
Analyzing Cognitive Patterns in Gifted Children Using MRI and
Morphometric Similarity Networks
Shuning Han
1,2 a
, Feng Duan
3 b
, Gemma Vilaseca
4,5 c
, N
´
uria Vilar
´
o
5 d
, Cesar F. Caiafa
6 e
,
Zhe Sun
2,7 f
and Jordi Sol
´
e-Casals
1,8 g
1
Data and Signal Processing Research Group, University of Vic-Central University of Catalonia, Vic, Catalonia, Spain
2
Image Processing Research Group, RIKEN Center for Advanced Photonics, RIKEN, Wako-Shi, Saitama, Japan
3
Tianjin Key Laboratory of Brain Science and Intelligent Rehabilitation, Nankai University, Tianjin, China
4
Psychological Department, Oms and Prat school, Fundaci
´
o Catalunya - La Pedrera, Manresa, Catalonia, Spain
5
Oms Foundation, Manresa, Catalonia, Spain
6
Instituto Argentino de Radioastronom
´
ıa-CCT La Plata, CONICET / CIC-PBA / UNLP, Argentina
7
Faculty of Health Data Science, Juntendo University, Urayasu, Chiba, Japan
8
Department of Psychiatry, University of Cambridge, Cambridge, U.K.
Keywords:
Gifted Children, Structural Magnetic Resonance Imaging, Morphometric Similarity Network, Connection
Density, Anatomical Modularity, Topological Features.
Abstract:
Advances in non-invasive neuroimaging, such as structural magnetic resonance imaging (sMRI), have en-
abled the construction of structural brain networks (SBNs), allowing in vivo mapping of anatomical connec-
tions. This study investigates brain network structural differences linked to different intelligence levels in
children by individual morphometric similarity networks (MSNs) derived from sMRI data. Through group-
and individual-level analyses, we aim to uncover key topological features associated with cognitive perfor-
mance and to identify a suitable connection density for SBN analysis. Connection density strongly affects
global and nodal topological features, with a range of p = 0.05 to 0.15 recommended for stable and optimal
results. Gifted individuals exhibit stronger intra-hemispheric and intra-modular connectivity, a more balanced
distribution of left-to-right intra-hemispheric connections, and lower mean versatility, supporting efficient and
stable cognitive processing. Moreover, anatomical modularity analyses based on von Economo indicate that
higher cognitive performance is linked to enhanced connectivity in specific areas (such as secondary sensory
area, motor to association area and secondary sensory to limbic area), alongside selective reduction in cer-
tain modular connections (such as motor to insular area, association to secondary sensory area and motor to
secondary sensory area). Furthermore, topological features, including participation coefficient and local effi-
ciency, are linked to cognitive performance. These findings provide valuable insights into the SBNs underlying
cognitive levels in children.
1 INTRODUCTION
Understanding the neural basis of cognitive abilities
has long been a key goal in cognitive neuroscience.
Despite considerable progress, significant gaps re-
main, particularly in understanding the structural and
functional differences in the brains of gifted individ-
a
https://orcid.org/0009-0004-0792-5484
b
https://orcid.org/0000-0002-2179-2460
c
https://orcid.org/0000-0002-7533-2355
d
https://orcid.org/0000-0002-7273-6039
e
https://orcid.org/0000-0001-5437-6095
f
https://orcid.org/0000-0002-6531-0769
g
https://orcid.org/0000-0002-6534-1979
uals. A promising approach to bridging this gap is
the study of brain networks, which explores the con-
nectivity and interactions among different brain re-
gions. Recent advances in non-invasive neuroimag-
ing techniques, such as structural magnetic resonance
imaging (sMRI) and diffusion MRI, have enabled re-
searchers to map these anatomical connections in vivo
(Genon et al., 2022; Lo et al., 2011). These techniques
allow for the construction of structural brain networks
(SBNs), where the brain is modeled as a graph com-
posed of nodes (brain regions) and edges (connections
between them), providing a foundation for applying
graph theory to explore the brain’s organization and
its relationship to cognitive abilities (Faskowitz et al.,
Han, S., Duan, F., Vilaseca, G., Vilaró, N., Caiafa, C. F., Sun, Z. and Solé-Casals, J.
Analyzing Cognitive Patterns in Gifted Children Using MRI and Morphometric Similarity Networks.
DOI: 10.5220/0013169200003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 729-740
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
729

2022).
Two main approaches are used to construct SBNs:
tractography from diffusion-weighted imaging (DWI)
and structural covariance networks (SCNs) derived
from sMRI. SCNs traditionally compute the covari-
ance of a single morphometric feature, such as corti-
cal thickness, between regions across a group of sub-
jects (Sol
´
e-Casals et al., 2019). While this method
has been invaluable in understanding group-level
brain organization, recent advances have introduced
individual-level SCNs, allowing for a more detailed,
personalized view of brain structure by incorporating
multiple morphometric features (Kong et al., 2015;
Li et al., 2017; Yu et al., 2018; Seidlitz et al., 2018;
Li et al., 2021; Sebenius et al., 2023; Sun et al.,
2024a,b). A notable development in this field is
the morphometric similarity network (MSN) (Seidlitz
et al., 2018), which constructs individual brain net-
works using multiple structural features derived from
sMRI. MSNs are based on the pairwise correlations
of morphometric feature vectors between brain re-
gions, facilitating a more granular investigation of
the brain’s structural organization. This method has
shown promise in revealing biologically relevant pat-
terns in brain networks.
Previous research has uncovered distinct patterns
in SBNs associated with various biological charac-
teristics, such as gender (Sun et al., 2015), cogni-
tive ability (Park and Friston, 2013) and neurode-
generative diseases progression (Sun et al., 2024b).
Despite these advances, key questions remain unan-
swered. How do the topological properties of MSNs
relate to cognitive performance, particularly in chil-
dren? How do modular and hemispheric specializa-
tions contribute to neural efficiency in gifted individ-
uals? How do different connection densities affect
brain network properties? These questions are criti-
cal for understanding the structural underpinnings of
cognitive abilities.
This study addresses these gaps by constructing
MSNs from sMRI data and analyzing their topo-
logical features in relation to cognitive performance
in both gifted and control groups. Specifically, we
conduct a comprehensive group comparison analy-
sis, focusing on modular connections based on von
Economo (VE) (Von Economo, 1929), the effects
of different connection densities, intra-/inter-modular
connections based on VE and intra-/inter-hemispheric
connections. We also explore how nodal, global topo-
logical features and specific VE-Region connections
are linked to cognitive performance. By combining
group-level and individual-level analyses, this study
offers new insights into the neural mechanisms of
giftedness, emphasizing the role of anatomical mod-
ularity, hemispheric specialization, and topological
features in efficient cognitive processing.
The rest of this paper is structured as follows: Sec-
tion 2 outlines the dataset and methods employed.
Section 3 presents the results of our analyses, which
are discussed in Section 4. Finally, Section 5 con-
cludes the study.
2 MATERIALS AND METHODS
2.1 Data
2.1.1 Participants
In this study, we use the dataset consisting of sMRI
and cognitive tests from 29 healthy right-handed male
participants with no history of psychiatric or neu-
rological disorders (Sol
´
e-Casals et al., 2019). The
raw (anonymized) MRI data and cortical thickness
data are accessible in the OpenNeuro repository at
https://openneuro.org/datasets/ds001988.
The participants are categorized into two groups:
a control group (CG, 14 subjects) and a gifted group
(GG, 15 subjects). Details of the participants’ infor-
mation can be found in Table 1. The table indicates no
significant age differences but significant difference
in full-scale IQ between the groups.
