
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast
Cells in Microstructure Environment
Khola Naseem
1,2 a
, Nabeel Khalid
1,2 b
, Lea Bertgen
3 c
, Johannes M Herrmann
3 d
,
Andreas Dengel
1,2 e
and Sheraz Ahmed
1 f
1
German Research Center for Artificial Intelligence (DFKI) GmbH, Kaiserslautern 67663, Germany
2
RPTU Kaiserslautern–Landau, 67663 Kaiserslautern, Germany
3
Cell Biology, University of Kaiserslautern, RPTU, Germany
Keywords:
Instance Segmentation, Deep Learning, Yeast Cell, Microstructure Environment, Traps, Time-Lapse
Fluorescence Microscopy, Synthetic Biology.
Abstract:
Cell segmentation is a crucial task, especially in microstructured environments commonly used in synthetic
biology. Segmenting cells in these environments becomes particularly challenging when the cells and the sur-
rounding traps share similar characteristics. While deep learning-based methods have shown success in cell
segmentation, limited progress has been made in segmenting yeast cells within such complex environments.
Most current approaches rely on traditional machine learning techniques. To address this challenge, the study
proposed a transfer-based instance segmentation approach to tackle both cell and trap segmentation in mi-
crostructured environments. The attention-based mechanism in the model’s backbone enables a more precise
focus on key features, leading to improved segmentation accuracy. The proposed approach outperforms exist-
ing state-of-the-art methods, achieving a 5% improvement in terms of Intersection over Union (IoU) for the
segmentation of both cells and traps in microscopic images.
1 INTRODUCTION
Yeast cells have been studied for decades in life
sciences due to their well-characterized genome,
membrane-bound organelles (like other eukaryotic
cells), genetic traceability, ease of gene manipulation,
availability, and overall simplicity of use(Lee, 2021).
In 1957, the ultrastructure of yeast cells was first ex-
plored(Osumi, 2012), and since then advancements in
imaging and molecular techniques have significantly
improved our ability to study them. The classical
baker’s yeast, Saccharomyces cerevisiae, is one of the
most widely used hosts for homologous and heterol-
ogous biopharmaceutical synthesis, protein produc-
tion, and gene manipulation(Mart
´
ınez et al., 2012).
Beyond basic research, yeast cells are extensively
a
https://orcid.org/0000-0003-4785-2588
b
https://orcid.org/0000-0001-9274-3757
c
https://orcid.org/0000-0003-1278-3279
d
https://orcid.org/0000-0003-2081-4506
e
https://orcid.org/0000-0002-6100-8255
f
https://orcid.org/0000-0002-4239-6520
used in industrial biotechnology, playing a crucial
role in fermentation processes for brewing, baking,
biofuel production, and winemaking(Onyema et al.,
2023). In recent years, yeast cell research has gained
important in various fields, including genetic engi-
neering, pharmaceutical research, synthetic biology,
and food science.
Data obtained from microscopes provide sig-
nificant insights into various biological processes.
Time-lapse fluorescence microscopy (TLFM) is an
advanced tool that allows the investigation of dy-
namic cellular processes in living, intact cells(Nasser
and Boudier, 2019). The extensive, standardized,
and quantitative data generated by TLFM help ad-
vance our understanding of biomolecular functions
and serve as a valuable resource for designing accu-
rate and advanced biomolecular systems. A typical
TLFM experiment using high-throughput microflu-
idics can generate thousands of specimen images,
making manual annotation and segmentation a chal-
lenging task(Mahmoud, 2019). To address this, vari-
ous automated segmentation techniques have been de-
veloped.
Naseem, K., Khalid, N., Bertgen, L., Herrmann, J. M., Dengel, A. and Ahmed, S.
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast Cells in Microstructure Environment.
DOI: 10.5220/0013169400003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 2, pages 407-417
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
407

Cell segmentation is a critical step in biomedi-
cal microscopy image analysis (Long, 2020). Af-
ter accurate cell segmentation, several downstream
tasks can be performed, such as cell tracking (Scherr
et al., 2020), (Lugagne et al., 2020; Wen et al.,
2021), cell counting (Loh et al., 2021; Ferreira and
Silveira, 2024), cell type classification (Witmer and
Bhanu, 2018), and cell phenotype analysis (Pratapa
et al., 2021), among others. In modern microscopy
studies, automated segmentation is essential for high-
throughput analysis. Numerous approaches have been
proposed to perform cell segmentation (Khalid et al.,
2024; Durkee et al., 2021; Edlund et al., 2021; Khalid
et al., 2023). However, it remains a challenging task
due to factors such as varying cell shapes, overlapping
cells, and inconsistent intensity levels in microscopic
images. The difficulty is compounded in complex en-
vironments like microstructured environments, where
precise control over mechanical properties, spatial ar-
rangement, and chemical gradients is possible. More-
over, it is crucial to distinguish cells from other struc-
tures, such as debris or traps, particularly in the con-
figurations discussed in this paper, where the appear-
ance of the cells and traps is quite similar.
In this paper, a transformer-based network is
proposed for cell-trap segmentation. The proposed
pipeline, YeastFormer, utilizes ViTDet (Li et al.,
2022) as the backbone and Cascade Mask R-CNN
(Cai and Vasconcelos, 2019) for instance-based seg-
mentation. ViTDet is responsible for feature extrac-
tion, while Cascade Mask R-CNN manages instance-
level segmentation tasks. A key strength of the model
lies in its capability to effectively extract both fine-
grained local details and global contextual informa-
tion, which are essential for accurate cell-trap seg-
mentation. A detailed explanation of the proposed
pipeline can be found in Section 3. Additionally, An
Anchor-based ResNeSt (Edlund et al., 2021) model
was fine-tuned and tested on the dataset. It was pre-
trained on the LiveCell dataset, one of the largest cell
segmentation datasets, and the other on the COCO
dataset. The proposed method outperformed it on
cell segmentation, further demonstrating its robust-
ness and effectiveness across various evaluations. The
major contributions of this paper include:
• Proposing a robust transformer-based network
that combines ViTDet (Li et al., 2022) for feature
extraction and Cascade Mask R-CNN (Cai and
Vasconcelos, 2019) for instance segmentation.
• Achieving a 5% improvement in IoU compared to
state-of-the-art techniques.
• Demonstrating the efficacy of the model by com-
paring it with other state-of-the-art techniques in
cell segmentation.
2 LITERATURE REVIEW
Many approaches have been proposed for cell detec-
tion and segmentation using both traditional computer
vision methods (Al-Hafiz et al., 2018; Mohammed
et al., 2013; Salem et al., 2016; He et al., 2022; Mand-
yartha et al., 2020) and deep learning-based tech-
niques (Khalid et al., 2023; Wang et al., 2023; Khalid
et al., 2022; Wang et al., 2022a). Traditional com-
puter vision approaches typically employ methods
such as intensity thresholding, region-based accumu-
lation, and deformable model fitting. However, these
approaches often rely on manual feature extraction
tailored to specific tasks, which limits their general-
izability. In contrast, deep learning (DL) approaches
have significantly advanced cell segmentation by of-
fering a data-driven methodology that requires less
domain-specific expertise. A major breakthrough oc-
curred with the introduction of U-Net (Ronneberger
et al., 2015), which won the ISBI 2015 cell track-
ing and segmentation challenge. This innovation has
greatly advanced biomedical research, leading to the
development of tools such as DeepCell (Van Valen
et al., 2016), CellPose (Stringer et al., 2021), Om-
nipose (Cutler et al., 2022), and Usiigaci (Tsai et al.,
2019).
