
Patient Trajectory Prediction: Integrating Clinical Notes with
Transformers
Sifal Klioui, Sana Sellami
a
and Youssef Trardi
Aix-Marseille Univ, LIS, CNRS Marseille, France
Keywords:
Trajectory Prediction, Transformers, Knowledge Integration, Deep Learning.
Abstract:
Patient trajectory prediction from electronic health records (EHRs) is challenging due to the non-stationarity of
medical data, the granularity of diagnostic codes, and the complexities of integrating multimodal information.
While structured data, like diagnostic codes, capture key patient details, unstructured data, such as clinical
notes, often hold complementary information overlooked by current approaches. We propose a transformer-
based approach that integrates clinical note embeddings with structured EHR data for patient trajectory predic-
tion. By combining these modalities, our model captures richer patient representations, improving predictive
accuracy. Experiments on MIMIC-IV datasets show our approach significantly outperforms traditional models
relying solely on structured data.
1 INTRODUCTION
The exponential growth of Electronic Health Records
(EHRs) has transformed patient care, providing un-
precedented access to longitudinal medical data while
introducing new analytical challenges. Healthcare
professionals must now navigate decades of patient
records, synthesizing extensive information to make
informed decisions about future health outcomes.
This paradigm shift has spurred the development of
automated systems to predict future diagnoses from
historical medical data, a cornerstone of personalized
and proactive medicine.
Machine learning, particularly deep learning, has
achieved significant advances in healthcare applica-
tions – from medical imaging to diagnostic predic-
tion – often rivaling or exceeding human expertise in
performance (Egger et al., 2022; Mall et al., 2023).
Building on these successes, researchers have applied
deep learning to sequential disease prediction – fore-
casting a patient’s next diagnosis (visit N+1) based on
prior visits (N) (Choi et al., 2016a; Rodrigues-Jr et al.,
2021; Shankar et al., 2023). However, modeling pa-
tient trajectories from EHR data involves addressing
several complex challenges:
• Non-stationarity of EHR data: Variability over
time undermines the generalizability of predictive
models.
a
https://orcid.org/0000-0001-8302-3053
• The high granularity of medical codes (e.g., more
than 70,000 in the International Classification
of Diseases, 10th revision, Clinical Modification
(ICD-10-CM
1
)) makes it difficult for prediction
models to explore and use these codes.
• Long-term dependencies: Capturing dependen-
cies across lengthy data sequences poses signif-
icant challenges for traditional recurrent neural
network (RNN) models.
• Integration of multimodal data: EHRs encompass
structured data (e.g., lab results) and unstructured
data (e.g., clinical notes), requiring sophisticated
fusion techniques.
Addressing these challenges is critical for devel-
oping robust and reliable systems capable of aid-
ing clinicians by delivering comprehensive forecasts
based on a patient’s clinical history.
This article focuses on enhancing the accuracy of
automated diagnostic systems by leveraging patients’
historical medical records. Traditional coding sys-
tems, such as the International Classification of Dis-
eases (ICD)
2
, often fail to capture the full richness of
clinical notes, resulting in a loss of valuable predic-
tive information. To address this limitation, we pro-
pose an approach that integrates clinical note embed-
1
https://www.cdc.gov/nchs/icd/icd-10-cm/index.html
2
https://www.who.int/standards/classifications/classific
ation-of-diseases
Klioui, S., Sellami, S. and Trardi, Y.
Patient Trajectory Prediction: Integrating Clinical Notes with Transformers.
DOI: 10.5220/0013166500003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 579-586
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
579

dings into transformer architectures, which tradition-
ally rely solely on medical codes. By enriching the
embeddings with contextual information, this method
reduces prediction errors and recovers valuable in-
sights often omitted in coding systems, thereby ad-
dressing challenges such as understanding the ratio-
nale behind prescriptions, procedures, and diagnoses.
The remainder of this article is structured as fol-
lows: Section 2 provides a review of related work
and outlines the key challenges. Section 3 details
our methodology, including the generation of embed-
dings and their integration into transformers. Section
4 presents experimental results and analysis. Finally,
Section 5 summarizes the findings and discusses po-
tential directions for future research.
2 STATE OF THE ART
Various methods, spanning both deep learning and
traditional approaches, have been developed to pre-
dict patient trajectories. Among the pioneering works,
Doctor AI (Choi et al., 2016a) utilizes a recurrent neu-
ral network (RNN)-based temporal model designed
for longitudinal time-stamped EHR data. Doctor AI
predicts both medical codes and the time until the next
visit. To address efficiency, LIG-Doctor (Rodrigues-
Jr et al., 2021) employs a minimal bidirectional re-
current network (MGRU) to handle the granularity of
ICD-9 codes. RETAIN (Choi et al., 2016b) intro-
duces an interpretable predictive model for healthcare
using a reverse-time attention mechanism, training
two RNNs in reverse chronological order to highlight
the importance of prior visits. Similarly, DeepCare
(Pham et al., 2017) utilizes Long Short-Term Mem-
ory (LSTM) networks to predict next-visit diagno-
sis codes, recommend interventions, and assess future
risk. Although models like LSTMs partially mitigate
the vanishing gradient problem, they all face chal-
lenges in modeling long-term dependencies. While
some approaches handle this issue better than oth-
ers, the problem persists as a significant frontier when
dealing with long sequences.
