
Comparison Between CNN and GNN Pipelines for Analysing the Brain
in Development
Antoine Bourlier
1,2
, Elodie Chaillou
2
, Jean-Yves Ramel
3
and Mohamed Slimane
1
1
LIFAT, Universit
´
e de Tours, 64 Av. Jean Portalis, Tours, France
2
INRAe, CNRS, Universit
´
e de Tours, Nouzilly, France
3
LISTIC, Universit
´
e de Savoie Mont-Blanc, Chamb
´
ery, France
Keywords:
Graph, Machine Learning, MRI, Segmentation.
Abstract:
In this study, we present a new pipeline designed for the analysis and comparison of non-conventional animal
brain models, such as sheep, without relying on neuroanatomical priors. This innovative approach combines
an automatic MRI segmentation with graph neural networks (GNNs) to overcome the limitations of traditional
methods. Conventional tools often depend on predefined anatomical atlases and are typically limited in their
ability to adapt to the unique characteristics of developing brains or non-conventional animal models. By
generating regions of interest directly from MR images and constructing a graph representation of the brain,
our method eliminates biases associated with predefined templates. Our results show that the GNN-based
pipeline is more efficient in terms of accuracy for an age prediction task (63.22%) compared to a classical
CNN architecture (59.77%). GNNs offer notable advantages, including improved interpretability and the
ability to model complex relational structures within brain data. Overall, our approach provides a promising
solution for unbiased, adaptable, and interpretable analysis of brain MRIs, particularly for developing brains
and non-conventional animal models.
1 INTRODUCTION
Automated methods have facilitated brain MRI anal-
ysis, especially in humans and conventional animal
models (Kaur and Gaba, 2021; Park and Friston,
2013). However, such tools are rarely available to
study developing brains or non-conventional animal
models like sheep. In these cases, brain structures
identification is often manual or based on automatic
segmentation using signal intensity and templates,
when available (Nitzsche et al., 2015; Ella et al.,
2017). These methods depend on biological priors
and the accuracy of atlases, which may not fully cap-
ture individual variations or patterns associated to dif-
ferent brain disorders. These issues are especially
challenging for developing brains, where contrast is
weak, development is uneven, and some structures are
not visible (Li et al., 2019). Additionally, using pre-
defined regions may limit the discovery of new brain
correlates.
To address segmentation bias, convolutional neu-
ral networks (CNNs) have been introduced, as they
work directly on MR images without segmentation
(Srinivasan et al., 2024; Coupeau et al., 2022). CNNs
provide valuable whole-brain or abnormality-level in-
formation but require large labelled datasets to learn
low-level image features for classification or segmen-
tation tasks. This makes them unsuitable for the de-
veloping brain or non-conventional animal models
with small cohorts.
In this paper, we present a pipeline to address
both segmentation biases and CNN limitations. We
propose generating regions of interest (ROIs) with-
out relying on neuro-anatomical priors. We use voxel
intensities to create segmented images via 2 differ-
ent segmentation algorithms. Additionally, we use
graph neural networks (GNNs) to identify and anal-
yse anatomical patterns, as GNNs model the brain as
interconnected patches, capturing complex relation-
ships between regions (Cui et al., 2021; Li et al.,
2021; Ravinder et al., 2023). By avoiding prede-
fined atlases, our method allows more flexible, image-
driven exploration of the brain. This approach is par-
ticularly useful for studying non-conventional animal
models of brain development, such as sheep. It offers
the potential to discover new structures and patterns
in both human and animal studies, enhancing under-
Bourlier, A., Chaillou, E., Ramel, J.-Y. and Slimane, M.
Comparison Between CNN and GNN Pipelines for Analysing the Brain in Development.
DOI: 10.5220/0013173500003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
475-482
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
475

standing and improving the diagnosis and treatment
of developing brain disorders.
This paper focuses on the specific task related to
study the developing brain. We tested various GNN
architectures and compared them to a classical CNN
approach. The comparison addresses the challenge of
predicting brain age in non-conventional animal mod-
els without neuro-anatomical priors. The next sec-
tions discuss the limitations of classical brain MRI
processing methods to study developing brain disor-
ders and non-conventional animal models. We de-
tail our pipeline, including segmentation, graph gen-
eration, and GNN prediction. Then, we present the
experimental comparison with a CNN approach, fol-
lowed by a discussion of its advantages and limita-
tions.
2 RELATED WORKS
2.1 Classical Approaches Used on Brain
MRI
The 2 main tasks studied in the literature are im-
age/brain segmentation (Coupeau et al., 2022) and
image/brain classification (Srinivasan et al., 2024;
Kaur and Gaba, 2021; Poriya, 2023). In both cases,
machine learning algorithms provide powerful tools
such as CNN and graph convolutional network (GCN)
layers that compute and find on its own the most rele-
vant features. Whether CNN or GNN is chosen, both
require specific key such as pre-processing and patch
extraction. The aim of the MRI pre-processing (de-
noising, normalisation etc.) is to adjust the inten-
sity values of the images to a standard range in or-
der to ensure consistency. A resampling step is used
to align images to a common voxel size or resolution
and a registration step can be added to align images
to a standard anatomical space, often using a template
brain. To remove non-brain tissues (e.g., skull, scalp)
cropping and skull-stripping are performed on MRI
images.
