
Deep Learning for ECG-Derived Respiration Using the Fantasia Dataset
Lana Dominkovi
´
c
1
, Biljana Mileva Boshkoska
1,2
and Aleksandra Rashkovska
1,2
1
Faculty of Information Studies, Ljubljanska cesta 31a, Novo Mesto, Slovenia
2
Jo
ˇ
zef Stefan Institute, Jamova cesta 39, Ljubljana, Slovenia
lana.caldarevic@student.fis.unm.si, {biljana.mileva, aleksandra.rashkovska}@fis.unm.si
Keywords:
ECG-Derived Respiration, Biosignal Analysis, Deep Learning, Signal Processing, Convolutional
Autoencoder, Fantasia, Respiratory Signal Estimation, Healthcare Monitoring.
Abstract:
In this paper, we explore a deep learning approach for extracting respiratory signals from electrocardiogram
(ECG) data using the Fantasia dataset. We implemented a fully convolutional neural network model, inspired
by the U-Net architecture, and designed to estimate respiratory signals from ECG data. The model incorpo-
rates convolutional layers, ReLU activations, batch normalization, max pooling, and up-sampling layers. Our
deep learning model achieved an average correlation coefficient (CC) of 0.51 and Mean Squared Error (MSE)
of 0.046, outperforming four out of six baseline signal processing algorithms based on the CC metric, and
outperforming all signal processing algorithms based on the MSE metric. These findings demonstrate the ef-
fectiveness of deep learning in improving the accuracy and robustness of ECG-derived respiration (EDR). The
research highlights the potential of advanced machine learning models for non-invasive respiratory monitoring
and paves the way for future studies focused on exploring more complex architectures and broader datasets to
further enhance performance and generalizability.
1 INTRODUCTION
ECG-derived respiration (EDR) is an emerging tech-
nique that extracts respiratory signals from electro-
cardiogram (ECG) data, offering valuable respiratory
information without the need for additional sensors.
This method is particularly advantageous in health-
care, where reducing the complexity of monitoring
devices while maintaining comprehensive physiolog-
ical data is critical. EDR enables continuous, non-
invasive patient monitoring by providing both cardiac
and respiratory information from a single source. The
motivation for EDR stems from the increasing de-
mand for cost-effective and multi-functional health-
care solutions. Multi-functional body sensors capa-
ble of capturing several physiological signals, such as
ECG and respiration, represent a significant advance-
ment in personalized healthcare (Trobec et al., 2014).
In recent years, artificial neural networks have
achieved state-of-the-art results across numerous do-
mains, including healthcare, where deep learning
models excel at capturing complex data representa-
tions. Architectures like autoencoders, convolutional
neural networks, and recurrent neural networks have
been successfully applied to medical tasks such as
noise reduction, arrhythmia detection, and predictive
analytics (Is¸ın and Ozdalili, 2017; Hire
ˇ
s et al., 2022;
Chiang et al., 2019). Unlike traditional signal pro-
cessing techniques, deep learning models can uncover
complex, non-linear patterns in time-series data, mak-
ing them particularly well-suited for extracting full
respiratory waveforms from ECG data. Deep learn-
ing methods also adapt better to variations in patient
physiology and signal noise, resulting in more ro-
bust and accurate outputs. This allows for enhanced
biosignal processing and the derivation of richer in-
sights, which are critical for patient monitoring and
personalized healthcare.
The goal of this paper is to apply deep learning
techniques to derive respiratory signals from ECG
data, using the Fantasia dataset as the primary re-
source. By building on the baselines established
through traditional signal processing methods in pre-
vious studies, particularly (Dominkovi
´
c et al., 2024),
this work aims to demonstrate the effectiveness of
deep learning in improving the accuracy and robust-
ness of EDR. Ultimately, the objective is to surpass
the performance of existing traditional signal process-
ing approaches and advance the use of deep learning
in biosignal analysis for enhanced respiratory moni-
toring.