Several approaches have also been proposed for
segmenting yeast cells in microscopic images (Salem
et al., 2021; Kruitbosch et al., 2022; Kong et al., 2020;
Lugagne et al., 2020; Haja and Schomaker, 2022).
Studying yeast cells in microstructured environments
allows for a tightly controlled setup, ensuring that the
cells remain within the microscope’s focal plane. Re-
searchers have extensively investigated yeast cells in
such environments (Liu et al., 2020; Gao et al., 2020;
Wang et al., 2022b).
Deep learning techniques have also been applied
to study cells in microstructured environments (Lu-
gagne et al., 2020; Tognato et al., 2023). In (Prange-
meier et al., 2020), the authors proposed a simple
and faster attention-based cell detection transformer
(CellDETR), which performs instance segmentation
in microstructured environments. CellDETR achieves
comparable results to Mask R-CNN, with a cell class
Jaccard index of 0.84, but with fewer parameters and
faster inference times. Synthetic data generation for
yeast cells in microstructured environments is dis-
cussed in (Reich et al., 2021), where a MultiStyle-
GAN is proposed for synthetic data generation based
on prior knowledge. In (Prangemeier et al., 2022), U-
Net was used for segmentation, while Mask R-CNN is
employed for instance segmentation of cells in com-
plex microstructured environments.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
408
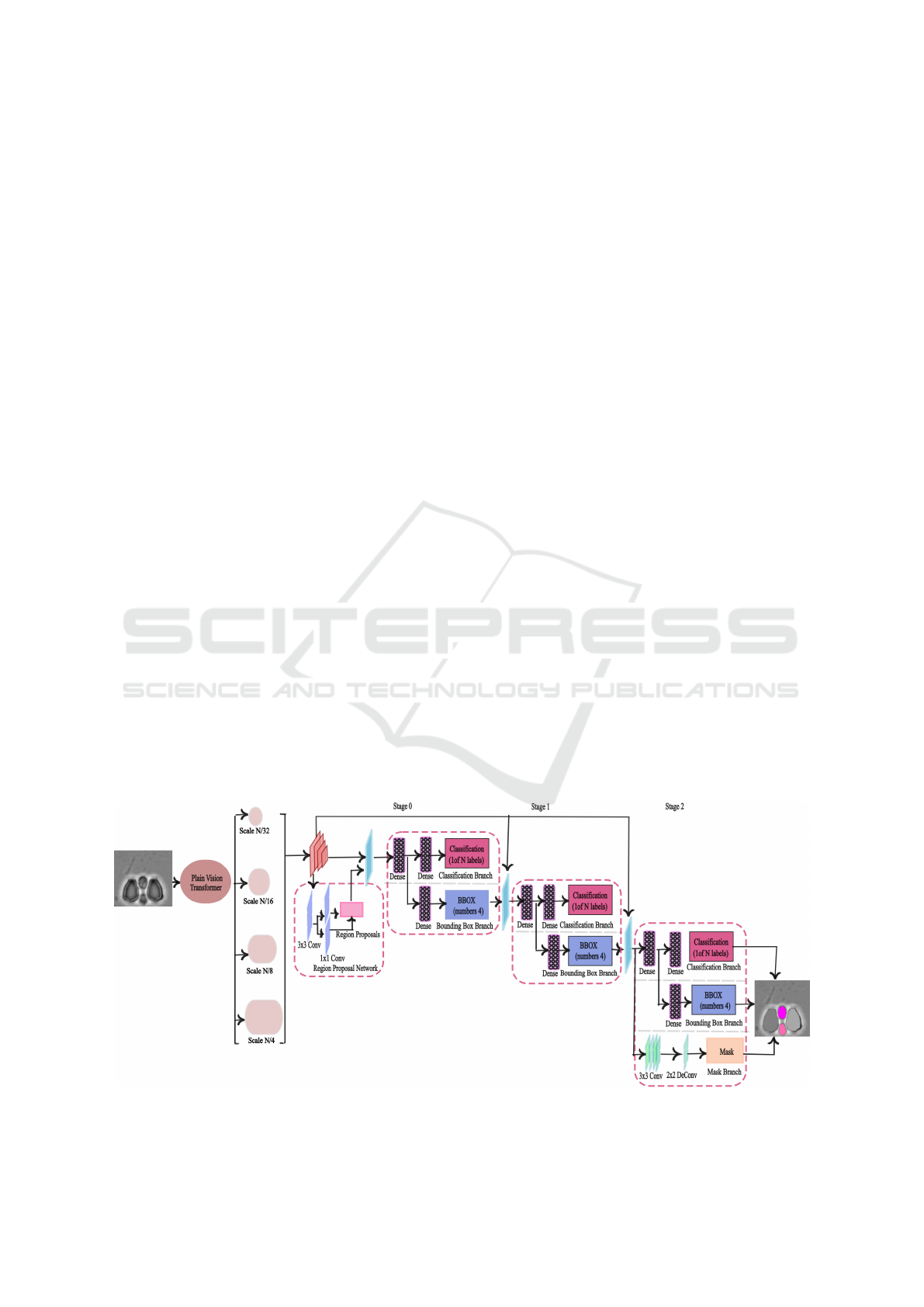
3 METHODOLOGY
Figure 1 provides a system overview of the proposed
pipeline for yeast cell segmentation. The proposed
network consists of the ViT Detector (ViTDet) back-
bone, Regional Proposal Network(RPN), and Cas-
cade Mask RCNN as the prediction head, respec-
tively.
3.1 Backbone Network
The backbone of the network is responsible for ex-
tracting features from the input data. It serves as
the core feature extractor, capturing both high-level
and low-level features from the input data. ViT De-
tector(ViTDet) has been used as the backbone of
the proposed network. ViT Detector (ViTDet) is a
specialized version of the Vision Transformer (ViT)
(Alexey, 2020), designed specifically for object de-
tection tasks. In ViTDet, the input image is divided
into patches, which are embedded into vectors and
passed through a stack of transformer blocks to cap-
ture both contextual and spatial relationships in a scal-
able manner. Using a global attention mechanism,
the backbone enhances feature extraction, producing
highly refined feature maps. The final feature map
is then fed into a Simple Feature Pyramid (SPF), a
streamlined version of traditional Feature Pyramids
(Lin et al., 2017), commonly used in object detection
tasks to handle objects of varying sizes. In the SPF, a
series of convolutions or deconvolutions is applied in
parallel, generating multi-scale feature maps at scales
of 1/32, 1/16, 1/8, 1/4 from an initial feature map at a
scale of 1/16.
3.2 Regional Proposal Network(RPN)
The features extracted from the backbone are passed
to the Region Proposal Network (RPN) (Ren et al.,
2016). The RPN is typically a fully convolutional
network (FCN), and its purpose is to identify regions
where objects may exist. This is achieved by draw-
ing anchor boxes on the input image and compar-
ing them to the ground truth using Intersection over
Union (IoU). If the IoU exceeds the 0.7 threshold, the
anchor box is assigned to the foreground and linked to
one of the ground truth boxes. If the IoU is less than
0.3, the anchor is considered background, otherwise,
it is ignored. After determining the anchor boxes,
the distance between the anchor boxes and the ground
truth is calculated. At this stage, Non-Maximum Sup-
pression (NMS) (Cai and Vasconcelos, 2018) is ap-
plied to retain only the best regions by removing over-
lapping or redundant proposals.