2.1.1 Segmentation
The aim of segmentation is to delineate and identify
anatomic ROI in the brain MRI, segmented images
being used to create a graph representation. Segmen-
tation is performed manually or using atlases based
on individual brain images or templates (Van Essen
and Drury, 1997; Yang et al., 2020; Fil et al., 2021).
They are used for ROI identification, delineating vari-
ous regions and structures or serve as reference points
to align individual MRI scans to a common space, fa-
cilitating consistent and accurate analysis across sub-
jects.
2.1.2 CNN Design
A CNN typically consists of multiple layers, includ-
ing convolutional layers, pooling layers, and fully
connected layers. Using advanced CNN architec-
tures, such as AlexNet (Krizhevsky et al., 2012), or
ResNet (He et al., 2016), enhances the model’s abil-
ity to learn complex patterns and achieve high per-
formance in various MRI analysis tasks, including
classification, clustering etc. Incorporating biologi-
cal priors through brain atlases can further refine these
methods, offering improved accuracy and consistency
in brain MRI analysis. Concerning brain age predic-
tion, several CNN architectures have been proposed
including VGG, ResNet, and DenseNet (Cole et al.,
2017; Jiang et al., 2020).. More advanced CNN archi-
tectures including attention mechanisms have since
been developed to boost the representational power
and improve prediction accuracy (Lam et al., 2020;
Cheng et al., 2021).
2.1.3 Graph Design
Nodes of a graph are usually defined at a region level
(i.e., one node per brain structure). The way the re-
gions are chosen is really linked with the aim of the
study and could represent neurons, anatomical struc-
tures, brain tissues, voxels, etc. From an anatomi-
cal point of view, node features provide information
about the position of the region (coordinates of the
center of gravity, orientation etc.), the shape (volume,
sphericity etc.), the signal intensity, which is useful to
understand the composition of the tissue, etc. Graph
theory features can also be used (centrality, strength
etc.). Several ways exist to build the edges of the
graph and characterise them depending on the aim of
the study. Generally, three types of edges are distin-
guished: structural, functional, and effective connec-
tions (Fedorov et al., 2012). The possibilities for edge
features are also numerous: Euclidean distances, tract
lengths, connection costs, etc. (Bullmore and Bassett,
2011; Sporns, 2018). Possibly, the trickiest compo-
nent in creating the brain network lies in edge cre-
ation. We could create a fully connected graph but for
an interpretable and efficient representation, we aim
to reduce the edge density, so that only the significant
connections are displayed. This is achieved by intro-
ducing a threshold and removing edges that do not
meet the required criteria. How to define the threshold
is still an active research question: typical approaches
use customised, statistical, or expert-based criteria.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
476

2.1.4 GNN Design
Once the brain has been modelled as a graph, GNNs
are trained to analyse these graphs, and to capture
complex relationships and variations in brain organ-
isation (Li et al., 2021; Ravinder et al., 2023; Srini-
vasan et al., 2024; Coupeau et al., 2022). For age
prediction, GNN architectures have also been pro-
posed to better exploit the inter-region relationships
like a GNN architecture that processes diffusion-
MRI-derived brain connectivity data while consider-
ing brain network topological locality (Sporns, 2007).
Many methods like multi-hop graph attention or
graph Transformer framework require graph struc-
tures such as tractography networks or the registra-
tion of multi-modal images based on a standard brain
template (Lim et al., 2024; Cai et al., 2023).
2.2 Approaches for Brain in
Development and Non-Conventional
Animal Model
The analysis of brains in development and/or from
non-conventional animal models, rises several chal-
lenges and considerations that differ from the analy-
sis of adult human brains. The primary differences in-
clude anatomical variations, the lack of standardised
atlases, tools and often smaller datasets.
For example, brain segmentation of non-conventional
animal models is often done manually. This pro-
cess is time-consuming, requires neuro-anatomical
expertise and introduces a high level of bias due
to inter-individual variation between operators (Fe-
dorov et al., 2012). An alternative to the manual
segmentation, is the atlas-based registration method.
The concept involves creating a template, achieved
by registering and normalising multiple brain MR
images into a common space using affine transfor-
mations, followed by segmentation of the template
and transferring the segmentation to each MR image
(De Vico Fallani et al., 2017). While this method is
beneficial for segmenting numerous images simulta-
neously, post-processing is essential to ensure accu-
rate correspondence between the segmentation and in-
dividual anatomy.
There are also some automatic and incremental seg-
mentation algorithms available that incorporate bio-
logical priors (Galisot et al., 2022).
3 THE PROPOSED PIPELINE
3.1 From 3D MR Images to Graphs
We propose a generic pipeline for transforming
3D MR images of brains in development of non-
conventional animal models into graphs. The objec-
tive is to generate graphs incorporating a maximum of
information from the brains and, to let the GNN select
the useful information inside these graphs (Figure 1).
