing.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V.,
Thirion, B., Grisel, O., Blondel, M., Prettenhofer,
P., Weiss, R., Dubourg, V., Vanderplas, J., Passos,
A., Cournapeau, D., Brucher, M., Perrot, M., and
Duchesnay, E. (2011). Scikit-learn: Machine learning
in Python. Journal of Machine Learning Research,
12:2825–2830.
Quinlan, J. R. (1986). Induction of decision trees. Machine
Learning, 1(1):81–106.
Quinlan, J. R. et al. (1992). Learning with continuous
classes. In 5th Australian joint conference on artifi-
cial intelligence, volume 92, pages 343–348. World
Scientific.
Rey, D. and Neuh
¨
auser, M. (2011). Wilcoxon-Signed-Rank
Test, pages 1658–1659. Springer Berlin Heidelberg,
Berlin, Heidelberg.
Singh, J. (2023). Computational complexity and analysis of
supervised machine learning algorithms. In Kumar,
R., Pattnaik, P. K., and R. S. Tavares, J. M., editors,
Next Generation of Internet of Things, pages 195–206,
Singapore. Springer Nature Singapore.
Torgo, L. (1997a). Functional models for regression tree
leaves. In ICML, volume 97, pages 385–393. Citeseer.
Torgo, L. (1997b). Kernel regression trees. In Poster papers
of the 9th European conference on machine learning
(ECML 97), pages 118–127. Prague, Czech Republic.
Wang, Y. and Witten, I. (1997). Induction of model trees
for predicting continuous classes. Induction of Model
Trees for Predicting Continuous Classes.
Wever, M., Mohr, F., and H
¨
ullermeier, E. (2018). Ensem-
bles of evolved nested dichotomies for classification.
In Aguirre, H. E. and Takadama, K., editors, Pro-
ceedings of the Genetic and Evolutionary Computa-
tion Conference, GECCO 2018, Kyoto, Japan, July
15-19, 2018, pages 561–568. ACM.
Wever, M.,
¨
Ozdogan, M., and H
¨
ullermeier, E. (2023). Co-
operative co-evolution for ensembles of nested di-
chotomies for multi-class classification. In Proceed-
ings of the Genetic and Evolutionary Computation
Conference, pages 597–605.
Wolpert, D. H. (1992). Stacked generalization. Neural Net-
works, 5(2):241–259.
Yoon, J., Zame, W. R., Banerjee, A., Cadeiras, M., Alaa,
A. M., and van der Schaar, M. (2018a). Personal-
ized survival predictions via trees of predictors: An
application to cardiac transplantation. PLOS ONE,
13(3):e0194985.
Yoon, J., Zame, W. R., and van der Schaar, M. (2018b).
ToPs: Ensemble learning with trees of predictors.
IEEE Transactions on Signal Processing, 66(8):2141–
2152.
APPENDIX
Hyperparameter Tuning
In this Appendix, we outline the hyperparameter tun-
ing procedure employed to optimise our predictors. It
is important to note that we perform hyperparameter
tuning for each cross-validation split and loss function
evaluated.
We implement our hyperparameter tuning proce-
dure using the state-of-the-art Bayesian Optimisation
framework SMAC3 (Lindauer et al., 2022). We em-
ploy the Hyperparameter Tuning facade, which uses
Random Forest as a surrogate model.
However, due to their substantial runtime, con-
ducting a comprehensive hyperparameter tuning pro-
cedure is not feasible within our constraints for both
PTEs and ToPs. Instead, we focus on tuning the hy-
perparameters of their respective base learners on the
specific dataset at hand. Although the optimal base
learner parameters for standalone use may differ from
those for ensemble methods, we assume that they are
a reasonable approximation.
Furthermore, we choose min leaf samples =
100, slightly higher than (Torgo, 1997a) to mitigate
overfitting. As done in (Yoon et al., 2018b), we set
val1 size = 0.15 and val2 size = 0.1.
In Table 8 we present the search space we set
for hyperparameter optimisation. For hyperparame-
ters not mentioned in this table, we rely on the default
values as provided by the scikit-learn library.