3.3 Prediction Head
Cascade Mask R-CNN (Cai and Vasconcelos, 2019)
was used as the prediction head in the proposed
model. This architecture is an extension of Cascade
R-CNN, with an additional mask branch to improve
pixel-level predictions. As a multistage network, Cas-
cade Mask R-CNN refines predictions by progres-
sively increasing the IoU threshold at each stage. In
the first stage, an IoU threshold of 0.5 is applied,
and predictions that have over 50% overlap with the
ground truth are passed to the next stage. In the sec-
ond stage, the output from the first stage is treated
as new region proposals, and an IoU threshold of 0.6
is used to refine the predictions further. In the fi-
nal stage, an IoU of 0.7 is applied to enhance accu-
Figure 1: System overview of YeastFormer. The input image is passed to the network and an instance segmentation mask is
produced as output.
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast Cells in Microstructure Environment
409
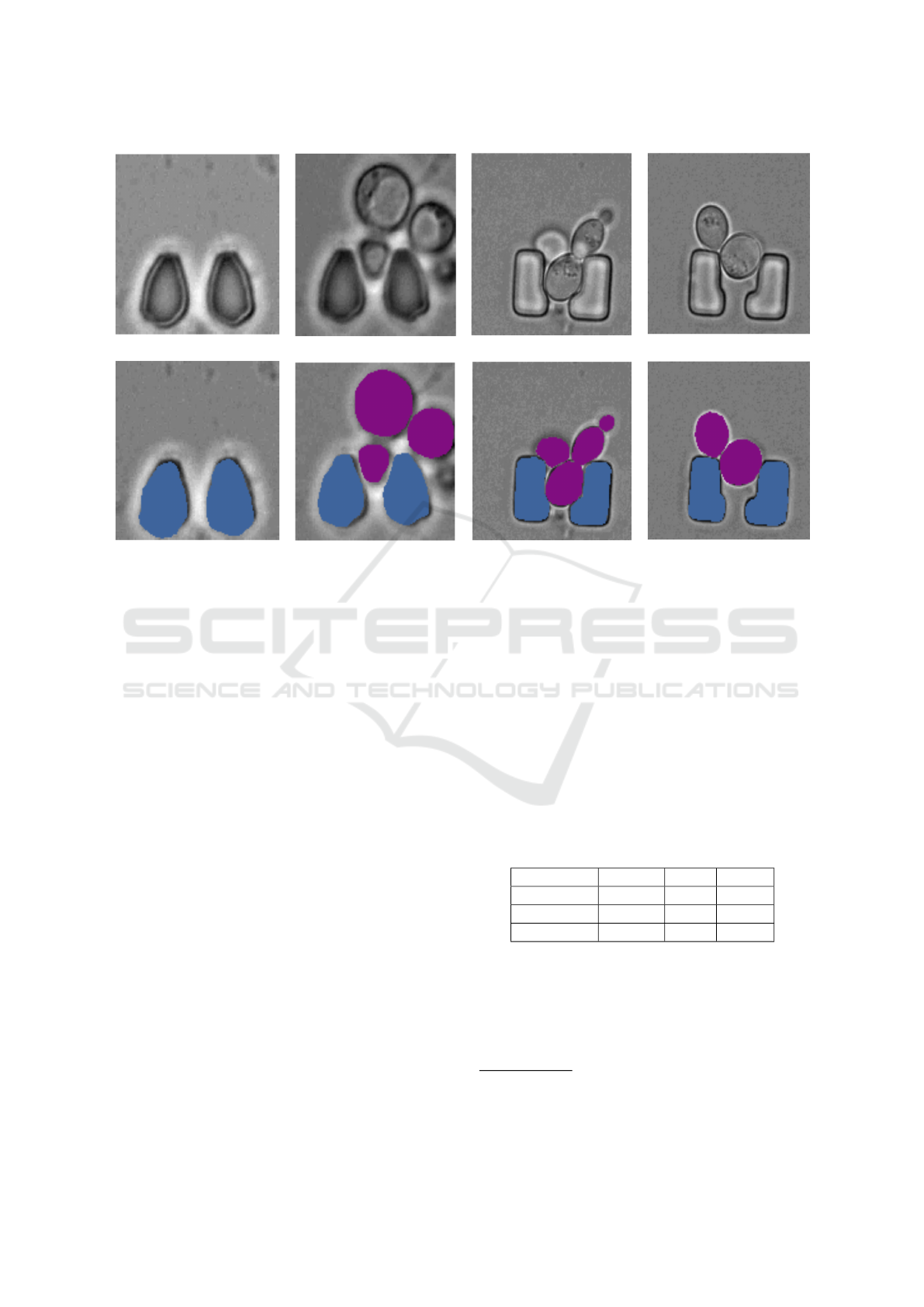
Figure 2: Sample images from the dataset: The top row shows the original images, and the bottom row shows the overlay of
the masks on the original images. Here, ∎ represents the cells and, ∎ represents the trap instances.
racy even more. In the proposed methodology, the
segmentation branch is added at the last stage of the
Cascade R-CNN. The box head classifies the object
within the ROI and fine-tunes the shape and position
of the box. A small fully convolution neural network
is used as the mask head to produce the segmentation
mask in pixel to pixel-to-pixel manner to attain the
instance segmentation mask.
4 DATASET
The dataset used in this study is designed for seg-
menting yeast cells within a microstructured environ-
ment(Reich et al., 2023). It consists of 493 densely
annotated brightfield microscopic images obtained
from various TLFM experiments. The dataset in-
cludes images with the most common yeast and trap
configurations, such as multiple cells, empty traps,
and single cells (with daughter cells). Two differ-
ent geometries of traps are included: L-shaped and
oval-shaped. Out of the 493 images, 398 contain reg-
ularly shaped traps, while the remaining 95 contain
L-shaped traps. The number of cells per image ranges
from zero to six. Figure 2 shows sample images
alongside their corresponding ground truth masks.
To enhance the versatility of the dataset, a diverse
range of variations is included, such as trap type, focal
shift, illumination levels, debris, and yeast morphol-
ogy. Cells and traps appear similar in the images be-
cause both have roughly circular shapes and charac-
teristic lengths. The dataset also captures challenging
edge cases, such as broken traps, which add complex-
ity to the segmentation task. Different instances of
cells and traps are marked with distinct colors, while
the background covering areas without cells or traps,
as well as debris and incomplete cells near the edges
is represented in gray. Table 1 provides an overview
of the number of images, as well as the instances of
cells and traps, across the training, validation, and test
sets. The dataset is publicly available
1
.
Table 1: Distribution of images, cells, and traps across train-
ing, validation, and test sets in the yeast cell dataset.
Split Images Cells Traps
Train 296 536 528
Validation 49 108 98
Test 148 270 291
5 EVALUATION METRICS
To evaluate the performance of the proposed network,
we employed the Jaccard index, the Panoptic Qual-
1
Dataset link:
https://github.com/ChristophReich1996/
Yeast-in-Microstructures-Dataset
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
410

ity (PQ) metric, and mean Average Precision (mAP)
as evaluation metrics. These metrics were chosen
because they comprehensively evaluate different as-
pects of segmentation quality, such as overlap accu-
racy, instance-wise segmentation, and class-specific
precision.