The main contributions of this work are: (i) ex-
564
Dominkovi
´
c, L., Boshkoska, B. M. and Rashkovska, A.
Deep Learning for ECG-Derived Respiration Using the Fantasia Dataset.
DOI: 10.5220/0013165300003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 564-570
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

tending the efforts in (Dominkovi
´
c et al., 2024) by ap-
plying deep learning methods to the Fantasia dataset,
(ii) presenting the initial results of these advanced
techniques, and (iii) demonstrating improved results
with deep learning models compared to signal pro-
cessing methods.
The rest of the paper is structured as follows: Sec-
tion 2 reviews the related work on applying deep
learning to EDR. In Section 3, we outline the exper-
imental data and data processing methods, provide
a brief overview of the signal-processing algorithms
used as baselines, describe the deep learning model
architecture, and detail the experimental setup (train-
ing, evaluation, and performance metrics). In Section
4, we present and discuss the results. We conclude
with the final remarks and directions for future work
in Section 5.
2 RELATED WORK
In our previous work (Dominkovi
´
c et al., 2024), the
RRest toolbox (Charlton et al., 2016) was used to ex-
tract respiratory signals from ECG data in the Fan-
tasia dataset. The study establishes baseline perfor-
mance metrics for feature-based and filter-based sig-
nal processing methods. Feature-based methods fo-
cus on identifying and extracting specific features in
the ECG waveform, such as amplitude or frequency
variations, which are influenced by respiration. In
contrast, filter-based methods isolate the frequency
components within the ECG signal which correspond
to the respiratory cycle. The results from the baseline
methods offer reliable benchmarks for evaluating fu-
ture deep learning approaches, providing a foundation
for comparing advanced models that aim to improve
the accuracy and robustness of ECG-derived respira-
tion.
In this study, we utilize the Fantasia dataset, which
has been previously also used to develop signal pro-
cessing methods for cardiac and respiratory monitor-
ing. For example, it was employed to evaluate an al-
gorithm for estimating EDR by comparing it to vari-
ous signal processing approaches for respiratory sig-
nal extraction (Schmidt et al., 2015). Additionally, it
was utilized to create a system for combined cardiac
and respiratory monitoring (Brandwood et al., 2023),
extracting both heart rate and respiration data from a
single-lead ECG signal.
Most existing studies primarily focus on estimat-
ing the respiratory rate rather than extracting the com-
plete respiratory waveform. This limited scope over-
looks the potential wealth of information that can be
derived from full respiratory signals, which could of-
fer deeper insights into various physiological condi-
tions. The current emphasis on respiratory rate es-
timation, while useful, does not fully exploit the ca-
pabilities of deep learning models in biosignal anal-
ysis. Research on extracting respiratory signals from
ECG and photoplethysmogram (PPG) signals using
deep learning is still in its infancy. Some promising
results of applying deep learning methods for respira-
tion extraction include the RespNet model (Ravichan-
dran et al., 2019), which employs a U-Net architec-
ture, which has demonstrated high precision in pre-
dicting respiration from PPG signals. Additionally, in
(Merdjanovska and Rashkovska, 2020), the RespNet
architecture was adapted for a custom dataset to ex-
tract respiration from ECG signals.
Future research should aim to extend beyond res-
piratory rate to extract full respiratory signals, which
could enhance diagnostic and monitoring capabili-
ties in healthcare. To this end, several promising
deep learning approaches can be explored. Recent
advancements in Transformer architectures and Gen-
erative Adversarial Networks (GANs) open up new
avenues for innovative solutions. For example, Cy-
cle GANs have been effectively used to derive respi-
ratory rates from PPG signals (Aqajari et al., 2021),
and similar techniques could be adapted for extract-
ing respiratory signals from ECG data. Addition-
ally, the Reservoir Computing framework (Gauthier
et al., 2021), known for its efficiency in handling dy-
namic systems with faster training times and reduced
data requirements, offers a promising approach for
this problem. Exploring these advanced architectures
and techniques could lead to breakthroughs in extract-
ing detailed respiratory signals from ECG data, ulti-
mately advancing the state of healthcare monitoring
and diagnostics.