Table 1: Participant information.
Groups CG (Mean±SD) GG (Mean±SD)
Age 12.53 ± 0.77 12.03 ± 0.54
IQ 122.71 ± 3.89 148.80 ± 2.93
2.1.2 sMRI Data
MRI scans were performed on a 3T scanner, yielding
high-resolution T1-weighted images obtained via the
MPRAGE 3D protocol. In this study, we adopted the
sMRI preprocessing method outlined in prior research
(Sol
´
e-Casals et al., 2019). FreeSurfer v5.3 was used
for preprocessing to estimate cortical thickness from a
three-dimensional cortical surface model based on in-
tensity and continuity information (Fischl and Dale,
2000). Cortical reconstructions were independently
reviewed by two experienced researchers to ensure
adherence to quality control criteria. Each brain
was parcellated into 308 (R = 308) regions (approxi-
mately 500 mm
2
each) using the standard FreeSurfer
template (fsaverage) by a backtracking algorithm,
which subdivides the regions defined in the Desikan-
Killiany atlas (Desikan et al., 2006). The surface-
based (non-linear) registration by the FreeSurfer com-
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
730

mand mri surf2surf was then applied to warp the
parcellation from the standard template to each indi-
vidual’s native MPRAGE space (Ghosh et al., 2010).
The 308 parcellated regions are classified into
seven cytoarchitectonic cortical types based on the
VE classification (Von Economo, 1929). Concisely,
structural type1 encompasses regions with minimal
laminar differentiation, notably the primary motor
cortex/precentral gyrus. Structural type2 and type3
generally include association cortices, while struc-
tural type4 and type5 correspond to secondary and
primary sensory areas, respectively. The original VE
classification of structural types does not differenti-
ate between true six-layered isocortex and mesocor-
tex or allocortex, which exhibit distinct cytoarchitec-
tures and ontogenies. Consequently, we introduced
two additional subtypes: type6, the limbic cortex,
encompassing the entorhinal, presubicular, retrosple-
nial, and cingulate cortices and primarily constitutes
allocortex; and type7, the insular cortex, including
agranular, dysgranular and granular regions (Sol
´
e-
Casals et al., 2019; Seidlitz et al., 2018; V
´
a
ˇ
sa et al.,
2018; V
´
ertes et al., 2016). In this study, we con-
duct anatomical modularity analyses based on VE-
Regions.
2.2 Methods
In this study, we construct MSNs from sMRI data.
Group-level analyses are performed by comparing the
average MSNs of the CG and GG to identify key
differences in brain network topology. Additionally,
we conduct individual-level analyses to explore the
relationship between cognitive performance and the
structural organization of brain networks.
2.2.1 Network Construction
In this study, we construct brain networks for each
subject using the morphometric similarity network
(MSN), which represents the structural connectivity
between brain regions (Seidlitz et al., 2018). The
MSN converts each individual’s set of multimodal
MRI features into a morphometric similarity matrix
of pairwise inter-regional correlations of morphome-
tric feature vectors. In this study, as depicted in Fig.
1, a set of d (d = 5) morphometric features (308 × 5
for each sample) derived from any T1-weighted MRI
scans: total surface area (tSA), total gray matter vol-
ume (tGMV), average cortical thickness (aCT), in-
tegrated rectified mean curvature (iMC), integrated
rectified (Gaussian) curvature (iGC), was employed
to construct MSNs. It has been demonstrated that
the MSNs based on these five features are similar to
MSNs utilizing a broader array of features (Seidlitz
et al., 2018), with tSA, tGMV, aCT, and iGC identi-
fied as the most discriminative features (Zhang et al.,
2021). Following prior studies (Li et al., 2017; Sei-
dlitz et al., 2018), each feature vector is standardized
by the z-score values before the correlation calcula-
tion. Nodes in the MSN correspond to the 308 regions
defined by the atlas. Edges are constructed based on
the morphometric similarity between each possible
pair of regions, quantified using the Pearson’s correla-
tion coefficient (PCC) between their normalized mor-
phometric feature vectors. Each sample’s MSN is rep-
resented as a weighted, undirected graph, where the
308 × 308 adjacency matrix contains PCC values as
edge weights. A PCC value close to -1 denotes anti-
correlation between the pair of features, while a PCC
value close to 1 denotes strong correlation between
the pair of features (Heinsfeld et al., 2018). Hence,
the diagonal elements of MSN equal to 1. However,
we uniformly assign NaN values to the diagonal ele-
ments of MSNs.
2.2.2 Group Comparison Analyses for MSNs
As shown in Fig. 1, we compute the average brain
networks for the two groups, CG and GG. In this
section, we present a comparative analysis of these
group-level networks. The brain networks are first
reorganized into 7 VE-Regions. To identify signifi-
cant differences in connections between CG and GG
average networks, we use the Mann–Whitney U test,
a non-parametric statistical method suitable for small
samples and non-normal distributions. Additionally,
the analysis of the networks across different densities
is conducted using the minimum spanning tree (MST)
method to ensure full connectivity. Furthermore, we
investigate the nodal topological features of the net-
works, quantifying the differences between the two
groups through Euclidean distance. Moreover, we
examine intra-/inter-VE connections and intra-/inter-
hemispheric connections for CG and GG at a specific
connection density.
• VE-Region analysis for group brain networks.
Firstly, the brain networks are reordered according
to the 7 VE-Regions. The connections between VE-
Region k and l in a brain network A are defined as
A
kl
= A
i j
(i ∈ N
k
, j ∈ N
l
,i ̸= j)
(1)
where A
kl
represents the connections of brain net-
work between nodes in VE-Region k and nodes in
VE-Region l, with k,l = 1,2,...,7; N
k
and N
l
denote
the sets of nodes in VE-Regions k and l, respectively.
When k = l, A
kl
denotes intra-VE connections, while
for k ̸= l, it denotes inter-VE connections.
Analyzing Cognitive Patterns in Gifted Children Using MRI and Morphometric Similarity Networks
731

z-score
MSN
Group
average
CG
GG
Regional-level morphometric features
of each subject
(308×5 each)
MSNs (308×308 each,
PCCs between morphometric
features of each region)
Group average MSNs
(308×308 each)
Normalized morphometric features
of each subject
(308×5 each)
Figure 1: MSN construction and processing for group average MSNs. Five morphometric features (tSA, tGMV, aCT, iMC,
iGC) are extracted from each brain region, resulting in 308 × 5 features per sample. These features are normalized using the
z-score method. MSNs (308 × 308) are then computed by calculating the PCCs between the normalized features of all region
pairs. Finally, group average MSNs are generated for both CG and GG.
The average VE connection or VE connection
strength E (7 × 7) of a brain network A can be de-
fined as
E (k,l) =
1
n
kl
∑
A
kl
(2)
where E (k, l) represents the average connection be-
tween two VE-Regions k and l; n
kl
is the total number
of connections between nodes in VE-Regions k and l.
We assess the significant difference between CG
and GG in connections between/within each VE-
Region in the group average brain networks using
Mann–Whitney U test, as illustrated in Equation 3.
[P
kl
,zval
kl
] = U[vec(A
kl
G
),vec(A
kl
C
)] (3)
Here, A
kl
C
represents the connections between VE-
Regions k and l of average CG brain network, while
A
kl
G
represents the connections between VE-Regions
k and l of average GG brain network. A
kl
C
and A
kl
G
are
then converted to vectors for Mann–Whitney U test.