Deep Patient (Miotto et al., 2016), in contrast,
adopts an unsupervised learning approach using Stack
Denoising Autoencoders (SDA) to extract meaning-
ful feature representations from EHR data. However,
it does not account for temporal characteristics, a sig-
nificant limitation given the inherent sequential nature
of patient trajectories. Traditional methods, including
Markov chains (Severson et al., 2020), Bayesian net-
works (Longato et al., 2022), and Hawkes processes
(Lima, 2023), have also been applied to patient trajec-
tory prediction. Yet, these approaches face scalabil-
ity challenges and computational inefficiencies when
dealing with large datasets.
The introduction of transformers has marked a
significant advancement in this field. For instance,
Clinical GAN (Shankar et al., 2023) employs a
Generative Adversarial Network (GAN) framework
based on transformer architecture. In this model,
an encoder-decoder structure serves as the genera-
tor, while an encoder-only transformer acts as the
critic. This approach addresses exposure bias (Arora
et al., 2022), a common issue associated with teacher-
forcing training strategies. However, GAN-based
methods encounter their own challenges, including
training instability, non-convergence, and mode col-
lapse (Saad et al., 2024).
Despite these advancements, a significant gap re-
mains: most existing models rely solely on structured
EHR data, such as ICD and CCS codes, while neglect-
ing unstructured data like clinical notes. These notes
contain rich contextual information, including medi-
cal reasoning and patient-specific nuances, which are
critical for accurately capturing the complexity of pa-
tient trajectories. Addressing this limitation is essen-
tial to further improving predictive performance and
enhancing the practical utility of these models.
Moreover, comparing results between different
studies poses several challenges:
• Dataset Variation: Studies utilize different
datasets (e.g., MIMIC-III vs. MIMIC-IV), which
encompass varying patient populations and time
periods (Johnson et al., 2016; Johnson et al.,
2020). This variation can lead to discrepancies in
results, as one dataset may present more challeng-
ing diagnoses to predict than another due to dif-
fering distributions. Consequently, such discrep-
ancies complicate the reliability of comparisons
between studies and may impact the applicability
of findings to clinical practice.
• Lack of Standardization: Inconsistencies in
dataset sizes, preprocessing steps (e.g., tokeniza-
tion and data cleaning (Edin et al., 2023)), and
evaluation metrics hinder direct comparisons. For
instance, test set sizes, such as the 5% test set (ap-
proximately 1700 visits) used by Shankar et al.
(Shankar et al., 2023), may not adequately repre-
sent patient diversity and complexity. Similarly,
variations in mapping schemes, such as apply-
ing the Clinical Classification Software Refined
(CCSR), lead to inconsistent code representations
and target labels.
These challenges underscore the importance of
careful consideration when comparing results across
different studies in this field. To enhance compara-
HEALTHINF 2025 - 18th International Conference on Health Informatics
580
bility and reproducibility in research on patient tra-
jectory prediction, it is crucial to standardize datasets,
preprocessing methods, and evaluation metrics.
3 PROPOSED METHODOLOGY
We describe our approach for predicting patient tra-
jectories using the MIMIC-IV datasets
3 4
, focus-
ing on comprehensive data preprocessing and clinical
note integration.
3.1 Data Preprocessing
Our preprocessing methodology encompassed six
critical operations: First, we extracted diagnoses,
procedures, and medications. Second, we selected
patients with at least two visits. Third, we ex-
cluded patients lacking all three types of medical
codes (Shankar et al., 2023). Fourth, we employed
CCSR (Clinical Classification Software Refined) to
map ICD-10-CM diagnoses into clinically significant
categories, balancing the specificity of ICD-9-CM
and ICD-10-CM coding schemes. Fifth, we removed
infrequent codes with a threshold of 5 (Edin et al.,
2023). Finally, we temporally ordered events to cre-
ate sequential patient trajectories.