3 MATERIALS AND METHODS
3.1 Data Description
We utilized the Fantasia dataset to establish baselines
for respiratory signal analysis using traditional sig-
nal processing methods and to compare with our deep
learning solution.
The Fantasia dataset is a publicly available re-
source provided by PhysioNet (Goldberger et al.,
2000). It contains long-term ECG and respiration sig-
nal recordings from 40 healthy subjects. The dataset
is evenly divided into two age groups: 20 younger
adults (ages 21-34) and 20 elderly adults (ages 68-
85). Each subject was monitored for approximately
two hours while lying in a supine position and watch-
Deep Learning for ECG-Derived Respiration Using the Fantasia Dataset
565

ing a movie Fantasia, ensuring stable conditions for
heart rate variability (HRV) and ECG-derived respi-
ration (EDR) analysis. The ECG signals were sam-
pled at 250 Hz, providing high temporal resolution
for detailed signal analysis. The data is raw and re-
quires preprocessing before use, offering flexibility in
applying various signal processing techniques to the
dataset. Here are the key details of the dataset:
• Number of Subjects: 40 (20 young, 20 elderly),
• Age: Young subjects (21-34 years), Elderly sub-
jects (68-85 years),
• Condition: Healthy,
• Sampling Frequency: 250 Hz,
• Duration: 120 minutes.
3.2 Data Preprocessing
The ECG and respiration signals are processed us-
ing the NeuroKit2 library (Makowski et al., 2021).
Missing values are interpolated using a linear method,
and the signals are cleaned to remove noise and arti-
facts. In addition, we utilized parameters from the re-
search conducted by (Merdjanovska and Rashkovska,
2020) to further process ECG and respiration sig-
nals. Specifically, we employed 32-second length
windows with a 16-second overlap, normalized the
signals to a [0, 1] range, and downsampled them to
1024 samples (32 Hz) as described in (Merdjanovska
and Rashkovska, 2020). The ECG is treated as the
input variable to the proposed model, while the respi-
ratory signal is the output variable, i.e., the signal we
are trying to derive.
3.3 Deep Learning Architecture
The deep learning architecture is a fully convolu-
tional neural network designed to estimate respira-
tory signals from ECG data and is shown in Figure 1.
This network’s architecture is inspired by the U-Net
model, commonly used for image segmentation tasks.
Our implementation is a simplified version of U-Net,
retaining the essential concept of shortcut connec-
tions but with fewer layers and parameters. This sim-
plification, initially proposed in (Merdjanovska and
Rashkovska, 2020), has been proven effective and we
adopted the same approach in our work.
The network comprises an encoder and a decoder,
both fully convolutional. The encoder captures fea-
ture representations of the ECG signal by applying
various filters. Specifically, the network has three lev-
els, each with an increasing number of filters: 4 in the
first level, 8 in the second, and 16 in the third. Each
filter is a 1D convolution filter with a length of 27.
In the encoder, each convolutional layer is followed
by ReLU activation and batch normalization, ensur-
ing stable and effective training. Max pooling layers
are used to down-sample the signal, reducing its di-
mensionality and capturing important features.
The decoder mirrors the encoder’s structure but
performs upsampling to reconstruct the signal. The
decoder layers also include convolutional layers with
ReLU activation and batch normalization. Addition-
ally, the network uses dropout layers, with a dropout
rate of 0.6, to prevent overfitting. Overall, the net-
work consists of several convolutional layers, pooling
layers, up-sampling layers, and dropout layers, result-
ing in a robust architecture for respiratory signal es-
timation from ECG data. The network was imple-
mented using TensorFlow and trained on the appro-
priate hardware to handle the computational require-
ments. The model contained 23,409 trainable param-
eters.