P
kl
denotes the probability of the difference between
CG and GG in connections between VE-Regions k
and l, with lower values (typically P
kl
≤ 0.05) indi-
cating a more significant difference; zval
kl
denotes the
value of the normal statistic between CG and GG in
connections between/within VE-Regions k and l, with
larger absolute values (typically zval
kl
≥ 1.96 corre-
sponding to P
kl
≤ 0.05) indicating a more significant
difference. Negative zval indicates smaller values in
the first group, while positive zval indicates larger
values. However, the interpretation of zval
kl
is lim-
ited here because A
kl
C
and A
kl
G
contain negative values,
making the zval
kl
statistic less meaningful.
• Analysis of group average brain networks
across different density.
We analyze the effects of density on brain net-
works using the MST (Van Wijk et al., 2010) to en-
sure that all graphs of different densities are node-
connected. An MST is a subgraph that connects all
nodes using exactly R − 1 edges, where R is the num-
ber of nodes in the network. We first find the MST and
then add edges to ensure the graph is node-connected.
The connection density p refers to the proportion of
edges presented in the network relative to the total
possible edges ((R−1)R/2, where R = 308). Notably,
when computing MST graphs for MSNs, negative val-
ues are treated as 0.
We denote M
p
as the MST network with con-
nection density p of a brain network A . Given
that an MST network M
p
is node-connected, the
minimum connection density p of MST network
is (R − 1)/[(R − 1)R/2] = 0.0065, where R = 308.
In this study, connection densities used are: p ∈
{
0.0065,0.01, 0.05,0.1, 0.15,0.2, 0.25,0.3, 0.35,0.4,
0.5,0.6, 0.7,0.8, 0.9,1
}
.
For a MST brain network M
p
, we extract the
nonzero values from upper triangle elements of M
p
,
denoted as t
p
, as shown in Equation 4.
t
p
= vec(M
p
(i, j) | 1 ≤ i < j ≤ R
and M
p
(i, j) ̸= 0)
(4)
We denote t
C
p
and t
G
p
as the nonzero values of upper
triangle elements of the CG and GG average brain
networks with connection density p, respectively.
Then, the Mann–Whitney U test is used to eval-
uate the significant difference between t
C
p
and t
G
p
, as
shown in Equation 5.
[P
p
,zval
p
] = U(t
G
p
,t
C
p
) (5)
P
p
denotes the probability of the difference between
t
G
p
and t
C
p
for specific p; zval
p
denotes the value of the
normal statistic between t
G
p
and t
C
p
.
• Nodal topological features in group networks
across different network densities.
Eight nodal topological features are adopted to
characterize the nodal topological organization of
brain networks. Each nodal topological feature is a
vector with a length equivalent to the number of re-
gions, which is R = 308 in this study.
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
732

1. The node degree F
nd
(i) refers to the number of
connections that node i has to other nodes in a
graph.
2. The node strength F
ns
(i) is the sum of connection
weights for node i.
3. The eigenvector centrality (Newman, 2010) of
node i, F
ec
(i), is equivalent to the i-th element in
the eigenvector corresponding to the largest eigen-
value of the adjacency matrix.
4. The participation coefficient F
pc
(i) (Guimera
and Nunes Amaral, 2005) quantifies how the con-
nections of node i are distributed across various
modules by representing the proportion of its con-
nectivity allocated to each module, identified here
using the Louvain community detection algorithm
(Blondel et al., 2008).
5. The node centrality F
nc
(i) (Brandes, 2001) quan-
tifies the proportion of shortest paths between all
node pairs in the network that pass through a given
index node i.
6. The local efficiency F
le
(Rubinov and Sporns,
2010) represents the global efficiency calculated
within each node’s neighborhood and is associ-
ated with the clustering coefficient.
7. The (weighted) clustering coefficient F
cc
(On-
nela et al., 2005) is defined as the average “inten-
sity” (geometric mean) of all triangles associated
with each node.
8. The nodal versatility F
nv
(i) (Shinn et al., 2017)
assesses the consistency with which a node i in
a modular decomposition is linked to a particu-
lar module. The identification of modules in this
research is derived by the Louvain community de-
tection algorithm (Blondel et al., 2008)).
For analyzing the nodal topological features of
group networks, the Euclidean distance is utilized to
quantify the disparity between CG and GG average
networks across each nodal topological feature at dif-
ferent densities p. Prior to computing the Euclidean
distance, each feature of the two groups is normal-
ized to [0,1] with the Min-Max method across all
p values. We use D
V
(p) represents Euclidean dis-
tance for a specific type of nodal topological feature
V ∈
{
nd, ns,ec, pc,nc,le,cc, nv
}
at a given connection
density p.
• Intra-/Inter-modular analysis based on VE-
Regions for the group brain networks.
We identify all intra-VE and inter-VE connec-
tions within a brain network A, denoted as A
intra
and A
inter
, respectively. Furthermore, we com-
pare the strength of intra-VE connections (S
intra
=
∑
A
kl
p
,k = l) and inter-VE connections (S
inter
=
∑
A
kl
p
,k ̸= l) for CG and GG at a specific connection
density (p = 0.1) processed by MST, with all connec-
tions positive.
• Intra-/Inter-hemispheric analysis for the group
brain networks.
For a brain network A , we obtain the intra-
hemispheric connections in left hemisphere (LL) and
right hemisphere (RR), as well as inter-hemispheric
correlations between left and right hemisphere (LR),
which can be denoted as A
LL
, A
RR
and A
LR
, respec-
tively. Moreover, we compare the strength of left
intra-hemispheric connections (S
LL
=
∑
A
LL
p
), right
intra-hemispheric connections (S
RR
=
∑
A
RR
p
) and
inter-hemispheric correlations (S
LR
=
∑
A
LR
p
) for CG
and GG at a specific connection density (p = 0.1) pro-
cessed by MST, with all connections positive.
2.2.3 Individual Cognitive Analyses for MSNs
This part explores the relationship between cognitive
performance and the structural organization of brain
networks, using two analytical approaches: the con-
nection strengths between VE-Regions and the global
topological features of brain networks. Both analyses
rely on the Spearman correlation to quantify the as-
sociations between network metrics and full-scale IQ
scores.
The Spearman correlation is a non-parametric
measure of rank correlation, evaluating the statistical
dependence between the rankings of two variables.
It determines how well the relationship between two
variables can be described using a monotonic func-
tion. The Spearman correlation coefficient (SCC) is
denoted as ρ in this work. We consider ρ > 0.2 as in-
dicating a significant positive monotonic relationship,
while ρ < −0.2 is interpreted as a significant negative
monotonic relationship.
The following in this part focuses on individual
cognitive analysis based on VE-Region connections
and global topological features of brain networks.
• Individual cognitive analysis based on VE-
Regions.
As defined in 2.2.2, E (k, l) represents the VE
connection strength between two VE-Regions k and
l within a brain network A. We introduce E
p
(k, l)
as the vector of the connection strength between the
two VE-Regions k and l across all subjects’ brain
networks at connection density p. To measure the
relationship between VE connections and cognitive
performance, we use ρ
p
(E
p
(k, l),C) to denote the
SCCs between each VE-Region connection strength
E
p
(k, l) and full-scale IQ scores C.
Analyzing Cognitive Patterns in Gifted Children Using MRI and Morphometric Similarity Networks
733

• Individual cognitive analysis with global topo-
logical features of brain networks.