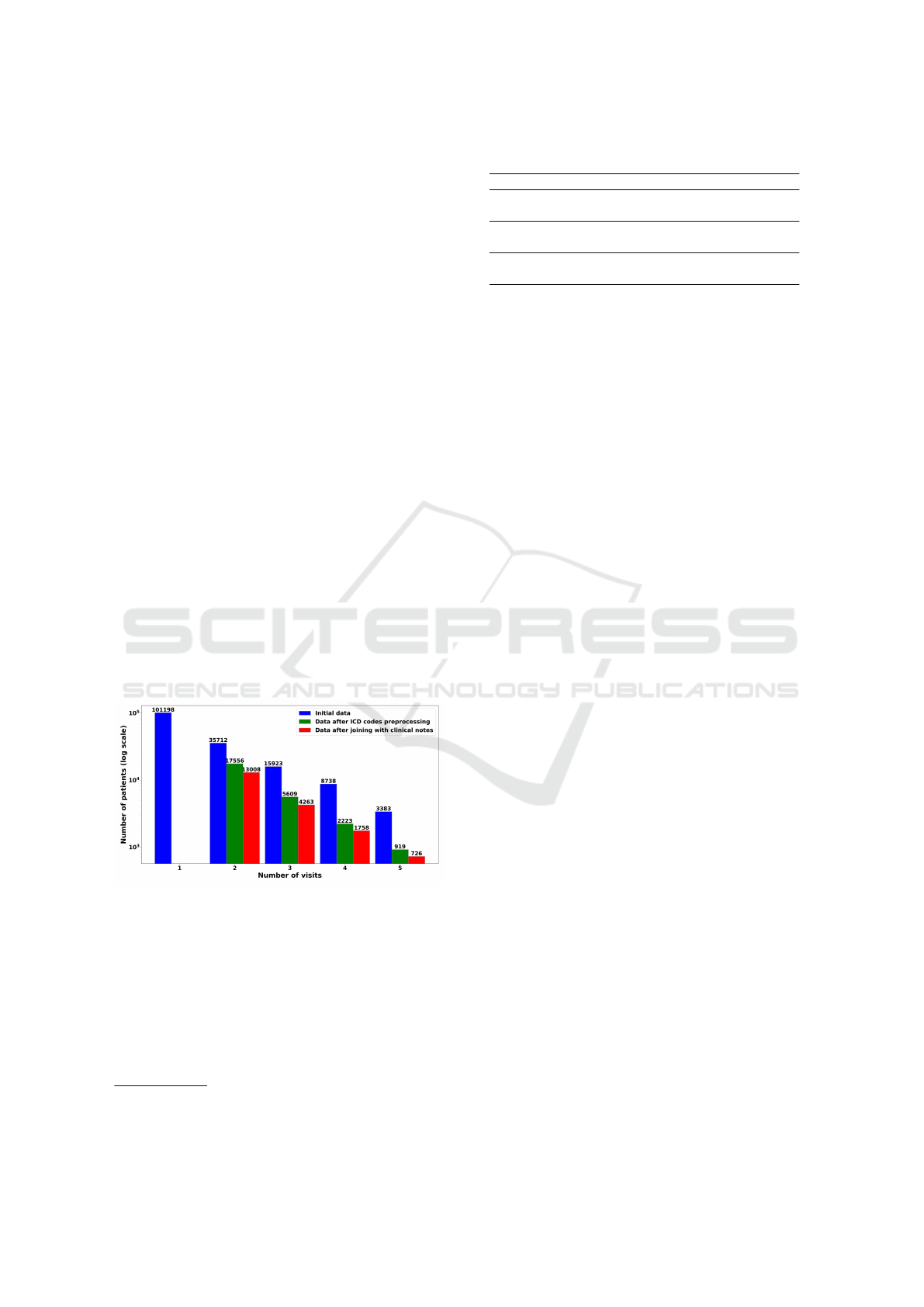
Table 1 presents code statistics before and after
processing. Figure 1 illustrates the predominance of
single-visit patients.
Figure 1: Sample distribution of patients by visit count.
Following (Alsentzer et al., 2019), we prepro-
cessed clinical notes by unifying medical abbrevia-
tions (e.g., ”hr”, ”hrs” to ”hours”), removing accents,
converting special characters, and normalizing text
to lowercase, these elements help mitigate variations
caused by subword tokenizers.
After these preprocessing steps, we obtain a
dataset of 37,000 source-target sequence pairs, ready
for model training.
3
https://physionet.org/content/mimiciv/2.1/
4
https://physionet.org/content/mimic-iv-note/2.2/
Table 1: Code statistics before and after processing.
Code Type At loading After preprocessing
Proc codes
8482 470
3.03 ± 2.81 2.99 ± 2.77
Diag codes
15763 762
12.50 ± 7.67 13.18 ± 8.58
Drug codes
1609 1609
24.12 ± 28.19 24.12 ± 28.19
Note: For each code type, the first row shows the num-
ber of distinct codes, and the second row shows the
mean ± standard deviation per visit.
3.2 Integration of Clinical Notes
To effectively utilize the information in clinical notes,
it is crucial to generate meaningful vector representa-
tions. BERT models (Devlin et al., 2018), particularly
Clinical BERT (Alsentzer et al., 2019), have demon-
strated strong capabilities in capturing semantic rep-
resentations in the medical domain. However, certain
limitations of Clinical BERT may affect its suitability
for our current context:
1. Limited Sequence Length: Clinical BERT was
pretrained on sequence lengths of 128 tokens,
which can hinder its ability to represent longer
texts like discharge summaries. Models trained
with larger context lengths, as shown in recent
studies (Wang et al., 2024), better capture long-
range dependencies and contextual information,
leading to improved performance.
2. Outdated Training Data: Clinical BERT was
pretrained on MIMIC-III, whereas our work uti-
lizes MIMIC-IV-NOTES 2.2, which includes
more recent and diverse clinical data. This mis-
match between the pretraining and target datasets
can lead to suboptimal adaptation to the language
patterns, terminology, and structure in the newer
data.
These limitations highlight the need for a model
that can better align with the characteristics of
MIMIC-IV-NOTES 2.2, ensuring more accurate and
contextually rich representations of clinical narra-
tives.