The Jaccard index (Hancock, 2004), also known
as Intersection over Union (IoU), was utilized to com-
pare the model with state-of-the-art techniques. It
measures the ratio of the intersection of pixels be-
tween the ground truth and the model output to their
union. This metric provides a clear understanding of
how well the predicted segmentation matches the ac-
tual segmentation. IoU is calculated using the formula
shown in Equation 1, where X represents the ground
truth and
ˆ
X denotes the prediction. Additionally, IoU
for specific classes is reported to facilitate direct com-
parisons with results from earlier approaches, further
validating the model’s performance.
IoU =
X ∩
̂
X
X ∪
̂
X
(1)
We also employed the standard COCO evaluation
protocol (Lin et al., 2014) for mean average preci-
sion (mAP). This metric evaluates model performance
at multiple IoU thresholds, specifically mAP50 and
mAP75, representing IoU thresholds of 50% and
75%, respectively. Additionally, results are reported
for different object size ranges, namely mAPs (small
objects) and mAPm (medium objects). This detailed
evaluation ensures a nuanced understanding of the
model’s performance across varying object scales.
The Panoptic Quality (PQ) metric (Kirillov et al.,
2019) was used to evaluate instance segmentation.
This metric is particularly well-suited for datasets that
can be treated as a specific case of panoptic segmen-
tation. In this context, the trap and cell are catego-
rized as thing classes, while the background is consid-
ered the sole stuff class. Consequently, the instance
segmentation predictions on this dataset can be effec-
tively assessed using PQ.
PQ =
∑
(p,g)∈T P
IoU(p, g)
∣T P∣
∣T P∣
∣T P∣+
1
2
∣FP∣+
1
2
∣FN∣
(2)
In PQ metric
1
∣T P∣
∑
(p,g)∈T P
IoU(p, g) is used to com-
pute the mean Intersection over Union (IoU) for all
matched predicted segments p and their correspond-
ing ground truth segments g. The PQ measures both
the instance-wise segmentation quality (SQ) and the
recognition quality (RQ)in a panoptic segmentation
context. The mean average precision (mAP) is calcu-
lated using the formula shown in Equation 3, where
n is the number of classes and (AP
i
) is the average
precision for the n classes.
mAP =
1
n
i=n
i=1
(AP
i
) (3)
6 EXPERIMENATAL SETUP
We conducted our evaluation using a range of dis-
tinct data settings to comprehensively assess the per-
formance, robustness, and limitations of segmenting
cells in a microstructured environment. The images
feature different types of traps, including L-shaped
and regular oval-shaped traps, with the number of
cells ranging from zero to six per image. Additionally,
we trained several networks on the dataset and com-
pared their performance to our network. We trained
and tested the Mask R-CNN model, following the
experimental setup from (Prangemeier et al., 2022).
Mask R-CNN enhances Faster R-CNN (Ren et al.,
2016) by integrating object detection with instance
segmentation. ROIAlign improves segmentation ac-
curacy by preserving spatial details, while FPN con-
structs a multi-scale feature pyramid to effectively
handle objects of different sizes.
Furthermore, we trained and tested the pre-trained
Anchor-based ResNeSt network. The Anchor-based
ResNeSt network was pre-trained on the LiveCell
dataset and utilized the aspect ratios of the COCO(Lin
et al., 2014) dataset (0.5, 1.0, and 2.0), with two addi-
tional aspect ratios of 3.0 and 4.0. The pixel means
and standard deviations were adjusted according to
the specifications of the LiveCell dataset. To handle
small objects effectively, anchor sizes of 8, 16, 32, 64,
and 128, were implemented.
The Segment Anything Model (SAM) (Kirillov
et al., 2023) was also fine-tuned and tested to perform
automated segmentation without requiring prompts.
The SAM model is a foundational model in com-
puter vision, leveraging a Masked Autoencoder with
a Vision Transformer for scalability. It features a
Prompt Encoder that generates prompt embeddings
and a Mask Decoder that maps image and prompt em-
beddings to the final segmentation mask. Drawing
inspiration from Transformer segmentation models,
SAM incorporates a learned output token embedding
into the prompt embedding. This output token plays a
key role in guiding the decoder by encapsulating crit-
ical information required for precise image segmen-
tation. We inputted the default embeddings from the
prompt encoder into SAM’s mask decoder. Empiri-
cal results confirm the effectiveness of this straight-
forward approach (Zhang and Liu, 2023; Gu et al.,
2024).
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast Cells in Microstructure Environment
411
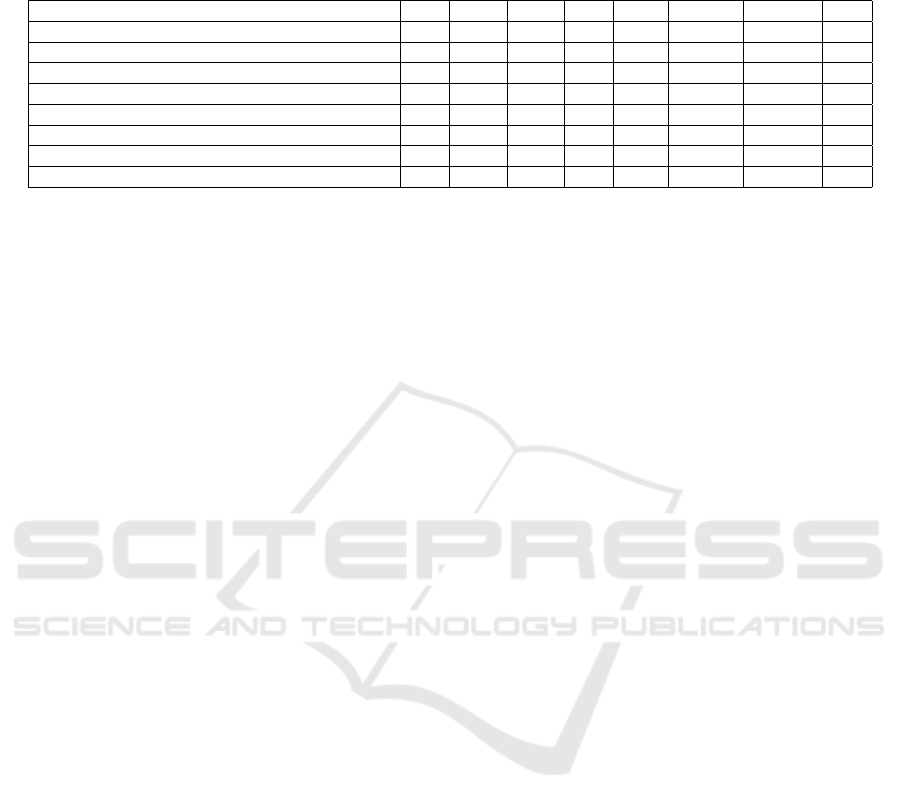
Table 2: Segmentation Performance – Average Precision (AP) is reported across various IoU thresholds and area ranges,
comparing IoU metrics for the cell and trap classes across multiple segmentation methods. Panoptic Quality (PQ) is included
to evaluate instance segmentation. The methods marked with an asterisk (*) utilized different data splits.