ICPRAM 2025 - 14th International Conference on Pattern Recognition Applications and Methods
68

Table 8: Hyperparameter search spaces.
Classifier Hyperparameter Search Space
Random Forest
n estimators {i|i ∈ N ∧ 10 ≤ i ≤ 1000}
criterion {“gini
′′
,“log loss
′′
}
max depth {i|i ∈ N ∧ 3 ≤ i ≤ 10}
AdaBoost
n estimators {i|i ∈ N ∧ 25 ≤ i ≤ 200}
learning rate [0.5,2.0]
Logistic Regression
penalty {“l2
′′
,“none
′′
}
C [0.5,5.0]
Proof of the Computational Complexity
Proof of Computational Complexity for One
Recursive Step
Statement: For a single recursive step of our
method the computational complexity is given by
O
nd
m
∑
i=1
(T
i
(n,d))
+
m
CD
∑
j=1
T
CD
j
(n,d) +
m
∑
i=1
(T
i
(n,d))
!!
(5)
Proof. Each recursive step consists of two main
parts. First, we create candidate splits of the feature
space as in ToPs. Second, we create candidate Class
Dichotomies.
The computational complexity of the splitting of
the feature space is shown in (Yoon et al., 2018b) to
be
O
nd
m
∑
i=1
T
i
(n,d)
!
. (6)
When creating a candidate Class Dichotomy for an
algorithm A
j
∈ A
CD
, we use the Random Pair method
to obtain the two meta-classes. In this process, we
require one execution of A
j
to obtain h
c
−
,c
+
, which
is used to determine the meta-classes. Subsequently,
another execution of A
j
is required to obtain the meta-
classifier h
Y
−
,Y
+
. Additionally, we need to select the
classifiers h
Y
−
and h
Y
+
. Both of these require one
execution each of every algorithm A ∈ A. In total, we
arrive at a complexity of
O
2 · T
CD
j
(n,d) +2 ·
m
∑
i=1
(T
i
(n,d))
!
= O
T
CD
j
(n,d) +
m
∑
i=1
(T
i
(n,d))
!
(7)
for the creation of a single Class Dichotomy. Since we
repeat this procedure for every A
j
∈ A
CD
we obtain
the complexity
O
m
CD
∑
j=1
T
CD
j
(n,d) +
m
∑
i=1
(T
i
(n,d))
!!
. (8)
Adding the complexity O(nd
∑
m
i=1
(T
i
(n,d))) of split-
ting the feature space we obtain
O
nd
m
∑
i=1
(T
i
(n,d))
+
m
CD
∑
j=1
T
CD
j
(n,d) +
m
∑
i=1
(T
i
(n,d))
!
. (9)
Proof of Computational Complexity for an Entire
Partition Tree Ensemble
Statement: For constructing an entire Partition
Tree Ensemble the computational complexity is
given by
O
n
nd
m
∑
i=1
(T
i
(n,d))
+
m
CD
∑
j=1
T
CD
j
(n,d) +
m
∑
i=1
(T
i
(n,d))
!
. (10)
Proof. To prove this, we must consider the max-
imum number of possible recursions. We know that
there cannot be more recursive calls than the number
of nodes in the tree. Furthermore, we understand that
every leaf node of our Partition Tree contains at least
one sample. Since there are n samples in total, the
tree can have at most n leaves. In Partition Trees, a
node only has children if we split it, in which case it
has two children. Therefore, Partition Trees are full
binary trees. Consequently, the number of recursive
calls is at most 2n − 1. This leads us to the overall
computational complexity of
O
2n − 1
nd
m
∑
i=1
T
i
(n,d) +
m
CD
∑
j=1
T
CD
j
(n,d)
+
m
∑
i=1
T
i
(n,d)
= O
n
nd
m
∑
i=1
T
i
(n,d) +
m
CD
∑
j=1
T
CD
j
(n,d)
+
m
∑
i=1
T
i
(n,d)
. (11)
Partition Tree Ensembles for Improving Multi-Class Classification
69