3.2.1 Clinical Mosaic
To address the limitations of existing models, we in-
troduce Clinical Mosaic, a model built on the Mo-
saic BERT architecture (Portes et al., 2024). This
architecture incorporates recent innovations, includ-
ing Attention with Linear Biases (ALiBi), which sup-
ports extrapolation to longer sequences, and Gated
Linear Units (GLU) (Shazeer, 2020), which enhance
the model’s ability to capture complex patterns and
Patient Trajectory Prediction: Integrating Clinical Notes with Transformers
581

relationships. Clinical Mosaic is pre-trained on
331794 clinical notes from the MIMIC-IV-NOTES
2.2 database, using distributed data parallelism across
7 A40 GPUs. Table 2 details the training parameters.
Table 2: Training parameters of the Clinical Mosaic model.
.
Parameter Value
Effective Batch Size 224
Training Steps 80,000
Sequence Length 512 tokens
Optimizer ADAMW
Initial Learning Rate 5e-4
Learning Rate Schedule Linear warmup for 33,000
steps, then cosine annealing
for 46,000 steps
Final Learning Rate 1e-5
Masking Probability 30%
During training, we track perplexity (PPL), a met-
ric quantifying prediction confidence for sequential
data. Mathematically, PPL is defined as:
PPL(X) = exp
(
−
1
t
t
∑
i=1
log p
θ
(x
i
| x
<i
)
)
where X = (x
1
, x
2
, . . . , x
t
) is the sequence, and
p
θ
(x
i
| x
<i
) is the probability assigned by the model
to the i-th element given the preceding elements.
Lower perplexity indicates better predictive perfor-
mance. Our model exhibited a consistent and smooth
decrease in perplexity, suggesting progressive im-
provement.
3.2.2 Clinical Reasoning Assessment
We assessed Clinical Mosaic’s clinical reasoning ca-
pabilities using the Medical Natural Language In-
ference (MedNLI) dataset (Romanov and Shivade,
2018). Derived from MIMIC-III clinical notes,
MedNLI comprises 14,049 premise-hypothesis pairs,
with the objective of classifying the relationship be-
tween each pair as entailment, contradiction, or neu-
tral.
The task evaluates critical aspects of clinical lan-
guage understanding, including semantic comprehen-
sion of medical terminology and logical reasoning in
clinical contexts, as well as the ability to discern nu-
anced relationships between clinical statements. Ta-
ble 3 compares Clinical Mosaic’s performance with
state-of-the-art models.
Clinical Mosaic achieved 86.5% accuracy, outper-
forming the original Clinical BERT (Alsentzer et al.,
2019) (84.1%), demonstrating enhanced clinical lan-
guage comprehension through our model optimiza-
tions.
Table 3: Comparison of performance of BERT variants and
Clinical Mosaic on downstream MedNLI tasks.
Model Accuracy
BERT 77.6%
BioBERT 80.8%
Discharge Summary BERT 80.6%
Clinical Discharge BERT 84.1%
Bio+Clinical BERT 82.7%
Clinical Mosaic 86.5%
3.2.3 Fusion of Clinical Representations
When generating clinical note embeddings using
Clinical Mosaic, each layer of the encoder produces
a different representation of the input sequences. Re-
cent research has shown that utilizing multiple layers
can enhance performance in various NLP tasks. No-
tably, Hosseini et al (Hosseini et al., 2023) demon-
strated that combining certain layers of BERT-based
models can yield substantially better sentence embed-
dings than using only the last layer, improving perfor-
mance without additional training. Inspired by these
findings, we hypothesized that aggregating represen-
tations from multiple layers would be beneficial for
our clinical tasks. To balance potential performance
gains with computational feasibility, we chose to use
the last 6 layers of BERT-Base. This pragmatic deci-
sion allowed us to explore the advantages of multi-
layer representations without exponentially increas-
ing the number of experiments required for testing all
possible combinations. By fixing our model to these
6 representation layers, we aimed to improve perfor-
mance over single-layer approaches while maintain-
ing efficiency in our clinical applications.
We then explored three embedding processing
strategies:
• Average Over Layers and Visits (MEAN): Cal-
culates average embeddings across 6 layers and
all visits, capturing global context and smoothing
noise.
• Average Only Over Layers (CONCAT): Aver-
ages embeddings across layers, reducing dimen-
sionality while maintaining multi-layer represen-
tations.
• Projection Method: Projects 6-layer embeddings
into a lower-dimensional space using linear lay-
ers with GeLU activation. This approach reduces
dimensionality while preserving critical informa-
tion, with concatenated projections enabling com-
plex inter-visit relationship learning (Figure 2).
After generating embeddings using one of the de-
scribed strategies, we integrate them along CCS code
embeddings as illustrated in Figure 3.