Method AP AP50 AP75 APs APm Cell IoU Trap IoU PQ
C-DETR A* (Prangemeier et al., 2020) - - - - - 83.0 85.0 -
C-DETR B* (Prangemeier et al., 2020) - - - - - 84.0 86.0 -
DISCO* (Prangemeier et al., 2020) - - - - - 70.0 - -
Mask R-CNN (SOTA) (Prangemeier et al., 2022) 65.4 98.4 85.4 56.5 71.1 84.0 89.0 -
Segment Anything (SAM)(Fine-tuned)(Kirillov et al., 2023) 44.8 94.0 83.8 47.8 50.0 63.1 89.9 88.1
Anchor-based ResNeSt(Edlund et al., 2021) 78.2 99.3 95.0 70.9 83.4 87.6 90.7 89.0
CellPose(Stringer et al., 2021) 44.4 94.5 82.2 - - - - -
YeastFormer (Proposed) 80.7 99.4 97.2 73.7 84.4 89.0 90.5 90.0
Note: CellPose does not provide classification capabilities to distinguish between different classes in a microstructure environment.
In our network, we employed aspect ratios of 0.5,
1.0, and 2.0, with the set of anchor sizes (32, 64, 128,
256, and 512), corresponding to these aspect ratios.
We used the AdamW optimizer (Loshchilov, 2017)
with an initial learning rate of 5 × 10
−5
and a decay
rate of 0.8. The model was trained for 6000 iterations
using an NVIDIA GeForce RTX 4090 GPU. All net-
works were implemented using the PyTorch frame-
work (Paszke et al., 2019). The best checkpoints are
selected based on the validation loss.
7 RESULTS AND DISCUSSION
Table 2 presents the segmentation AP scores aver-
aged across both classes. The proposed method out-
performed both competing approaches, achieving an
overall AP score of 80.7%. At IoU thresholds of 0.50
and 0.75, the model attained AP scores of 99.4% and
97.2%, respectively.
We provided a comparison with state-of-the-art
methods i.e. Mask R-CNN(Prangemeier et al.,
2022). We also compared our model’s results with
DISCO, Cell-DETR A, and Cell-DETR B (Prange-
meier et al., 2020), as presented in Table 2. DISCO
(Bakker et al., 2018) utilized traditional techniques,
such as template matching, Support Vector Machines
(SVM)(Suthaharan and Suthaharan, 2016), and ac-
tive contours. In contrast, Cell-DETR A and Cell-
DETR B employed an attention-based transformer
for Cell-Trap instance segmentation. However, our
approach’s results are not directly comparable with
these methods due to differences in the data splits
used across the models. For the experiments, we also
trained and tested the Anchor-based ResNeSt LIVE-
Cell model (Edlund et al., 2021) and the Segment
Anything Model (SAM).
The results presented in Table 2 show that our
model not only outperformed the state-of-the-art
Mask R-CNN model but also performed better in all
experiments. Our model achieved the highest cell IoU
of 89.0, outperforming Mask R-CNN by a margin
of 5%. The automatic mode of the Segment Any-
thing Model (SAM) was utilized without providing
any prompts to perform segmentation. However, the
results were suboptimal, indicating that SAM requires
prompts for better performance. The use of prompts,
however, requires the integration of expert knowl-
edge. In contrast, our proposed method operates in-
dependently of prompts and consistently outperforms
all compared models. Cellpose (Stringer et al., 2021)
performs class-agnostic segmentation on the dataset
used in this study, meaning it cannot distinguish be-
tween cell and trap instances. However, distinguish-
ing between these two is a key objective of our study.
The results of Cellpose are also reported in Table 2.
The proposed method outperforms CellPose on the
segmentation task by a wide margin of 36.3% in terms
of AP. These results show the potential of our pro-
posed approach for instance segmentation of cells and
traps in microstructure environment. In the current
setup, the Intersection over Union (IoU) for the cell
holds greater significance than the IoU for the trap.
Sample segmentation results of the different meth-
ods are shown in Figure 3. The results demon-
strate that our proposed model consistently outper-
forms other approaches in various challenging scenar-
ios. The first column displays the results from the
Mask R-CNN (SoTA) model, followed by the sec-
ond column showing outputs from the Anchor-based
ResNeSt model trained on the LiveCell dataset. The
third column presents the results from the Segment
Anything Model (SAM), while the fourth column
features the predictions generated by our proposed
method. Ground truth cell masks are depicted by
solid blue lines, with the model’s predictions shown
as dotted orange lines. Similarly, solid green lines
represent the trap ground truth, and dotted red lines
correspond to the model’s predictions. Our method
achieves the best IoU for both the cell and trap classes
across all images and performs well even near bound-
ary areas. In row (a), it is evident that all methods
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
412
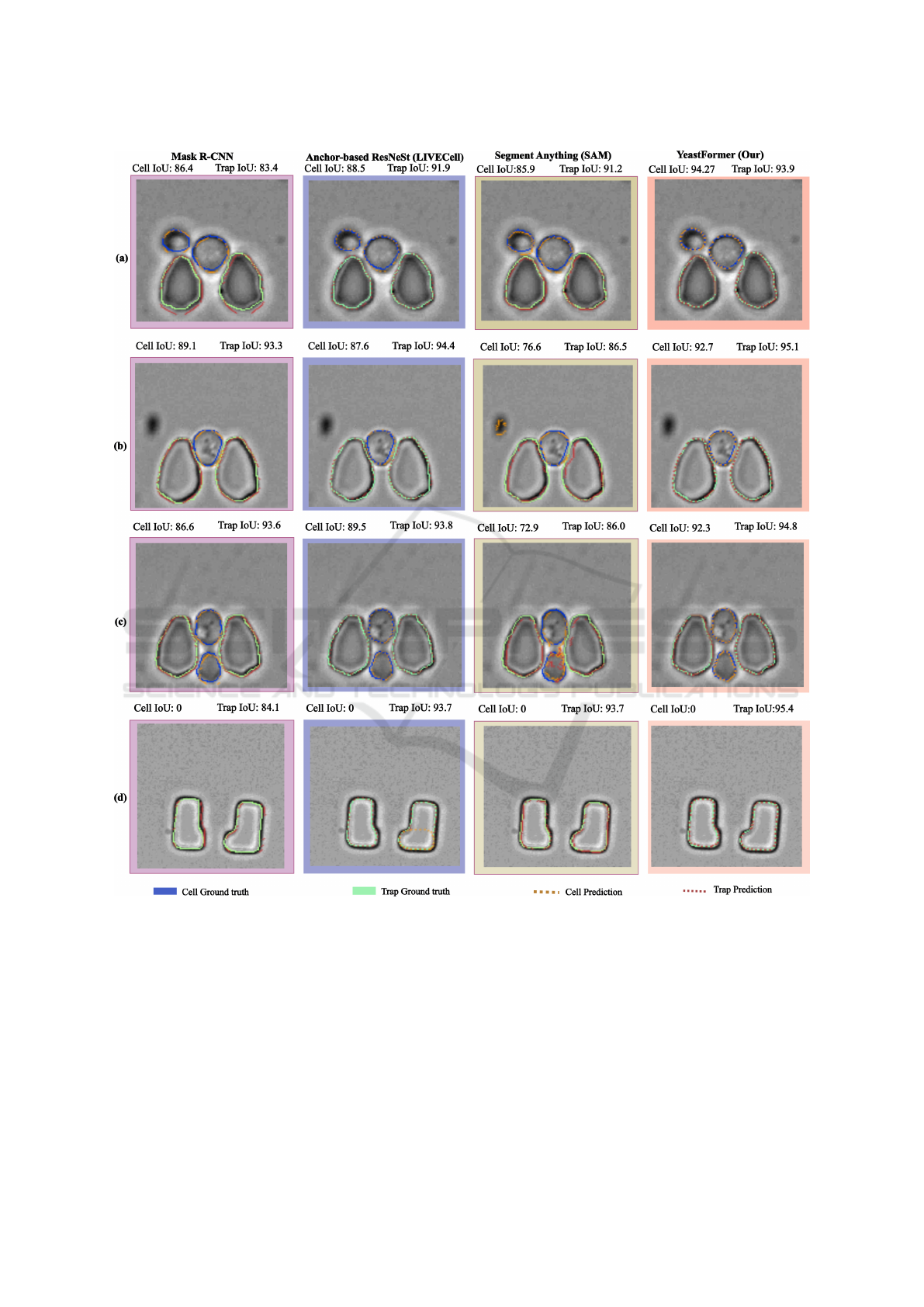
Figure 3: The inference results for several sample images in which our model performed sufficiently are presented. Ground
truth cell masks are outlined in solid blue, while model predictions are marked by dotted orange lines. Solid green lines
represent ground truth traps, and dotted red lines indicate the model’s trap predictions.