Ten network metrics are adopted to characterize the
global topological organization of structural brain net-
works:
1. The assortativity (Newman, 2002) f
a
is defined
as the correlation coefficient for the degrees of
neighboring nodes, which refers to the tendency
of nodes in a network to link with other similar
nodes.
2. The transitivity f
t
is the ratio of “triangles to
triplets” in the network.
3. A network’s global efficiency (Latora and Mar-
chiori, 2001) f
ge
is the reciprocal of the harmonic
mean of its path lengths.
4. The characteristic path length (Watts and Stro-
gatz, 1998) f
cpl
is the average shortest path length
between all possible pairs of nodes in a network.
Moreover, the characteristic path length of a net-
work is strongly positively correlated with the net-
work’s average strength.
5. The mean participation coefficient f
mpc
can
measure the global integration of a network.
6. The mean clustering coefficient f
mcc
is the mean
value of the clustering coefficient of a network.
7. The mean versatility f
mv
is the mean value of
nodal versatility which can measure the global in-
tegration of a network.
8. The ratio of left to right intra-hemispheric con-
nections is denoted as f
lr
=
∑
A
LL
p
∑
A
RR
p
, where A
LL
p
and
A
RR
p
are denoted as the intra-hemispheric connec-
tions in the left and right hemisphere of a MST
network with density p, respectively.
9. The ratio of intra- to inter-hemispheric connec-
tions is denoted as f
ii
=
∑
A
LL
p
+
∑
A
RR
p
∑
A
LR
p
, where A
LR
p
is denoted as the inter-hemispheric connections of
a MST network with density p.
10. The ratio of intra- to inter-VE connections is
denoted as f
V E
=
∑
A
kl
p
,k=l
∑
A
kl
p
,k̸=l
, where A
kl
p
represents
connections between two VE-Regions k and l in a
MST network with density p.
In this research, we aim to enhance our compre-
hension of the relationship between these topological
features and cognitive abilities. We analyze Spearman
correlations between the full-scale IQ scores and each
global topological feature of the brain networks.
Here we utilize ρ
p
( f
p
v
,C) to denote the
SCC between each global topological fea-
ture f
p
v
and full-scale IQ scores C, where
v ∈
{
a,t,ge,cpl, mpc, mcc,mv,lr,ii,V E
}
represents
various global topological features.
3 RESULTS
3.1 Group Analysis Results for MSNs
As illustrated in Section 2.2.2, we conducted group
comparison analyses of MSNs, and the results are
shown in Fig. 2.
3.1.1 Results of VE-Region Analysis for Group
Average Brain Networks
• Heatmap Comparison for Group Average
Brain Networks.
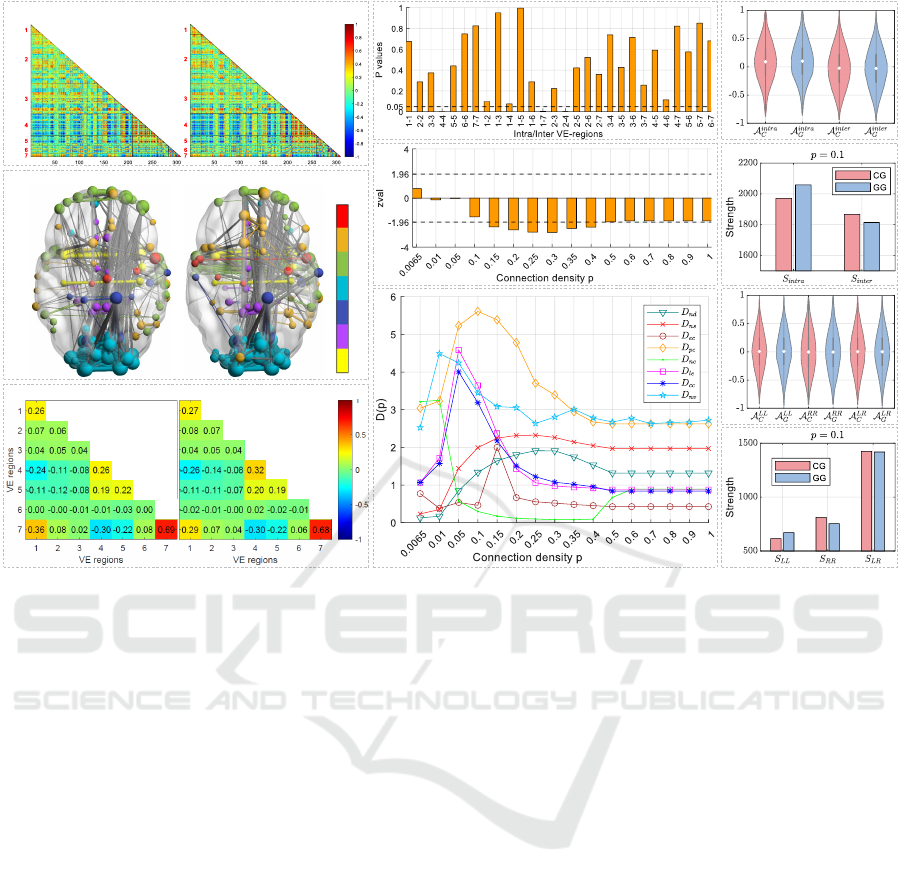
Fig. 2 a shows the heatmaps of group average net-
works for VE-ordered MSNs. In each heatmap, the
value range represented by colors varies from -1 to 1,
and warmer colors indicate higher values. Typically,
MSNs encompass both positive and negative values.
Notably, it appears that there is no discernible visual
distinction between the CG and GG in the heatmaps
of the group average MSNs.
• Comparison of Top 1% Group Brain Net-
works.
Fig. 2 b displays the top 1% (absolute) of group
average brain networks, as visualized using Brain-
Net Viewer (Xia et al., 2013). The seven-colored
nodes represent the grouped VE-Regions, and the
color links denote intra-VE connections, while grey
links denote inter-VE connections. The size of nodes
reflects the degree in the network. Top 1% of a brain
network (R = 308) contains [(R −1)R/2] ×1% = 474
connections. Notably, discernible visual differences
exist between the CG and GG average MSNs for
the top 1% of connections: GG shows more intra-
VE connections and fewer inter-VE connections com-
pared to CG.
• Results of Average VE-Region Connections of
Group Brain Networks.
As detailed in Section 2.2.2, we compute the aver-
age VE connection E , for the average brain network
of each group. The results are shown in Figure 2 c.
In the figure, each cell in the heatmaps represents the
average connection between VE-Regions k and l, de-
noted as E (k,l). The seven diagonal values corre-
spond to the average of connections within each intra-
VE-Region, while the values in the lower triangle rep-
resent the average of connection between each inter-
VE-Region pair. From the visualization, the group
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
734
CG
GG
a.
b.
c.
1
2
3
4
5
6
7
CG
GG
CG
GG
d.
f.
e.
i.
h.
g.
j.
Figure 2: Comparison of CG and GG average brain networks. a. Heatmaps of the CG (left) and GG (right) average networks,
organized by the seven VE-Regions. b. Comparison of the top 1% (absolute) of CG (left) and GG (right) average brain
networks, labeled with seven VE-Regions. c. Average VE-Region connection E of each group average brain network. d. P
values of the Mann–Whitney U test for CG and GG across each VE-Region. e. zval
p
results of the Mann–Whitney U test
between t
C
p
and t
G
p
for group average MSNs across different connection densities p. f. Euclidean distance between CG and
GG average networks on each nodal topological feature across different connection densities p. g. Violins for intra-/inter-VE
connections of group average MSNs. h. Strength of intra-VE (S
intra
) and inter-VE (S
inter
) connections for CG and GG at a
connection density of p = 0.1. i. Violins for intra-/inter-hemispheric connections of group average MSNs. j. Strengths of
hemispheric connections, S
LL
, S
RR
and S
LR
, at a connection density of p = 0.1.
average MSNs reveal more pronounced differences
between the CG and GG in the average VE-Region
connections in VE 4-4, 4-6, 5-5, 1-7 and 2-4 (differ-
ences ≥ 0.3). Specifically, GG exhibits stronger av-
erage connections in VE 4-4 and 4-6, while showing
weaker average connections in VE 5-5, 1-7 and 2-4.