HEALTHINF 2025 - 18th International Conference on Health Informatics
582
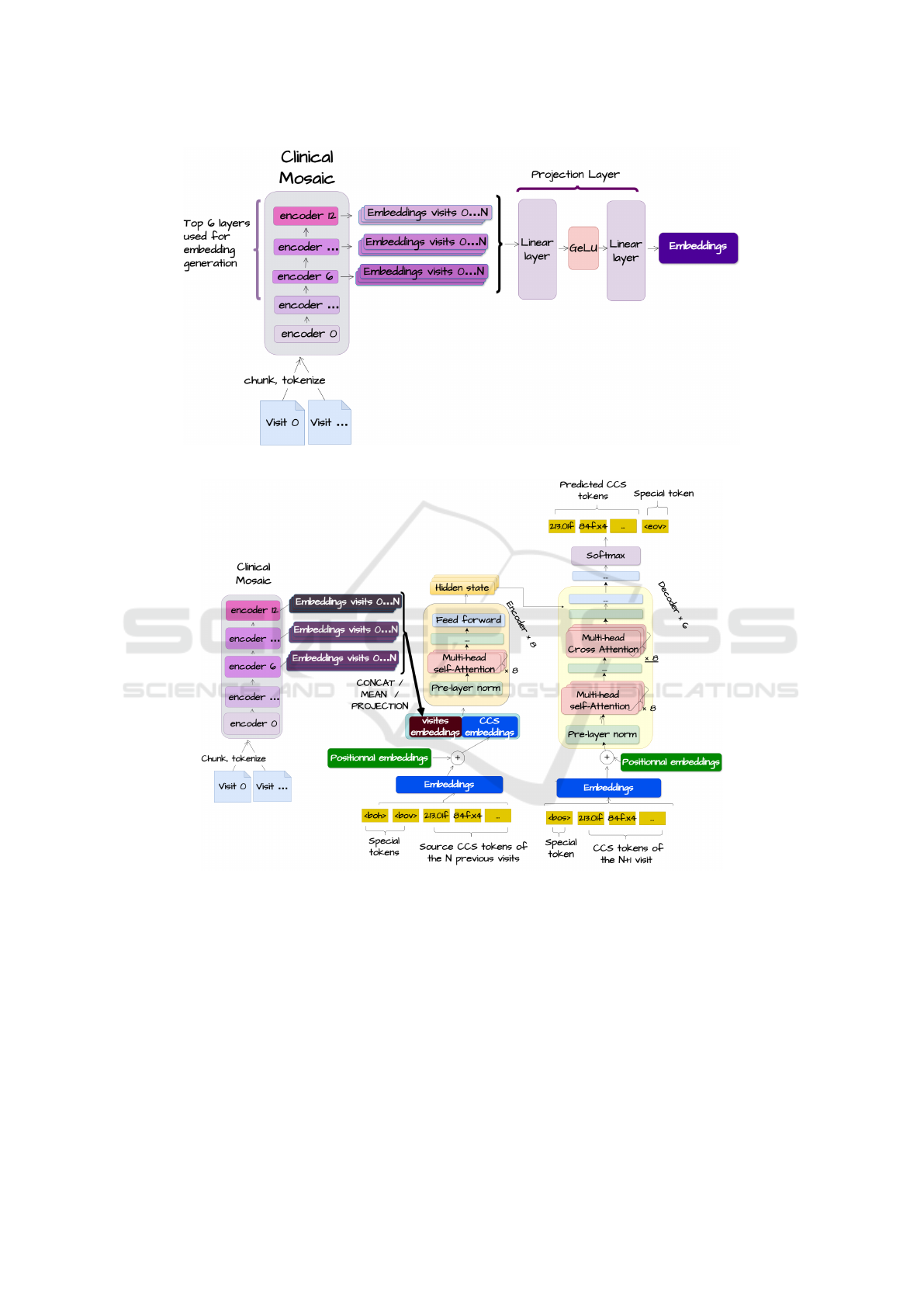
Figure 2: Approach using a projection layer.
Figure 3: Architecture for integrating notes.
This integration uses the transformer architecture,
where CCS codes can attend to clinical note embed-
dings via a self-attention mechanism, creating a uni-
fied representation. The transformer’s decoder, us-
ing causal cross-attention, uses this representation to
predict diagnoses for future visits. This approach
allows the model to effectively combine structured
(CCS codes) and unstructured (clinical notes) data,
offering a comprehensive view of the patient’s clinical
history and aiming to improve predictive performance
for patient trajectories.
4 EXPERIMENTS
This section outlines our experimental evaluation of
the clinical note integration approach.
4.1 Metrics
We used Mean Average Precision (MAP@K) and
Mean Average Recall (MAR@K) at K=20, 40, 60
(equations 1 and 2) to assess model performance.
These metrics, suitable for order-sensitive recom-
Patient Trajectory Prediction: Integrating Clinical Notes with Transformers
583

Table 4: Performance of different models using MAP@k and MAR@k. Values are presented as mean(standard deviation in
the last decimal place).