struggle near the boundaries of cells and traps. Mask
R-CNN fails to accurately detect both cells and traps,
while the Anchor-based ResNeSt and SAM also face
difficulties in boundary areas, leading to cell over-
segmentation. In row (b), SAM incorrectly identifies
an artifact as a cell, significantly lowering the IoU,
and fails to accurately segment traps. In row (c), the
other networks continue to struggle with boundary de-
lineation, as SAM is still unable to draw precise cell
boundaries and over-segments cells. Meanwhile, in
row (d), the Anchor-based ResNeSt detects a cell in
the trap area where no cell is present. These observa-
tions highlight the robustness and superior accuracy
of our model in handling complex and diverse situa-
tions compared to other methods.
For analysis, we present the results of our model
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast Cells in Microstructure Environment
413

Figure 4: The inference results for several sample images in which our model performed insufficiently are presented. Ground
truth cell masks are outlined in solid blue, while model predictions are marked by dotted orange lines. Solid green lines
represent ground truth traps, and dotted red lines indicate the model’s trap predictions.
on samples where it did not perform well, as shown in
Figure 4. The first column illustrates results from the
state-of-the-art Mask R-CNN (SoTA) model, while
the second column presents outputs from the Anchor-
based ResNeSt model. The third column features pre-
dictions from the Segment Anything Model (SAM),
and the fourth column showcases the outputs of our
proposed approach. Ground truth cell masks are rep-
resented by solid blue lines, with model predictions
depicted as dotted orange lines. Similarly, solid green
lines denote trap ground truth, and dotted red lines
represent trap predictions by the models. Both types
of traps are included in the images, providing a com-
prehensive assessment of segmentation performance.
Our model demonstrates consistent difficulties
with trap prediction, reflected in a low Intersection
over Union (IoU) score for the trap class. In row (a),
it is clear that our approach struggles to delineate the
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
414

lower boundary of the cell at the image’s edge, as well
as the trap boundary. SAM also fails in this scenario,
unable to detect the cell near the border of the im-
age. In row (b), the issue persists, with our model
struggling to accurately capture boundaries at the im-
age edge. Row (c) depicts a scenario where no cell
is present, leading to a cell IoU of 0 for all meth-
ods. Here, our model achieves the lowest trap IoU
among the approaches. In row (d), the model over-
segments the cell located between two traps and fails
to detect the traps accurately, further highlighting its
limitations in handling complex spatial arrangements.
Overall, our proposed approach faces challenges
when cell boundaries are ambiguous or when edge
cases occur near image borders. These limitations
point to areas for potential improvement in segmen-
tation accuracy and robustness. Linking the model’s
outputs to biologically meaningful metrics or insights,
such as cell counts, size distributions, or spatial re-
lationships, helps biologists directly interpret predic-
tions in the context of their experimental hypotheses
or workflows.
Vision Transformers are generally more compu-
tationally demanding than CNNs due to their self-
attention mechanism, which calculates interactions
between every pair of image patches (Maur
´
ıcio et al.,
2023). This capability allows Vision Transformers to
effectively capture global dependencies across the en-
tire image. In contrast, CNNs utilize convolutional
operations that scale more efficiently in terms of com-
putational complexity, although they may struggle to
capture global dependencies in larger images as ef-
fectively as Vision Transformers. This study demon-
strated that fine-tuning Vision Transformers achieved
better results compared to fine-tuning CNNs. How-
ever, this performance improvement came with a
trade-off: an increase in inference time for Vision
Transformers relative to CNNs.
8 CONCLUSIONS
In this research, we introduce a novel framework for
instance, the segmentation of yeast cells within a mi-
crostructured environment using a transformer-based
technique. Our proposed approach enables precise
detection and segmentation of individual yeast cells
and traps, even in situations where they share similar
characteristics, allowing for improved analysis of cell
morphology. Through extensive experimentation, we
demonstrate that this framework successfully differ-
entiates between instances of yeast cells and traps in
microscopic images, achieving robust performance.
To show the adaptability of the model to different in
vivo and in vitro microstructured environments, we
tested it on various trap types. The proposed tech-
nique holds significant potential to assist biologists
in analyzing yeast cell behavior within controlled en-
vironments, providing valuable insights into cellular
dynamics. Future work will aim to enhance model
performance further, ensuring greater accuracy and
reliability in diverse scenarios. Efforts will also focus
on extending this method to more microstructured en-
vironmental settings, broadening its applicability to a
wider range of experimental conditions. Additionally,
reducing computational demands will be a key objec-
tive, making the approach more practical and accessi-
ble for real-world applications.
ACKNOWLEDGEMENTS
This research was supported by a Grant from
the German-Israeli Foundation for Scientific Re-
search and Development (GIF, Grant number I-1561-
412.13/2023 to AD and JMH).
REFERENCES
Al-Hafiz, F., Al-Megren, S., and Kurdi, H. (2018). Red
blood cell segmentation by thresholding and canny de-
tector. Procedia Computer Science, 141:327–334.
Alexey, D. (2020). An image is worth 16x16 words: Trans-
formers for image recognition at scale. arXiv preprint
arXiv: 2010.11929.
Bakker, E., Swain, P. S., and Crane, M. M. (2018). Morpho-
logically constrained and data informed cell segmen-
tation of budding yeast. Bioinformatics, 34(1):88–96.
Cai, Z. and Vasconcelos, N. (2018). Cascade r-cnn: Delving
into high quality object detection. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 6154–6162.
Cai, Z. and Vasconcelos, N. (2019). Cascade r-cnn: High-
quality object detection and instance segmentation.
IEEE transactions on pattern analysis and machine
intelligence, 43(5):1483–1498.