• Significant Difference Evaluation Between CG
and GG Across Each VE-Region Connections.
As described in Section 2.2.2, we evaluate the sig-
nificant difference between the connections of A
kl
C
and A
kl
G
for each VE-k and VE-l. Figure 2 d provide a
visual representation of P
kl
values resulting from the
Mann–Whitney U test. It is apparent that the group
average MSNs exhibit significant differences between
the CG and GG in the VE connections 4-4, 1-7 and 2-
4, with P
kl
< 0.05. Additionally, notable differences
are observed in VE connections 1-2, 1-4 and 4-6,
with P
kl
≳ 0.05. These findings align closely with the
“Results of average VE-Region connections of group
brain networks,” which also identify pronounced dif-
ferences between CG and GG in VE-Region connec-
tions 4-4, 4-6, 1-7 and 2-4 (differences ≥ 0.3).
3.1.2 Analysis Results of Group Average Brain
Networks Across Different Density
As described in Section 2.2.2, we conducted an anal-
ysis to examine the effects of network density on the
difference between average CG and GG brain net-
works by comparing t
C
p
and t
G
p
across different con-
nection densities p. Fig. 2 e displays the zval
p
results
of the Mann–Whitney U test between t
C
p
and t
G
p
for
group average MSNs across different connection den-
sities p. The zval
p
values for MSN exceed 1.96 for p
values ranging from 0.15 to 0.4, reaching a maximum
of 2.80 at p = 0.3, indicating significantly larger con-
nection values in the CG average MSN compared to
the GG average MSN. Notably, the zval
p
values stabi-
lize as p ≥ 0.5. This stability arises from the roughly
equal numbers of positive and negative connections in
the MSNs, as observed in Section 3.1.4, with negative
Analyzing Cognitive Patterns in Gifted Children Using MRI and Morphometric Similarity Networks
735

connections being treated as zero. This also explains
the stability observed in the nodal topological feature
analysis when p ≥ 0.5 in the following Section 3.1.3.
3.1.3 Analysis Results of Nodal Topological
Features in Group Networks Across
Various Network Densities
Figure 2 f presents the Euclidean distance D
V
(p) be-
tween the CG and GG average networks on each
nodal topological feature at different densities p, in-
cluding: D
nd
(teal triangles), D
ns
(red crosses), D
ec
(red circles), D
pc
(orange diamonds), D
nc
(green
dots), D
le
(purple squares), D
cc
(blue asterisks), D
pc
(cyan stars).
Notably, the trends of D
nd
vs. D
ns
and D
cc
vs. D
le
exhibit similar patterns as p varies. Additionally, the
values of D
ns
and D
le
are slightly higher than those of
D
nd
and D
cc
, respectively. The details are as follows:
• Both D
nd
and D
ns
values start low at very small p
and show a slight increase as p increases, with a
relatively stable trend as p = 0.15 ∼ 0.3.
• Both D
le
and D
cc
values start low, rise sharply,
peak at p = 0.05, and then decrease rapidly.
• D
pc
values exhibit an early sharp increase, peak-
ing at p = 0.1, followed by a gradual decline.
Despite the decline, they maintain higher values
compared to other features.
• D
nv
values start high, peak early (p = 0.01), and
then decline steadily as density increases, still
maintaining higher values compared to other fea-
tures.
• D
nc
values show an initial peak at lower densities
(p = 0.0065 and p = 0.01), followed by a rapid
decline, stabilizing at very low values.
• D
ec
remain very low values except for a noticeable
increase at p = 0.15.
3.1.4 Results of Intra-/Inter-modular
Connections Based on VE-Regions for
Group Brain Networks
Figure 2 g shows violin plots representing the intra-
VE and inter-VE connections for CG and GG average
MSNs. Each violin includes a white dot representing
the average value. The light red violins correspond
to A
intra
C
and A
inter
C
for CG, while the light blue vio-
lins represent A
intra
G
and A
inter
G
for GG. The connection
values in the group average MSNs range from −1 to
1. Moreover, the average value of intra-VE connec-
tions is consistently higher than that of inter-VE con-
nections in both CG and GG average MSNs. How-
ever, the differences in intra-VE and inter-VE connec-
tions between CG and GG are minimal in the original
group average MSNs.
Fig. 2 h illustrates the intra-VE connection
strength (S
intra
) and inter-VE connection strength
(S
inter
) for CG and GG at a connection density of
p = 0.1. Both CG and GG exhibit stronger intra-
VE connectivity compared to inter-VE connectivity.
However, GG demonstrates stronger intra-VE con-
nectivity and weaker inter-VE connectivity compared
to CG.
3.1.5 Results of Intra-/Inter-hemispheric
Analysis for Group Average Brain
Networks
Fig. 2 i illustrates the violin plots depicting the intra-
/inter-hemispheric connections of each group average
brain network. The dot in each violin represents the
average value. The light red violins represent the LL,
RR, and LR connections in the average CG brain net-
work, while the light blue violins represent those in
the average GG brain network. It can be observed that
the differences in hemispheric connections between
CG and GG are minimal in the original group average
MSNs.
Fig. 2 j illustrates the hemispheric connection
strengths, S
LL
, S
RR
, and S
LR
, at a connection density
of p = 0.1. The results show that both CG and GG
exhibit stronger right intra-hemispheric connections
than left intra-hemispheric connections, and stronger
inter-hemispheric connections than intra-hemispheric
ones. However, GG shows higher S
LL
, weaker S
RR
,
and slightly weaker S
LR
compared to CG.
3.2 Results of Individual Cognitive
Analysis for MSNs
As outlined in Section 2.2.3, we analyzed the relation-
ship between cognitive performance and the structural
organization of individual MSNs using Spearman cor-
relation, with the results presented in Fig. 3. As stated
in Section 3.1, stability in MSNs occurs when the con-
nection density p ≥ 0.5. Therefore, our analysis fo-
cuses on connection densities p ≤ 0.6 in this section.