3.4 Training and Evaluation Procedure
The training process employs an inter-patient evalua-
tion scheme (Merdjanovska and Rashkovska, 2021),
which is a more realistic approach to partitioning the
dataset. In this procedure, data from individual pa-
tients is kept distinct between the train, test and the
validation sets. This ensures that data from the same
patient do not overlap across sets, which would other-
wise lead to data leakage and inflate the model’s per-
formance metrics. By adopting an inter-patient split,
the evaluation process becomes more representative
of real-world scenarios, where the model is expected
to generalize to completely unseen patients.
Specifically, 90% of the patients were allocated to
the training and testing set, while the remaining 10%
were reserved for validation. The validation set is
used during the training process for hyperparameter
tuning to assess the performance of different model
configurations and to guide the selection of hyperpa-
rameter. During hyperparameter tuning, hyperparam-
eters such as learning rate, L2 regularization factor,
dropout rate, number of filters, kernel size, and batch
size are set. Hyperparameter tuning was conducted to
find the best combination of these parameters, ensur-
ing optimal performance.
For model evaluation, we used cross-validation, a
standard technique in machine learning that ensures
each record in the dataset is used as a test sample
exactly once. This approach helps in verifying the
model’s ability to generalize to new, unseen data.
Specifically, we implemented 5-fold cross-validation,
meaning that in each iteration, the model was trained
HEALTHINF 2025 - 18th International Conference on Health Informatics
566

Figure 1: Model architecture.
on 80% of the dataset set which was initially set aside
for training and testing, and tested on the remaining
20%. The average performance across all folds was
taken as the final evaluation metric. The model was
trained for up to 200 epochs with a batch size of 256,
using the Adam optimizer for its efficiency and effec-
tiveness. The learning rate for Adam was set to 3e-4.
The performance of the model was evaluated us-
ing two metrics: Mean Squared Error (MSE) and
Mean Cross-Correlation (CC). MSE calculates the av-
erage of the squared differences between estimated
values and actual values, with lower MSE values in-
dicating better accuracy. It is calculated as:
MSE =
1
n
n
∑
i=1
(r
i
− ˆr
i
)
2
where r
i
represents the true respiratory signal at time
i, ˆr
i
represents the predicted respiratory signal at time
i, and n is the total number of samples.
The CC, also known as the Pearson Correlation
Coefficient, measures the linear similarity between
the true respiratory signal and the predicted respira-
tory signal. It is defined as:
CC =
∑
n
i=1
(r
i
− ¯r)(ˆr
i
−
¯
ˆr)
p
∑
n
i=1
(r
i
− ¯r)
2
·
∑
n
i=1
(ˆr
i
−
¯
ˆr)
2
where r
i
and ˆr
i
are the true and predicted respiratory
signals, respectively, ¯r is the mean of the true respira-
tory signal:
¯r =
1
n
n
∑
i=1
r
i
,
and
¯
ˆr is the mean of the predicted respiratory signal:
¯
ˆr =
1
n
n
∑
i=1
ˆr
i
.
The CC values range from −1 (perfect negative cor-
relation) to +1 (perfect positive correlation). A CC
value of 0 indicates no correlation between the true
and predicted signals. Specifically, higher CC values
indicate a closer match between the estimated and ref-
erence signals, reflecting better performance
The average CC and MSE were measured across
each test fold to provide a comprehensive assessment
of the model performance. MSE was also used as the
loss function during training.