Model
K = 20 K = 40 K = 60
MAR MAP MAR MAP MAR MAP
Projection 0.425(5) 0.556(21) 0.439(4) 0.556(21) 0.439(4) 0.556(21)
Concat 0.420(6) 0.569(6) 0.425(5) 0.571(6) 0.425(5) 0.571(6)
Mean 0.416(6) 0.538(84) 0.423(6) 0.567(17) 0.423(6) 0.567(17)
Clinical GAN
1
0.410(5) 0.558(11) 0.414(5) 0.559(12) 0.414(5) 0.559(12)
Transformer Only 0.398(23) 0.565(23) 0.405(25) 0.566(23) 0.405(25) 0.566(23)
LIG-Doctor
2
0.267(48) 0.474(94) 0.361(42) 0.431(87) 0.420(37) 0.402(80)
Doctor AI
3
0.233(5) 0.206(46) 0.233(5) 0.207(47) 0.233(5) 0.207(47)
1
(Shankar et al., 2023),
2
(Rodrigues-Jr et al., 2021),
3
(Choi et al., 2016a)
Note: Values are presented as mean(standard deviation). For example, 0.425(5) represents 0.425±0.005.
mendation tasks, allow direct comparisons with prior
studies, though some previous works used only one
metric (Rodrigues-Jr et al., 2021).
MAP@K =
1
|Q|
|Q|
∑
u=1
1
min(m, K)
K
∑
k=1
P(k) · rel(k) (1)
MAR@K =
1
|Q|
|Q|
∑
u=1
1
m
K
∑
k=1
rel(k) (2)
Where |Q| is the number of target sequences, m is
the number of relevant items in a target sequence, K
is the rank limit, P(k) is the precision at rank k, and
rel(k) is a function that equals 1 if the item at rank k
is relevant, 0 otherwise.
4.2 Baselines
We compare our approach with state-of-the-art mod-
els and with the Transformer model without clinical
notes integration (Vaswani et al., 2017). The models
were reproduced using the Pytorch framework, fol-
lowing their associated codes and publications. All
models were evaluated using 5-fold cross-validation
and 95% confidence intervals. The source code is
made available for reproducibility [1].
Below, we provide an overview of the baseline
models:
• LIG-Doctor (Rodrigues-Jr et al., 2021): A bidi-
rectional GRU model with embedding and hidden
dimensions of 714. It uses a projection layer to
merge bidirectional contexts, followed by a soft-
max layer. Trained for up to 100 epochs (con-
verging in 13) with a batch size of 512 using the
Adadelta optimizer.
• Doctor AI (Choi et al., 2016a): RNN-based model
with embedding and hidden dimensions of 2000, a
dropout rate of 0.5, and trained for 20 epochs with
a batch size of 384 using the Adadelta optimizer.
• Clinical GAN (Shankar et al., 2023): Includes a 3-
layer, 8-head encoder-decoder generator (hidden
dimension 256) and a 1-layer, 4-head transformer
encoder discriminator. Trained for 100 epochs
(converging in 11) with a batch size of 8 using
Adam for the generator, SGD for the discrimina-
tor, and a Noam scheduler.
4.3 Results
The performance of different models is summarized
in Table 4.
Injecting clinical note embeddings significantly
improves performance, especially in terms of
MAR@K (see Figure 4). However, this improvement
may be constrained by the limited dataset size (37k
samples), which could hinder the model’s ability to
learn to fully utilize these embeddings.
Among embedding injection methods:
• The Mean strategy produces the lowest MAR@K
scores, likely due to excessive information com-
pression leading to loss of critical details. Despite
this, it is the most computationally efficient ap-
proach, adding only one vector, which is advan-
tageous given the O(N
2
) complexity of the trans-
former’s attention mechanism.
• The Projection method achieves the best
MAP@K scores as shown in Figure 5 but lags
behind in MAR@K. This can be attributed to the
method’s focus on dimensionality reduction using
learnable parameters that is unable to recover the
full information of the embeddings.
• The Concat approach, which averages embed-
ding layers, achieves the highest MAR@K while
maintaining competitive MAP@K scores. This
method enhances information richness by pre-
serving critical details and enabling the model to
process independent elements from different vis-
its selectively.