3.2.1 Results of Individual Cognitive Analysis
Based on VE-Regions
As described in 2.2.3, we analyzed the Spearman
correlations between full-scale IQ scores and each
VE-Region connection strength E
p
(k, l) across dif-
ferent connection densities p. The SCC results
ρ
p
(E
p
(k, l),C) are shown in Fig. 3 a. Each line corre-
sponds to a different connection density p = 0.0065 ∼
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
736
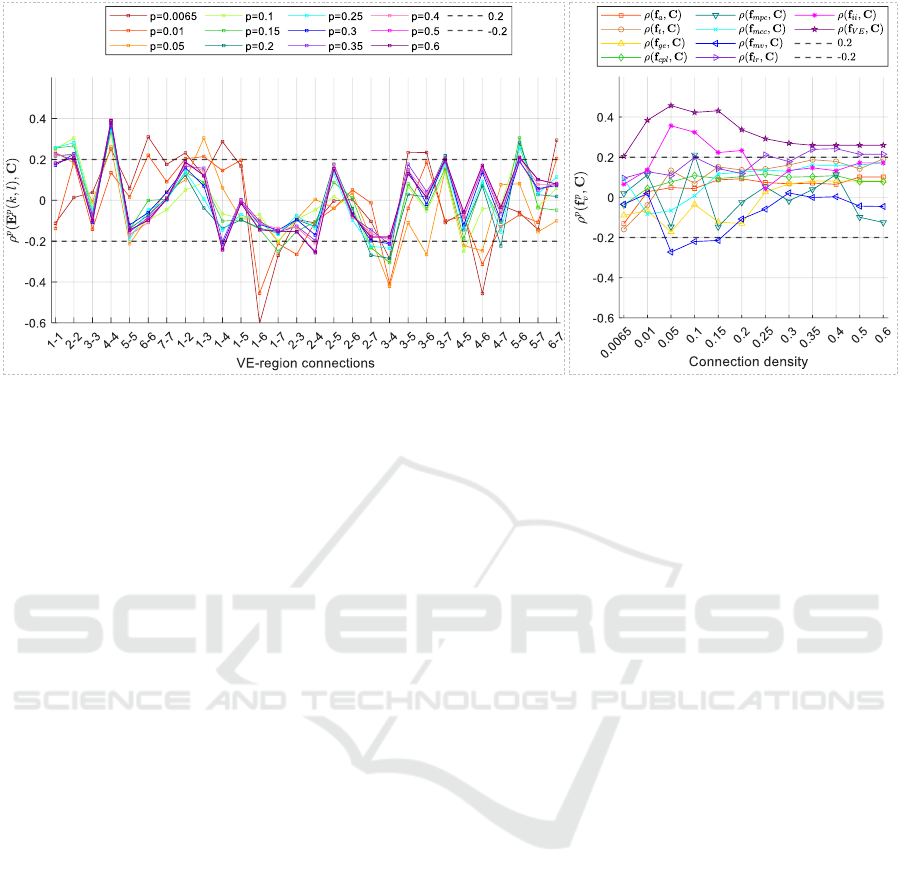
a. b.
Figure 3: Results of individual cognitive analysis for MSNs. a. SCCs ρ
p
(E
p
(k, l),C) between each VE-Region connection
strength E
p
(k, l) and full-scale IQ scores across different connection densities p. b. SCCs ρ
p
( f
p
v
,C) between various global
topological features (v) of MSNs and full-scale IQ scores (C) across different connection densities p.
0.6. The x-axis represents the various VE-Region
connections, while the y-axis denotes the SCC values.
It can be observed that:
• For lower connection densities (p ≤ 0.05), SCCs
exhibit more pronounced variations and instabil-
ity compared to those observed at higher connec-
tion densities (p ≥ 0.1) for the corresponding VE-
Region connections, particularly in 1-1, 6-6, 1-4,
1-6, 3-6 and 4-6. However, there are higher SCCs
in VE-Region connections of 1-6, 3-4, and 4-6 as
p ≤ 0.05.
• As the connection density p ≥ 0.1, the SCCs for
each VE-Region connection tend to stabilize, with
significant positive correlations for VE-Region
connections 1-1, 2-2, 4-4, 3-7, 5-6, 1-2 and 4-
6, significant negative correlations for VE-Region
connections 5-5, 1-4, 1-7, 2-4, 2-7, 3-4, 4-5 and
4-7.
3.2.2 Results of Individual Cognitive Analysis
with Global Topological Features
As described in 2.2.3, we analyzed the Spearman cor-
relations between each global topological feature of
brain networks and the full-scale IQ scores across dif-
ferent connection densities p. The results are dis-
played in Fig. 3 b. The x-axis represents different
values of p from 0.0065 to 1, while the y-axis shows
the SCCs for each feature. It can be observed that the
values of ρ
p
( f
p
v
,C) vary with different global topo-
logical features for MSNs:
• IQ is significantly positively correlated with the
ratio of intra- to inter-hemispheric connections
( f
ii
) for 0.05 ≤ p ≤ 0.2 (with the highest ρ = 0.36
at p = 0.05), and with the ratio of intra- to inter-
VE connections ( f
V E
) across all p values (with
the highest ρ = 0.46 at p = 0.05). These find-
ings suggest that individuals with higher IQ tend
to exhibit stronger intra-hemispheric and intra-
modular connections while displaying weaker
inter-hemispheric and inter-modular connections.
• IQ is significantly positively correlated with ra-
tio of left to right intra-hemispheric connec-
tions f
lr
when p = 0.1, indicating stronger left
intra-hemispheric connections in individuals with
higher IQ.
• IQ is significantly negatively correlated with
mean versatility f
mv
when 0.05 ≤ p ≤ 0.15, sug-
gesting less versatility in MSNs of individuals
with high cognitive performance.
• Correlations between IQ and mean participation
coefficient f
mpc
exhibit sharp fluctuations as con-
nection density p changes for MSNs.
• There are no significant correlations between IQ
and other global topological features, includ-
ing assortativity f
a
, transitivity f
t
, global effi-
ciency f
ge
, characteristic path length f
cpl
(aver-
age strength), mean weighted clustering coeffi-
cient f
mcc
.
• Generally, the correlations ρ
p
( f
p
v
,C) tend to
achieve higher values when 0.05 ≤ p ≤ 0.15 in
more cases.
Analyzing Cognitive Patterns in Gifted Children Using MRI and Morphometric Similarity Networks
737

4 DISCUSSION
In this study, we used sMRI data to compute the
MSNs of children with different intelligence levels.
We compared group-level MSNs of CG and GG to
identify key differences and analyzed individual-level
data to link cognitive performance with brain network
structure. The results of our experiments revealed
several important findings:
• The effects of connection density.
The effects of connection density on brain networks
reveal distinct trends in both group and individual
analyses. (1) Group-level analysis: The signifi-
cant differences between the CG and GG networks
become more pronounced when p = 0.15 ∼ 0.4.
The Euclidean distances between the nodal topolog-
ical features of two groups show high sensitivity to
changes in density when p ≤ 0.05, peaking between
p = 0.05 ∼ 0.25 in most cases. (2) Individual-
level analysis: For VE-Region connections, lower
connection densities (p ≤ 0.05) tend to show more
pronounced variations in correlations with IQ, while
higher densities stabilize these correlations. For
global topological features, higher SCCs are achieved
when p = 0.05 ∼ 0.15. Generally, a connection den-
sity of p = 0.05 ∼ 0.15 is recommended in MSN cog-
nitive analysis for stable and optimal results. Thus,
we chose a connection density of p = 0.1 for group-
level analyses of intra-/inter-modular and intra-/inter-
hemispheric connections.
• The relationship between VE-Region (modu-
lar) connections and cognitive performance.
Both the group and individual analyses reveal that
there are stronger connections in VE 4-4 (secondary
sensory area), 1-2 (motor to association area) and 4-
6 (secondary sensory to limbic area) for individuals
with higher IQ scores. Conversely, weaker connec-
tions in VE 1-7 (motor to insular area), 2-4 (associa-
tion to secondary sensory area) and 1-4 (motor to sec-
ondary sensory area) are observed in individuals with
higher IQ scores. These findings suggest that higher
cognitive performance is linked not only to enhanced
connectivity in specific sensory, motor, and limbic ar-
eas but also to a selective reduction in certain modular
connections, potentially contributing to greater neural
efficiency.