Cutler, K. J., Stringer, C., Lo, T. W., Rappez, L., Strous-
trup, N., Brook Peterson, S., Wiggins, P. A., and
Mougous, J. D. (2022). Omnipose: a high-precision
morphology-independent solution for bacterial cell
segmentation. Nature methods, 19(11):1438–1448.
Durkee, M. S., Abraham, R., Clark, M. R., and Giger, M. L.
(2021). Artificial intelligence and cellular segmenta-
tion in tissue microscopy images. The American jour-
nal of pathology, 191(10):1693–1701.
Edlund, C., Jackson, T. R., Khalid, N., Bevan, N., Dale,
T., Dengel, A., Ahmed, S., Trygg, J., and Sj
¨
ogren, R.
(2021). Livecell—a large-scale dataset for label-free
live cell segmentation. Nature methods, 18(9):1038–
1045.
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast Cells in Microstructure Environment
415

Ferreira, E. and Silveira, G. (2024). Classification and
counting of cells in brightfield microscopy images: an
application of convolutional neural networks. Scien-
tific Reports, 14(1):9031.
Gao, Z., Xu, J., Chen, K., Wang, S., Ouyang, Q., and Luo,
C. (2020). Comparative analysis of yeast replicative
lifespan in different trapping structures using an inte-
grated microfluidic system. Advanced Materials Tech-
nologies, 5(12):2000655.
Gu, H., Dong, H., Yang, J., and Mazurowski, M. A. (2024).
How to build the best medical image segmentation al-
gorithm using foundation models: a comprehensive
empirical study with segment anything model. arXiv
preprint arXiv:2404.09957.
Haja, A. and Schomaker, L. R. (2022). A fully automated
end-to-end process for fluorescence microscopy im-
ages of yeast cells: From segmentation to detection
and classification. In Proceedings of 2021 Interna-
tional Conference on Medical Imaging and Computer-
Aided Diagnosis (MICAD 2021) Medical Imaging and
Computer-Aided Diagnosis, pages 37–46. Springer.
Hancock, J. (2004). Jaccard Distance (Jaccard Index, Jac-
card Similarity Coefficient).
He, F., Mahmud, M. P., Kouzani, A. Z., Anwar, A., Jiang,
F., and Ling, S. H. (2022). An improved slic algorithm
for segmentation of microscopic cell images. Biomed-
ical Signal Processing and Control, 73:103464.
Khalid, N., Froes, T. C., Caroprese, M., Lovell, G., Trygg,
J., Dengel, A., and Ahmed, S. (2023). Pace: Point
annotation-based cell segmentation for efficient mi-
croscopic image analysis. In International Confer-
ence on Artificial Neural Networks, pages 545–557.
Springer.
Khalid, N., Koochali, M., Leon, D. N. L., Caroprese, M.,
Lovell, G., Porto, D. A., Trygg, J., Dengel, A., and
Ahmed, S. (2024). Cellgenie: An end-to-end pipeline
for synthetic cellular data generation and segmenta-
tion: A use case for cell segmentation in microscopic
images. In Annual Conference on Medical Image Un-
derstanding and Analysis, pages 387–401. Springer.
Khalid, N., Koochali, M., Rajashekar, V., Munir, M., Ed-
lund, C., Jackson, T. R., Trygg, J., Sj
¨
ogren, R., Den-
gel, A., and Ahmed, S. (2022). Deepmucs: a frame-
work for co-culture microscopic image analysis: from
generation to segmentation. In 2022 IEEE-EMBS In-
ternational Conference on Biomedical and Health In-
formatics (BHI), pages 01–04. IEEE.
Kirillov, A., He, K., Girshick, R., Rother, C., and Doll
´
ar, P.
(2019). Panoptic segmentation. In Proceedings of the
IEEE/CVF conference on computer vision and pattern
recognition, pages 9404–9413.
Kirillov, A., Mintun, E., Ravi, N., Mao, H., Rolland, C.,
Gustafson, L., Xiao, T., Whitehead, S., Berg, A. C.,
Lo, W.-Y., et al. (2023). Segment anything. In Pro-
ceedings of the IEEE/CVF International Conference
on Computer Vision, pages 4015–4026.
Kong, Y., Li, H., Ren, Y., Genchev, G. Z., Wang, X.,
Zhao, H., Xie, Z., and Lu, H. (2020). Automated
yeast cells segmentation and counting using a parallel
u-net based two-stage framework. OSA Continuum,
3(4):982–992.
Kruitbosch, H. T., Mzayek, Y., Omlor, S., Guerra, P., and
Milias-Argeitis, A. (2022). A convolutional neural
network for segmentation of yeast cells without man-
ual training annotations. Bioinformatics, 38(5):1427–
1433.
Lee, B. H. (2021). Advanced Fermentation and Cell Tech-
nology, 2 Volume Set. John Wiley & Sons.
Li, Y., Mao, H., Girshick, R., and He, K. (2022). Explor-
ing plain vision transformer backbones for object de-
tection. In European conference on computer vision,
pages 280–296. Springer.
Lin, T.-Y., Doll
´
ar, P., Girshick, R., He, K., Hariharan, B.,
and Belongie, S. (2017). Feature pyramid networks
for object detection. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 2117–2125.
Lin, T.-Y., Maire, M., Belongie, S., Hays, J., Perona, P.,
Ramanan, D., Doll
´
ar, P., and Zitnick, C. L. (2014).
Microsoft coco: Common objects in context. In Com-
puter Vision–ECCV 2014: 13th European Confer-
ence, Zurich, Switzerland, September 6-12, 2014, Pro-
ceedings, Part V 13, pages 740–755. Springer.
Liu, P., Liu, H., Yuan, D., Jang, D., Yan, S., and Li, M.
(2020). Separation and enrichment of yeast saccha-
romyces cerevisiae by shape using viscoelastic mi-
crofluidics. Analytical Chemistry, 93(3):1586–1595.
Loh, D. R., Yong, W. X., Yapeter, J., Subburaj, K., and
Chandramohanadas, R. (2021). A deep learning ap-
proach to the screening of malaria infection: Auto-
mated and rapid cell counting, object detection and
instance segmentation using mask r-cnn. Computer-
ized Medical Imaging and Graphics, 88:101845.
Long, F. (2020). Microscopy cell nuclei segmentation with
enhanced u-net. BMC bioinformatics, 21(1):8.
Loshchilov, I. (2017). Decoupled weight decay regulariza-
tion. arXiv preprint arXiv:1711.05101.
Lugagne, J.-B., Lin, H., and Dunlop, M. J. (2020). Delta:
Automated cell segmentation, tracking, and lineage
reconstruction using deep learning. PLoS computa-
tional biology, 16(4):e1007673.
Mahmoud, L. N. K. (2019). A dictionary-based denois-
ing method toward a robust segmentation of noisy and
densely packed nuclei in 3D biological microscopy
images. PhD thesis, Sorbonne Universit
´
e.
Mandyartha, E. P., Anggraeny, F. T., Muttaqin, F., and Ak-
bar, F. A. (2020). Global and adaptive thresholding
technique for white blood cell image segmentation. In
Journal of Physics: Conference Series, volume 1569,
page 022054. IOP Publishing.
Mart
´
ınez, J. L., Liu, L., Petranovic, D., and Nielsen, J.