HEALTHINF 2025 - 18th International Conference on Health Informatics
584
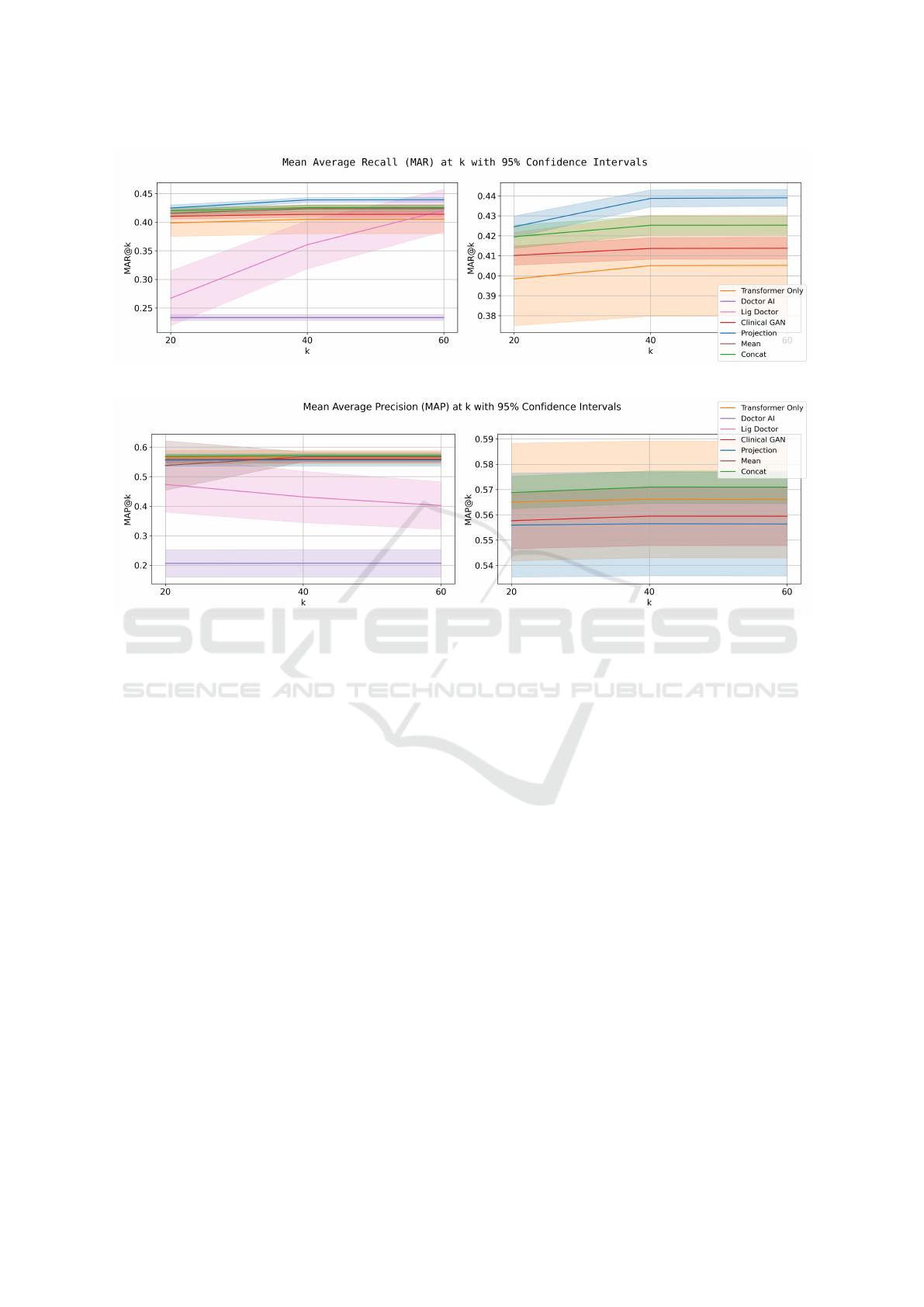
Figure 4: Mean average recall @ 20, 40, and 60 for different models.
Figure 5: Mean average precision @ 20, 40, and 60 for different models.
LIG-Doctor performs as a classification model,
using a linear layer to predict diagnoses without gen-
erating predictions in a specific order. To calculate
metrics like MAP@K, logits are sorted post hoc, but
this approach limits performance improvement as K
increases. Additionally, its classification-based setup
prevents repetitive predictions, contributing to higher
MAR@K scores.
Doctor AI, relying on a single GRU layer, shows
lower performance compared to other models. Its
performance could improve with increased hidden di-
mensions or additional GRU layers to better handle
the expanded prediction space.
Clinical GAN performs well on MAP@K but
struggles with MAR@K, indicating difficulty in gen-
erating a diverse set of relevant predictions.
5 CONCLUSION
In this study, we tackled the challenge of predicting
patient trajectories by integrating clinical note embed-
dings into transformer models, combining structured
electronic medical records (EMRs) data with rich, un-
structured clinical notes. This approach provides a
more holistic view of patient histories, enhancing pre-
dictive accuracy.
Experimental results on the MIMIC-IV datasets
demonstrated that our method significantly outper-
forms models relying solely on structured data, under-
scoring the value of unstructured medical information
in improving healthcare predictions.
Future work will focus on multimodal data in-
tegration (medical imaging, genomics) and refining
unordered prediction handling in non-autoregressive
models.
ACKNOWLEDGEMENTS
This work was carried out as part of the PICOMALE
project, which is funded by AMIDEX.
REFERENCES
Alsentzer, E., Murphy, J. R., Boag, W., Weng, W.-H.,
Jin, D., Naumann, T., and McDermott, M. (2019).
Publicly available clinical bert embeddings. arXiv
preprint arXiv:1904.03323.
Patient Trajectory Prediction: Integrating Clinical Notes with Transformers
585

Arora, K., Asri, L. E., Bahuleyan, H., and Cheung, J. C. K.
(2022). Why exposure bias matters: An imitation
learning perspective of error accumulation in language
generation. arXiv preprint arXiv:2204.01171.
Choi, E., Bahadori, M. T., Schuetz, A., Stewart, W. F., and
Sun, J. (2016a). Doctor ai: Predicting clinical events
via recurrent neural networks. In Machine learning
for healthcare conference, pages 301–318.