• Analyses on intra-/inter-modular and intra-
/inter-hemispheric connections
In the group average analysis, both CG and GG
demonstrate stronger intra-VE connections compared
to inter-VE connections, as well as stronger right
intra-hemispheric connections compared to left intra-
hemispheric connections, aligning with (Jiang et al.,
2019). Moreover, both group and individual anal-
yses reveal that individuals with higher IQ tend
to display stronger intra-VE and intra-hemispheric
connections, alongside weaker inter-VE and inter-
hemispheric connections. As stronger MSN con-
nectivity reflects greater morphometric similarity, in-
dividuals with higher IQ demonstrate higher mor-
phometric similarity within VE-Regions and hemi-
spheres, alongside greater differentiation between
VE-Regions and between the left and right hemi-
spheres. These findings suggest a more integrated
intra-hemispheric and intra-VE-Region organization,
which may facilitate more efficient cognitive pro-
cessing (Sol
´
e-Casals et al., 2019; Santarnecchi et al.,
2015; Krupnik et al., 2021). Additionally, the anal-
ysis shows that a higher ratio of left-to-right intra-
hemispheric connections in MSNs is associated with
enhanced IQ, suggesting that individuals with higher
IQ exhibit a more balanced distribution of connec-
tions between the left and right intra-hemispheric net-
works. Interestingly, this finding contrasts with (Sol
´
e-
Casals et al., 2019), which reported stronger right
intra-hemispheric connections for the GG in tradi-
tional SCNs.
• The relationship between topological features
and cognitive performance.
Our analysis reveals significant associations between
certain topological features of brain networks and IQ.
(1) Group-level analysis: Sensitivity to connection
density varies across nodal topological features. The
participation coefficient F
pc
, nodal versatility F
nv
, lo-
cal efficiency F
le
and clustering coefficient f
cc
are
sensitive to density variations, and achieve higher
Euclidean distance when p = 0.05 ∼ 0.1. Both the
node degree F
nd
and node strength F
ns
stably achieve
high Euclidean distance as p = 0.15 ∼ 0.3. However,
the node centrality F
nc
and eigenvector centrality F
ec
show little utility for cognitive analysis, as they do
not perform well in distinguishing cognitive groups.
(2) Individual-level analyses: IQ is negatively cor-
related with mean versatility f
mv
at p = 0.05 ∼ 0.1,
contrasting with (Sol
´
e-Casals et al., 2019), which
found higher mean versatility for the GG in traditional
SCNs. Other features, such as the mean participation
coefficient, show fluctuating correlations across den-
sities are less reliable for MSN cognitive analysis.
This study has some limitations. First, the small
sample size may reduce the generalizability of the
findings, underscoring the need for larger cohorts in
future research. Second, we neglect the negative value
in MSNs in connection density related analyses, po-
tentially affecting the interpretation of network struc-
tures. Third, while the study focuses on structural
networks, integrating functional imaging data could
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
738

provide deeper physiological and cognitive insights.
5 CONCLUSIONS
In this study, we constructed MSNs from sMRI data
to investigate brain network characteristics in chil-
dren with different intelligence levels. Group-level
analyses were performed by comparing the average
MSNs of the CG and GG to identify key differences in
brain network topology. Additionally, we conducted
individual-level analyses to explore the relationship
between cognitive performance and the structural or-
ganization of brain networks.
Our results show that variations in connection
density have a significant impact on both global and
nodal topological features, with each exhibiting dis-
tinct trends. A connection density of p = 0.05 ∼ 0.15
is recommended in MSN cognitive analysis for sta-
ble and optimal results. Additionally, gifted indi-
viduals exhibit stronger intra-hemispheric and intra-
modular connectivity, weaker inter-hemispheric and
inter-modular connectivity, a more balanced distri-
bution of left-to-right intra-hemispheric connections,
and lower mean versatility, which may be associ-
ated with more efficient and stable cognitive process-
ing. Moreover, the analyses on anatomical modular-
ity of VE indicate that higher cognitive performance
is linked not only to enhanced connectivity in spe-
cific modules (such as secondary sensory area, mo-
tor to association area, and secondary sensory to lim-
bic area) but also to a selective reduction in certain
modular connections (such as motor to insular area,
association to secondary sensory area, and motor to
secondary sensory area), potentially contributing to
greater neural efficiency. Furthermore, key topolog-
ical features, such as participation coefficient, nodal
versatility, local efficiency and clustering coefficient,
are linked to cognitive performance at specific con-
nection density. However, other features, such as
the mean participation coefficient (showing fluctuat-
ing correlations across densities), assortativity, char-
acteristic path length, and the mean weighted clus-
tering coefficient (showing no significant correlations
with IQ), are less reliable for MSN cognitive analysis.
In conclusion, our findings highlight the effects
of connection density and demonstrate how modu-
lar and hemispheric connectivity, along with specific
topological features, relate to children’s cognitive per-
formance. These insights pave the way for future re-
search to further explore the neural mechanisms un-
derlying cognitive abilities with brain networks. By
using larger, more diverse samples and longitudinal
designs, future studies could enhance our understand-
ing of how brain network topology evolves with cog-
nitive development and deepen our knowledge of the
neural mechanisms that underpin human intelligence.
ACKNOWLEDGEMENTS
This work was carried out as part of the doctoral pro-
gramm in Experimental Sciences and Technology at
the University of Vic - Central University of Catalo-
nia. F.D. work was partially supported by the National
Natural Science Foundation of China (Key Program)
(No. 11932013), and the Tianjin Science and Tech-
nology Plan Project (No. 22PTZWHZ00040). C.F.C
work was partially supported by grants PICT 2020-
SERIEA-00457 and PIP 112202101 00284CO (Ar-
gentina).
REFERENCES
Blondel, V. D., Guillaume, J.-L., Lambiotte, R., and Lefeb-
vre, E. (2008). Fast unfolding of communities in large
networks. Journal of statistical mechanics: theory
and experiment, 2008(10):P10008.
Brandes, U. (2001). A faster algorithm for between-
ness centrality. Journal of mathematical sociology,
25(2):163–177.
Desikan, R. S., S
´
egonne, F., Fischl, B., Quinn, B. T., Dick-
erson, B. C., Blacker, D., Buckner, R. L., Dale, A. M.,
Maguire, R. P., Hyman, B. T., et al. (2006). An au-
tomated labeling system for subdividing the human
cerebral cortex on mri scans into gyral based regions
of interest. Neuroimage, 31(3):968–980.
Faskowitz, J., Betzel, R. F., and Sporns, O. (2022). Edges in
brain networks: Contributions to models of structure
and function. Network Neuroscience, 6(1):1–28.
Fischl, B. and Dale, A. M. (2000). Measuring the thickness
of the human cerebral cortex from magnetic resonance
images. Proceedings of the National Academy of Sci-
ences, 97(20):11050–11055.
Genon, S., Eickhoff, S. B., and Kharabian, S. (2022). Link-
ing interindividual variability in brain structure to be-
haviour. Nature Reviews Neuroscience, 23(5):307–
318.
Ghosh, S. S., Kakunoori, S., Augustinack, J., Nieto-
Castanon, A., Kovelman, I., Gaab, N., Christodoulou,
J. A., Triantafyllou, C., Gabrieli, J. D., and Fischl,
B. (2010). Evaluating the validity of volume-based
and surface-based brain image registration for devel-
opmental cognitive neuroscience studies in children 4
to 11 years of age. Neuroimage, 53(1):85–93.
Guimera, R. and Nunes Amaral, L. A. (2005). Functional
cartography of complex metabolic networks. nature,
433(7028):895–900.