4 RESULTS AND DISCUSSION
The performance of our deep learning solution com-
pared to signal processing methods, extracted from
(Dominkovi
´
c et al., 2024), is shown in Table 1, with
Deep Learning for ECG-Derived Respiration Using the Fantasia Dataset
567
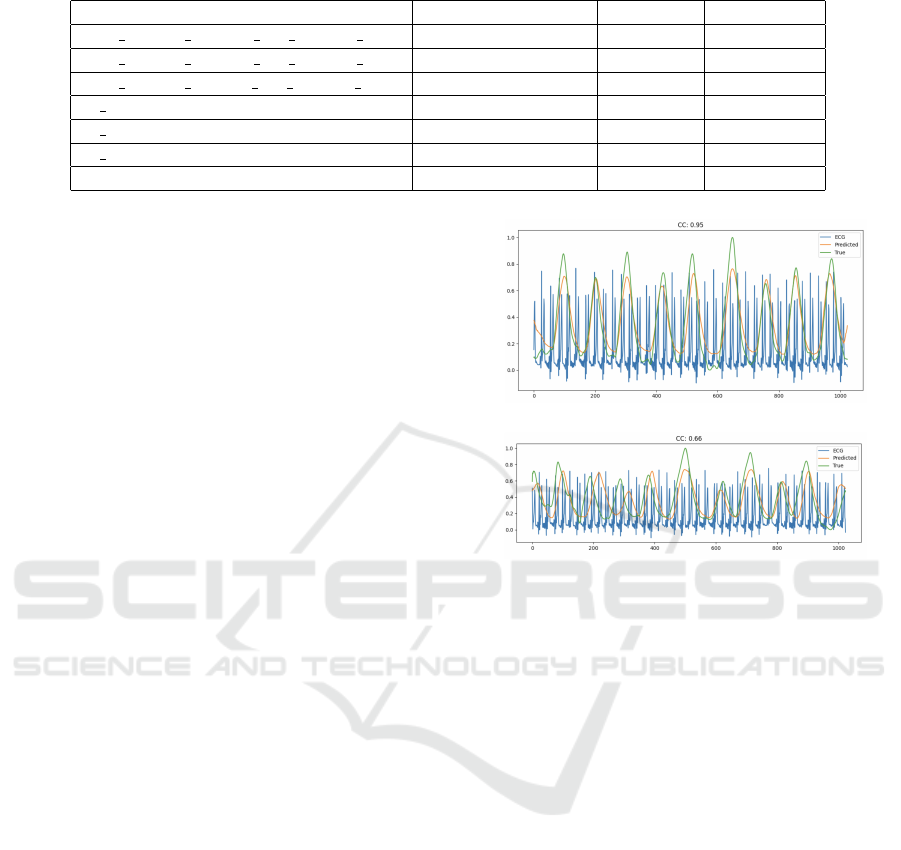
Table 1: Performance of signal processing and deep learning methods.
Method Type Mean CC Mean MSE
ELF RSlinB FMeam FPt RDtGC EHF RRest feature-based 0.59 0.073
ELF RSlinB FMebw FPt RDtGC EHF RRest feature-based 0.50 0.069
ELF RSlinB FMefm FPt RDtGC EHF RRest feature-based 0.56 0.070
flt BFi RRest filter-based 0.37 0.083
flt Wam RRest filter-based 0.38 0.093
flt Wfm RRest filter-based 0.44 0.081
U-net Deep Learning 0.51 0.046
the best performance metrics highlighted in bold. The
performance of our deep learning method resulted in
an average correlation coefficient (CC) of 0.51 and an
average Mean Squared Error (MSE) of 0.046.
Given that CC is a more critical metric for this
type of problem, the results highlight both strengths
and areas for improvement. Specifically, using CC
as the primary metric, the method outperformed 4
out of 6 signal processing algorithms. Moreover, it
demonstrated superior performance compared to all
filter-based algorithms and outperformed one of the
feature-based methods based on the CC metric. This
result is in agreement with the findings made in the
related study (Merdjanovska and Rashkovska, 2020),
where a similar U-net architecture on different cus-
tom dataset also outperformed the filter-based meth-
ods based on the CC metric, but was worse than the
feature-based.
While the model achieved superior performance
over all signal processing algorithms based on the
MSE metric, the CC results indicate room for refine-
ment. These findings emphasize the need to prioritize
optimization of CC in future work to ensure the deep
learning approach more consistently outperforms tra-
ditional methods across all relevant metrics.
For more visual representation, in Figure 2, we
show examples of measured and ECG-derived respi-
ratory signal with high correlation (CC = 0.95) and
lower correlation (CC = 0.66), alongside the ECG
signal. In the first example, there is a strong align-
ment between the actual and derived respiration sig-
nals, indicating good performance. However, in the
second example, more discrepancies are noticeable,
highlighting areas where the derived signal deviates
from the actual respiration.