Choi, E., Bahadori, M. T., Sun, J., Kulas, J., Schuetz, A.,
and Stewart, W. (2016b). Retain: An interpretable
predictive model for healthcare using reverse time at-
tention mechanism. Advances in neural information
processing systems, 29.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
Edin, J., Junge, A., Havtorn, J. D., Borgholt, L., Maistro,
M., Ruotsalo, T., and Maaløe, L. (2023). Automated
medical coding on mimic-iii and mimic-iv: A crit-
ical review and replicability study. In Proceedings
of the 46th International ACM SIGIR Conference on
Research and Development in Information Retrieval,
pages 2572–2582.
Egger, J., Gsaxner, C., Pepe, A., Pomykala, K. L., Jonske,
F., Kurz, M., Li, J., and Kleesiek, J. (2022). Medical
deep learning—a systematic meta-review. Computer
methods and programs in biomedicine, 221:106874.
Hosseini, M., Munia, M., and Khan, L. (2023). BERT
has more to offer: BERT layers combination yields
better sentence embeddings. In Bouamor, H., Pino,
J., and Bali, K., editors, Findings of the Association
for Computational Linguistics: EMNLP 2023, pages
15419–15431, Singapore. Association for Computa-
tional Linguistics.
Johnson, A., Bulgarelli, L., Pollard, T., Horng, S.,
Celi, L. A., and Mark, R. (2020). Mimic-iv.
PhysioNet. Available online at: https://physionet.
org/content/mimiciv/1.0/(accessed August 23, 2021),
pages 49–55.
Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H.,
Feng, M., Ghassemi, M., Moody, B., Szolovits, P.,
Anthony Celi, L., and Mark, R. G. (2016). Mimic-
iii, a freely accessible critical care database. Scientific
data, 3(1):1–9.
Lima, R. (2023). Hawkes processes modeling, inference,
and control: An overview. SIAM Review, 65(2):331–
374.
Longato, E., Morieri, M. L., Sparacino, G., Di Camillo, B.,
Cattelan, A., Menzo, S. L., Trevenzoli, M., Vianello,
A., Guarnieri, G., Lionello, F., et al. (2022). Time-
series analysis of multidimensional clinical-laboratory
data by dynamic bayesian networks reveals trajecto-
ries of covid-19 outcomes. Computer Methods and
Programs in Biomedicine, 221:106873.
Mall, P. K., Singh, P. K., Srivastav, S., Narayan, V., Pa-
przycki, M., Jaworska, T., and Ganzha, M. (2023).
A comprehensive review of deep neural networks
for medical image processing: Recent developments
and future opportunities. Healthcare Analytics, page
100216.
Miotto, R., Li, L., Kidd, B. A., and Dudley, J. T. (2016).
Deep patient: an unsupervised representation to pre-
dict the future of patients from the electronic health
records. Scientific reports, 6(1):1–10.
Pham, T., Tran, T., Phung, D., and Venkatesh, S.
(2017). Predicting healthcare trajectories from med-
ical records: A deep learning approach. Journal of
biomedical informatics, 69:218–229.
Portes, J., Trott, A., Havens, S., King, D., Venigalla, A.,
Nadeem, M., Sardana, N., Khudia, D., and Frankle,
J. (2024). Mosaicbert: A bidirectional encoder opti-
mized for fast pretraining. Advances in Neural Infor-
mation Processing Systems, 36.
Rodrigues-Jr, J. F., Gutierrez, M. A., Spadon, G., Brandoli,
B., and Amer-Yahia, S. (2021). Lig-doctor: Efficient
patient trajectory prediction using bidirectional mini-
mal gated-recurrent networks. Information Sciences,
545:813–827.
Romanov, A. and Shivade, C. (2018). Lessons from natural
language inference in the clinical domain. In Riloff,
E., Chiang, D., Hockenmaier, J., and Tsujii, J., edi-
tors, Proceedings of the 2018 Conference on Empiri-
cal Methods in Natural Language Processing, pages
1586–1596, Brussels, Belgium. Association for Com-
putational Linguistics.
Saad, M. M., O’Reilly, R., and Rehmani, M. H. (2024). A
survey on training challenges in generative adversar-
ial networks for biomedical image analysis. Artificial
Intelligence Review, 57(2):19.
Severson, K. A., Chahine, L. M., Smolensky, L., Ng, K.,
Hu, J., and Ghosh, S. (2020). Personalized input-
output hidden markov models for disease progression
modeling. In Machine learning for healthcare confer-
ence, pages 309–330. PMLR.
Shankar, V., Yousefi, E., Manashty, A., Blair, D., and Tee-
gapuram, D. (2023). Clinical-gan: Trajectory fore-
casting of clinical events using transformer and gen-
erative adversarial networks. Artificial Intelligence in
Medicine, 138:102507.
Shazeer, N. (2020). Glu variants improve transformer. arXiv
preprint arXiv:2002.05202.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in neural
information processing systems, 30.
Wang, X., Salmani, M., Omidi, P., Ren, X., Reza-
gholizadeh, M., and Eshaghi, A. (2024). Beyond
the limits: A survey of techniques to extend the con-
text length in large language models. arXiv preprint
arXiv:2402.02244.
HEALTHINF 2025 - 18th International Conference on Health Informatics
586