Heinsfeld, A. S., Franco, A. R., Craddock, R. C., Buch-
weitz, A., and Meneguzzi, F. (2018). Identification of
Analyzing Cognitive Patterns in Gifted Children Using MRI and Morphometric Similarity Networks
739

autism spectrum disorder using deep learning and the
ABIDE dataset. NeuroImage: Clinical, 17:16–23.
Jiang, X., Shen, Y., Yao, J., Zhang, L., Xu, L., Feng, R., Cai,
L., Liu, J., Chen, W., and Wang, J. (2019). Connec-
tome analysis of functional and structural hemispheric
brain networks in major depressive disorder. Transla-
tional psychiatry, 9(1):136.
Kong, X.-z., Liu, Z., Huang, L., Wang, X., Yang, Z., Zhou,
G., Zhen, Z., and Liu, J. (2015). Mapping individual
brain networks using statistical similarity in regional
morphology from mri. PloS one, 10(11):e0141840.
Krupnik, R., Yovel, Y., and Assaf, Y. (2021). Inner
hemispheric and interhemispheric connectivity bal-
ance in the human brain. Journal of Neuroscience,
41(40):8351–8361.
Latora, V. and Marchiori, M. (2001). Efficient behav-
ior of small-world networks. Physical review letters,
87(19):198701.
Li, J., Seidlitz, J., Suckling, J., Fan, F., Ji, G.-J., Meng, Y.,
Yang, S., Wang, K., Qiu, J., Chen, H., et al. (2021).
Cortical structural differences in major depressive dis-
order correlate with cell type-specific transcriptional
signatures. Nature communications, 12(1):1647.
Li, W., Yang, C., Shi, F., Wu, S., Wang, Q., Nie, Y., and
Zhang, X. (2017). Construction of individual mor-
phological brain networks with multiple morphomet-
ric features. Frontiers in Neuroanatomy, 11:34.
Lo, C.-Y. Z., He, Y., and Lin, C.-P. (2011). Graph theoret-
ical analysis of human brain structural networks. Re-
views in the Neurosciences, 22(5):551–563.
Newman, M. (2010). Networks: An Introduction. Oxford
University Press.
Newman, M. E. (2002). Assortative mixing in networks.
Physical review letters, 89(20):208701.
Onnela, J.-P., Saram
¨
aki, J., Kert
´
esz, J., and Kaski, K.
(2005). Intensity and coherence of motifs in weighted
complex networks. Physical Review E—Statistical,
Nonlinear, and Soft Matter Physics, 71(6):065103.
Park, H.-J. and Friston, K. (2013). Structural and functional
brain networks: from connections to cognition. Sci-
ence, 342(6158):1238411.
Rubinov, M. and Sporns, O. (2010). Complex network mea-
sures of brain connectivity: uses and interpretations.
Neuroimage, 52(3):1059–1069.
Santarnecchi, E., Tatti, E., Rossi, S., Serino, V., and Rossi,
A. (2015). Intelligence-related differences in the
asymmetry of spontaneous cerebral activity. Human
brain mapping, 36(9):3586–3602.
Sebenius, I., Seidlitz, J., Warrier, V., Bethlehem, R. A.,
Alexander-Bloch, A., Mallard, T. T., Garcia, R. R.,
Bullmore, E. T., and Morgan, S. E. (2023). Robust
estimation of cortical similarity networks from brain
mri. Nature Neuroscience, 26(8):1461–1471.
Seidlitz, J., V
´
a
ˇ
sa, F., Shinn, M., Romero-Garcia, R.,
Whitaker, K. J., V
´
ertes, P. E., Wagstyl, K., Reardon,
P. K., Clasen, L., Liu, S., et al. (2018). Morphometric
similarity networks detect microscale cortical organi-
zation and predict inter-individual cognitive variation.
Neuron, 97(1):231–247.
Shinn, M., Romero-Garcia, R., Seidlitz, J., V
´
a
ˇ
sa, F.,
V
´
ertes, P. E., and Bullmore, E. (2017). Versatility of
nodal affiliation to communities. Scientific Reports,
7(1):4273.
Sol
´
e-Casals, J., Serra-Grabulosa, J. M., Romero-Garcia,
R., Vilaseca, G., Adan, A., Vilar
´
o, N., Bargall
´
o, N.,
and Bullmore, E. T. (2019). Structural brain net-
work of gifted children has a more integrated and
versatile topology. Brain Structure and Function,
224(7):2373–2383.
Sun, K., Chen, G., Liu, C., Chu, Z., Huang, L., Li, Z.,
Zhong, S., Ye, X., Zhang, Y., Jia, Y., et al. (2024a). A
novel msn-ii feature extracted from t1-weighted mri
for discriminating between bd patients and mdd pa-
tients. Journal of Affective Disorders.
Sun, Y., Chen, P., Liu, Y., and Zhao, K. (2024b). Macroscale
brain structural network coupling is related to ad pro-
gression. In 2024 IEEE International Symposium on
Biomedical Imaging (ISBI), pages 1–4. IEEE.
Sun, Y., Lee, R., Chen, Y., Collinson, S., Thakor, N., Bez-
erianos, A., and Sim, K. (2015). Progressive gen-
der differences of structural brain networks in healthy
adults: a longitudinal, diffusion tensor imaging study.
PloS one, 10(3):e0118857.
Van Wijk, B. C., Stam, C. J., and Daffertshofer, A. (2010).
Comparing brain networks of different size and con-
nectivity density using graph theory. PloS one,
5(10):e13701.
V
´
a
ˇ
sa, F., Seidlitz, J., Romero-Garcia, R., Whitaker, K. J.,
Rosenthal, G., V
´
ertes, P. E., Shinn, M., Alexander-
Bloch, A., Fonagy, P., Dolan, R. J., et al. (2018). Ado-
lescent tuning of association cortex in human struc-
tural brain networks. Cerebral Cortex, 28(1):281–
294.
V
´
ertes, P. E., Rittman, T., Whitaker, K. J., Romero-Garcia,
R., V
´
a
ˇ
sa, F., Kitzbichler, M. G., Wagstyl, K., Fonagy,
P., Dolan, R. J., Jones, P. B., et al. (2016). Gene tran-
scription profiles associated with inter-modular hubs
and connection distance in human functional magnetic
resonance imaging networks. Philosophical Trans-
actions of the Royal Society B: Biological Sciences,
371(1705):20150362.
Von Economo, C. (1929). The cytoarchitectonics of the hu-
man cerebral cortex. H. Milford Oxford University
Press.
Watts, D. J. and Strogatz, S. H. (1998). Collective dynam-
ics of ‘small-world’networks. nature, 393(6684):440–
442.
Xia, M., Wang, J., and He, Y. (2013). Brainnet viewer:
a network visualization tool for human brain connec-
tomics. PloS one, 8(7):e68910.
Yu, K., Wang, X., Li, Q., Zhang, X., Li, X., and Li, S.
(2018). Individual morphological brain network con-
struction based on multivariate euclidean distances be-
tween brain regions. Frontiers in human neuroscience,
12:204.
Zhang, J., Feng, F., Han, T., Duan, F., Sun, Z., Caiafa, C. F.,
and Sol
´
e-Casals, J. (2021). A hybrid method to select
morphometric features using tensor completion and f-
score rank for gifted children identification. Science
China Technological Sciences, 64(9):1863–1871.
BIOSIGNALS 2025 - 18th International Conference on Bio-inspired Systems and Signal Processing
740