5 CONCLUSIONS
In this work, we developed a simplified convolutional
autoencoder inspired by the U-Net model to estimate
respiratory signals from ECG data. The model used
convolutional layers, ReLU activations, and batch
(a)
(b)
Figure 2: Examples of measured and ECG-derived respi-
ration with (a) high correlation CC = 0.95 and (b) lower
correlation CC = 0.66.
normalization to effectively capture and reconstruct
respiratory signals. Data was segmented into 32-
second windows, and the model was trained using the
Adam optimizer and Mean Squared Error (MSE) as
the loss function.
Using 5-fold cross-validation, the model achieved
an average correlation coefficient (CC) of 0.51 and an
average MSE of 0.046. Our deep learning approach
outperformed 4 out of 6 traditional signal processing
methods based on the CC metric, and all signal pro-
cessing methods based on the MSE metric.
The results of this study are derived from a sin-
gle dataset, and considering additional datasets to test
our method would enhance the value of the find-
ings. Therefore, in addition to leveraging the Fanta-
sia dataset, exploring other publicly available datasets
for ECG-derived respiration, such as the BIDMC (Pi-
mentel and et al., 2016) and CapnoBase (Karlen et al.,
2010) datasets, could provide valuable insights and
improve model generalization. The BIDMC dataset
includes comprehensive ECG and respiratory signals
recorded from ICU patients, making it a useful re-
source for developing robust models that can handle
noisy, real-world data. The CapnoBase dataset, which
HEALTHINF 2025 - 18th International Conference on Health Informatics
568

contains simultaneous recordings of ECG, respira-
tory signals, and other physiological measurements
from both healthy subjects and patients, offers an-
other rich source of data. By utilizing these datasets,
researchers can benchmark the performance of their
models across different populations and conditions,
ensuring that the methods developed are generaliz-
able and effective in diverse clinical scenarios. These
datasets provide an excellent opportunity to further
refine deep learning models for ECG-derived respi-
ration, offering a broader evaluation framework for
improving non-invasive respiratory monitoring.
By leveraging these datasets and establishing ro-
bust baselines with traditional signal processing meth-
ods, we provided a comprehensive comparison with
our deep learning approach. This demonstrated the ef-
fectiveness of advanced algorithms in respiratory sig-
nal estimation from ECG data. However, we have
not explored other machine learning approaches to
enhance the comparative analysis. Therefore, future
work will also include exploring different deep learn-
ing architectures, like Generative Adversarial Net-
works (GANs), or frameworks such as Reservoir
Computing, to further improve results, and also ex-
perimenting with different datasets for ECG-derived
respiration, to enhance generalizability and robust-
ness of the models. Finally, it would be valuable to in-
vestigate also the performance of the methods across
different age groups. For this purpose, the Fanta-
sia dataset presents a promising option, given its bal-
anced representation of both young and elderly sub-
jects, enabling a more comprehensive age-related per-
formance analysis.
ACKNOWLEDGEMENTS
Author Aleksandra Rashkovska acknowledges the fi-
nancial support from the Slovenian Research and
Innovation Agency (ARIS) under Grant No. P2-
0095. Biljana Mileva Boshkoska acknowledges EU
funding through Erasmus + KA220 project number
101132761 and the ARIS funding under Grant No.
P1-0383.
REFERENCES
Aqajari, S. A. H., Cao, R., Zargari, A. H. A., and Rahmani,
A. M. (2021). An End-to-End and accurate PPG-
based respiratory rate estimation approach using cycle
generative adversarial networks. In 2021 43rd Annual
International Conference of the IEEE Engineering in
Medicine & Biology Society (EMBC), pages 2029–
2032. IEEE.
Brandwood, B. M., Naik, G. R., Gunawardana, U., and
Gargiulo, G. D. (2023). Combined Cardiac and Res-
piratory Monitoring from a Single Signal: A Case
Study Employing the Fantasia Database. Sensors,
23(17):7401.