(2012). Pharmaceutical protein production by yeast:
towards production of human blood proteins by mi-
crobial fermentation. Current opinion in biotechnol-
ogy, 23(6):965–971.
Maur
´
ıcio, J., Domingues, I., and Bernardino, J. (2023).
Comparing vision transformers and convolutional
neural networks for image classification: A literature
review. Applied Sciences, 13(9):5521.
Mohammed, E. A., Mohamed, M. M., Naugler, C., and Far,
B. H. (2013). Chronic lymphocytic leukemia cell seg-
mentation from microscopic blood images using wa-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
416

tershed algorithm and optimal thresholding. In 2013
26th IEEE Canadian Conference on Electrical and
Computer Engineering (CCECE), pages 1–5. IEEE.
Nasser, L. and Boudier, T. (2019). A novel generic
dictionary-based denoising method for improving
noisy and densely packed nuclei segmentation in 3d
time-lapse fluorescence microscopy images. Scientific
reports, 9(1):5654.
Onyema, V., Amadi, O., Moneke, A., and Agu, R. (2023).
A brief review: Saccharomyces cerevisiae biodiver-
sity potential and promising cell factories for exploita-
tion in biotechnology and industry processes–west
african natural yeasts contribution. Food Chemistry
Advances, 2:100162.
Osumi, M. (2012). Visualization of yeast cells by elec-
tron microscopy. Journal of electron microscopy,
61(6):343–365.
Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J.,
Chanan, G., Killeen, T., Lin, Z., Gimelshein, N.,
Antiga, L., et al. (2019). Pytorch: An imperative style,
high-performance deep learning library. Advances in
neural information processing systems, 32.
Prangemeier, T., Reich, C., and Koeppl, H. (2020).
Attention-based transformers for instance segmenta-
tion of cells in microstructures. In 2020 IEEE Interna-
tional Conference on Bioinformatics and Biomedicine
(BIBM), pages 700–707. IEEE.
Prangemeier, T., Wildner, C., Franc¸ani, A. O., Reich, C.,
and Koeppl, H. (2022). Yeast cell segmentation
in microstructured environments with deep learning.
Biosystems, 211:104557.
Pratapa, A., Doron, M., and Caicedo, J. C. (2021). Image-
based cell phenotyping with deep learning. Current
opinion in chemical biology, 65:9–17.
Reich, C., Prangemeier, T., Franc¸ani, A. O., and Koeppl,
H. (2023). An instance segmentation dataset of yeast
cells in microstructures. In 2023 45th Annual In-
ternational Conference of the IEEE Engineering in
Medicine & Biology Society (EMBC), pages 1–4.
IEEE.
Reich, C., Prangemeier, T., Wildner, C., and Koeppl,
H. (2021). Multi-stylegan: Towards image-based
simulation of time-lapse live-cell microscopy. In
Medical Image Computing and Computer Assisted
Intervention–MICCAI 2021: 24th International Con-
ference, Strasbourg, France, September 27–October
1, 2021, Proceedings, Part VIII 24, pages 476–486.
Springer.
Ren, S., He, K., Girshick, R., and Sun, J. (2016). Faster
r-cnn: Towards real-time object detection with re-
gion proposal networks. IEEE transactions on pattern
analysis and machine intelligence, 39(6):1137–1149.
Ronneberger, O., Fischer, P., and Brox, T. (2015). U-
net: Convolutional networks for biomedical image
segmentation. In Medical image computing and
computer-assisted intervention–MICCAI 2015: 18th
international conference, Munich, Germany, October
5-9, 2015, proceedings, part III 18, pages 234–241.
Springer.
Salem, D., Li, Y., Xi, P., Phenix, H., Cuperlovic-Culf,
M., and Kaern, M. (2021). Yeastnet: Deep-
learning-enabled accurate segmentation of budding
yeast cells in bright-field microscopy. Applied Sci-
ences, 11(6):2692.
Salem, N., Sobhy, N. M., and El Dosoky, M. (2016). A com-
parative study of white blood cells segmentation using
otsu threshold and watershed transformation. Jour-
nal of Biomedical Engineering and Medical Imaging,
3(3):15.
Scherr, T., L
¨
offler, K., B
¨
ohland, M., and Mikut, R. (2020).
Cell segmentation and tracking using cnn-based dis-
tance predictions and a graph-based matching strat-
egy. PLoS One, 15(12):e0243219.
Stringer, C., Wang, T., Michaelos, M., and Pachitariu, M.
(2021). Cellpose: a generalist algorithm for cellular
segmentation. Nature methods, 18(1):100–106.
Suthaharan, S. and Suthaharan, S. (2016). Support vector
machine. Machine learning models and algorithms
for big data classification: thinking with examples for
effective learning, pages 207–235.
Tognato, R., Bronte Ciriza, D., Marag
`
o, O., and Jones, P.
(2023). Modelling red blood cell optical trapping by
machine learning improved geometrical optics calcu-
lations. Biomedical Optics Express, 14(7):3748–3762.
Tsai, H.-F., Gajda, J., Sloan, T. F., Rares, A., and Shen,
A. Q. (2019). Usiigaci: Instance-aware cell tracking in
stain-free phase contrast microscopy enabled by ma-
chine learning. SoftwareX, 9:230–237.
Van Valen, D. A., Kudo, T., Lane, K. M., Macklin, D. N.,
Quach, N. T., DeFelice, M. M., Maayan, I., Tanouchi,
Y., Ashley, E. A., and Covert, M. W. (2016). Deep
learning automates the quantitative analysis of indi-
vidual cells in live-cell imaging experiments. PLoS
computational biology, 12(11):e1005177.
Wang, C.-W., Huang, S.-C., Lee, Y.-C., Shen, Y.-J., Meng,
S.-I., and Gaol, J. L. (2022a). Deep learning for bone
marrow cell detection and classification on whole-
slide images. Medical Image Analysis, 75:102270.
Wang, Y., Wang, W., Liu, D., Hou, W., Zhou, T., and Ji, Z.
(2023). Genesegnet: a deep learning framework for
cell segmentation by integrating gene expression and
imaging. Genome Biology, 24(1):235.
Wang, Y., Zhu, Z., Liu, K., Xiao, Q., Geng, Y., Xu, F.,
Ouyang, S., Zheng, K., Fan, Y., Jin, N., et al. (2022b).
A high-throughput microfluidic diploid yeast long-
term culturing (dylc) chip capable of bud reorienta-
tion and concerted daughter dissection for replicative
lifespan determination. Journal of Nanobiotechnol-
ogy, 20(1):171.
Wen, C., Miura, T., Voleti, V., Yamaguchi, K., Tsutsumi,
M., Yamamoto, K., Otomo, K., Fujie, Y., Teramoto,
T., Ishihara, T., et al. (2021). 3deecelltracker, a deep
learning-based pipeline for segmenting and tracking
cells in 3d time lapse images. Elife, 10:e59187.
Witmer, A. and Bhanu, B. (2018). Multi-label classifica-
tion of stem cell microscopy images using deep learn-
ing. In 2018 24th International Conference on Pattern
Recognition (ICPR), pages 1408–1413. IEEE.
Zhang, K. and Liu, D. (2023). Customized segment any-
thing model for medical image segmentation. arXiv
preprint arXiv:2304.13785.
YeastFormer: An End-to-End Instance Segmentation Approach for Yeast Cells in Microstructure Environment
417