Charlton, P. H., Bonnici, T., Tarassenko, L., Clifton, D. A.,
Beale, R., and Watkinson, P. J. (2016). An assessment
of algorithms to estimate respiratory rate from the
electrocardiogram and photoplethysmogram. Physi-
ological measurement, 37(4):610.
Chiang, H., Hsieh, Y., Fu, S., Hung, K., Tsao, Y., and Chien,
S. (2019). Noise reduction in ecg signals using fully
convolutional denoising autoencoders. IEEE Access,
7:60806–60813.
Dominkovi
´
c, L., Boshkoska, B. M., and Rashkovska, A.
(2024). ECG-Derived Respiration on the Fantasia
Dataset using the Signal Processing RRest Toolbox.
In ITIS 2024 Book of Proceedings, page 95. Faculty of
Information Studies, Novo Mesto, Slovenia and Jo
ˇ
zef
Stefan Institute, Ljubljana, Slovenia.
Gauthier, D. J., Bollt, E. M., Griffith, A., and Barbosa, W.
a. S. (2021). Next generation reservoir computing.
Nature Communications, 12(1):1–8.
Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov,
P. C., Mark, R., Mietus, J. E., Moody, G. B., Peng,
C. K., and Stanley, H. E. (2000). PhysioBank, Phys-
ioToolkit, and PhysioNet: Components of a new re-
search resource for complex physiologic signals. Cir-
culation [Online], 101(23):e215–e220.
Hire
ˇ
s, M., Bugata, P., Gazda, M., Hre
ˇ
sko, D., Kan
´
asz, R.,
Vavrek, L., and Drot
´
ar, P. (2022). Brief overview of
neural networks for medical applications. Acta Elec-
trotechnica et Informatica, 22(2):34–44.
Is¸ın, A. and Ozdalili, S. (2017). Cardiac arrhythmia detec-
tion using deep learning. Procedia Computer Science,
120:268–275.
Karlen, W., Turner, M., Cooke, E., Dumont, G., and Anser-
mino, J. M. (2010). CapnoBase: Signal database and
tools to collect, share and annotate respiratory signals.
Makowski, D., Pham, T., Lau, Z. J., Brammer, J. C.,
Lespinasse, F., Pham, H., Sch
¨
olzel, C., and Chen, S.
H. A. (2021). NeuroKit2: A Python toolbox for neu-
rophysiological signal processing. Behavior Research
Methods, 53(4):1689–1696.
Merdjanovska, E. and Rashkovska, A. (2020). Respiration
Extraction from Single-Channel ECG using Signal-
Processing Methods and Deep Learning. In 2020 43rd
International Convention on Information, Communi-
cation and Electronic Technology (MIPRO), pages
848–852. IEEE.
Merdjanovska, E. and Rashkovska, A. (2021). Com-
prehensive survey of computational ECG analysis:
Databases, methods and applications. Review.
Pimentel, M. A. F. and et al. (2016). Towards a Robust
Estimation of Respiratory Rate from Pulse Oxime-
ters. IEEE Transactions on Biomedical Engineering,
64(8):1914–1923.
Ravichandran, V. et al. (2019). RespNet: A deep learning
model for extraction of respiration from photoplethys-
mogram. arXiv preprint arXiv:1902.04236.
Deep Learning for ECG-Derived Respiration Using the Fantasia Dataset
569

Schmidt, M., Krug, J. W., Schumann, A., B
¨
ar, K.-J., and
Rose, G. (2015). Estimation of a respiratory signal
from a single-lead ECG using the 4th order central
moments. Current Directions in Biomedical Engi-
neering, 1(1):61–64.
Trobec, R., Avbelj, V., and A.Rashkovska (2014). Multi-
functionality of wireless body sensors. The IPSI BgD
transactions on internet research, 10:23–27.
HEALTHINF 2025 - 18th International Conference on Health Informatics
570