3.2 Pre-Processing
Pre-processing includes skull-stripping and z-score
normalisation. Z-score normalisation is reported to be
more suitable for brain MRI analysis, particularly in
machine learning, because it maintains the alignment
of intensity peaks for white matter, grey matter, and
CSF (Schmid, 2023). After the image pre-processing,
the graph creation is divided into two parts: node cre-
ation and edge creation.
3.3 Nodes and Edges Creation
The creation of nodes corresponds to segmented ROIs
(Figure 2). Since our goal is to process brains in de-
velopment of growing non-conventional animal mod-
els, segmentation without biological priors offers an
alternative approach to analyse brain MR images and
constructing graphs. This method relies solely on im-
age information and the data is treated purely as a
conventional image rather than specifically as a brain
image. For this study, we have chosen to test a
histogram-based clustering algorithm and a ”split and
merge” algorithm. The histogram-based algorithm
splits the intensity range into N equal parts. One of
the main challenges is to determine the optimal pa-
rameters of the algorithm which depends on the study
objectives, the desired level of details etc. In the im-
age 1, the first segmentation is made with this algo-
rithm. The second algorithm is the ”split and merge”
algorithm, (Gonzalez and Woods, 2017) which oper-
ates in two distinct phases: ”split” and ”merge”. In the
first phase, the algorithm recursively divides the im-
age into smaller and homogeneous regions (”cubes”)
based on a user-defined homogeneity criterion and a
minimum region size. The homogeneity criterion is
the intensity amplitude of the region intensities. Then,
in the merge step, adjacent regions are combined if
their merged region meets another homogeneity crite-
rion. Each region of the segmentation is represented
as a node with associated normalised features : re-
gion’s volume divided by the brain’s volume, region’s
surface area divided by the brain’s surface area, mean
Comparison Between CNN and GNN Pipelines for Analysing the Brain in Development
477

Figure 1: Graph construction pipeline in which we tested different segmentation algorithms (histogram-based segmentation
and ”Split and merge” segmentation) and parameters. ”N” is the hyperparameter of the histogram-based algorithm. ”Split” and
”merge” are the hyperparameters of the ”split and merge” algorithm. ”p” is the number of edge features, 0 in our experiment.
intensity of the voxels inside the region, standard de-
viation of the intensity, region’s center of gravity in
the spherical coordinate system (radial distance from
the center of gravity of the brain, polar angle and az-
imuthal angle).
Then, there are many possibilities to define the
edges, and we propose to build an adjacency graph
by connecting two nodes if the geometric distance be-
tween them is below a threshold d (d is a hyperparam-
eter of the method). Depending on the task the user
wants to perform with the graphs, choosing the appro-
priate value for d might be another challenge.
3.4 Graph Analysis and Classification
As discussed before, GNNs are particularly well-
suited for brain MRI analysis due to their ability to
capture the intricate relationships between different
brain ROIs. Unlike CNNs, which primarily focus on
local voxel information, GNNs can model the brain at
a higher level where nodes represent different struc-
tures or parts of structures and edges represent con-
nections between them. This relational representation
can lead to more accurate and interpretable predic-
tions.
In the conducted experiments, the proposed GNN ar-
chitecture tries to harness these advantages to predict
the age of a sheep brain (by solving a graph classifi-
cation task with K classes of age range) (Figure 2).
3.4.1 Graph Convolution Layers
To propagate the information over the graph, we use
a set of 3 convolution layers. As described in the pre-
vious sections, multiple convolution layers are avail-
able. Our dataset is relatively small, consisting of ap-
proximately 200 graphs. Consequently, using a com-
plex neural network with sophisticated and large lay-
ers is not advisable. Therefore, in this study, we
utilised the GCNConv layer, which considers both the
node feature (F) and adjacency (A) matrices. The GC-
NConv layer computes updated node features by ag-
gregating information from neighbour nodes and their
connections. The input to the first layer consists of the
node features matrix (F), initialized with 7 features
per node, and the adjacency matrix (A) that encodes
the graph structure. The transformation from 7 fea-
tures to 32 or 64 features occurs progressively across
the layers, as follows:
• First set of parameters: The model begins by
transforming the 7 initial features into 8 features
using the first GCNConv layer (Figure 2). This
output is then passed to a second GCNConv layer,
which further transforms it to 16 features. Finally,
a third GCNConv layer transforms the features to
32 dimensions. Each layer applies a learned linear
transformation followed by an activation function
(ReLU in our case), enabling the network to pro-
gressively capture complex patterns in the data.
• Second set of parameters: Instead of incremental
changes, the model starts with 7 features and dou-
bles the number of dimensions at each layer: from
7 to 16, then to 32, and finally to 64. This more
aggressive dimensionality increase aims to test the
model’s ability to learn richer representations.
Dropout regularisation is applied and set to 0.5 to pre-
vent overfitting during training.
3.4.2 Pooling and Fully Connected Layers
A global mean pooling and a global max pooling were
tested to capture interesting information while taking
into account the small amount of data.
The pooled features are then passed through three
fully connected (FC) layers (fc1, fc2 and fc3) to per-
form the classification. The first FC layer maps the
features to a 128-dimensional space, the second FC
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
478
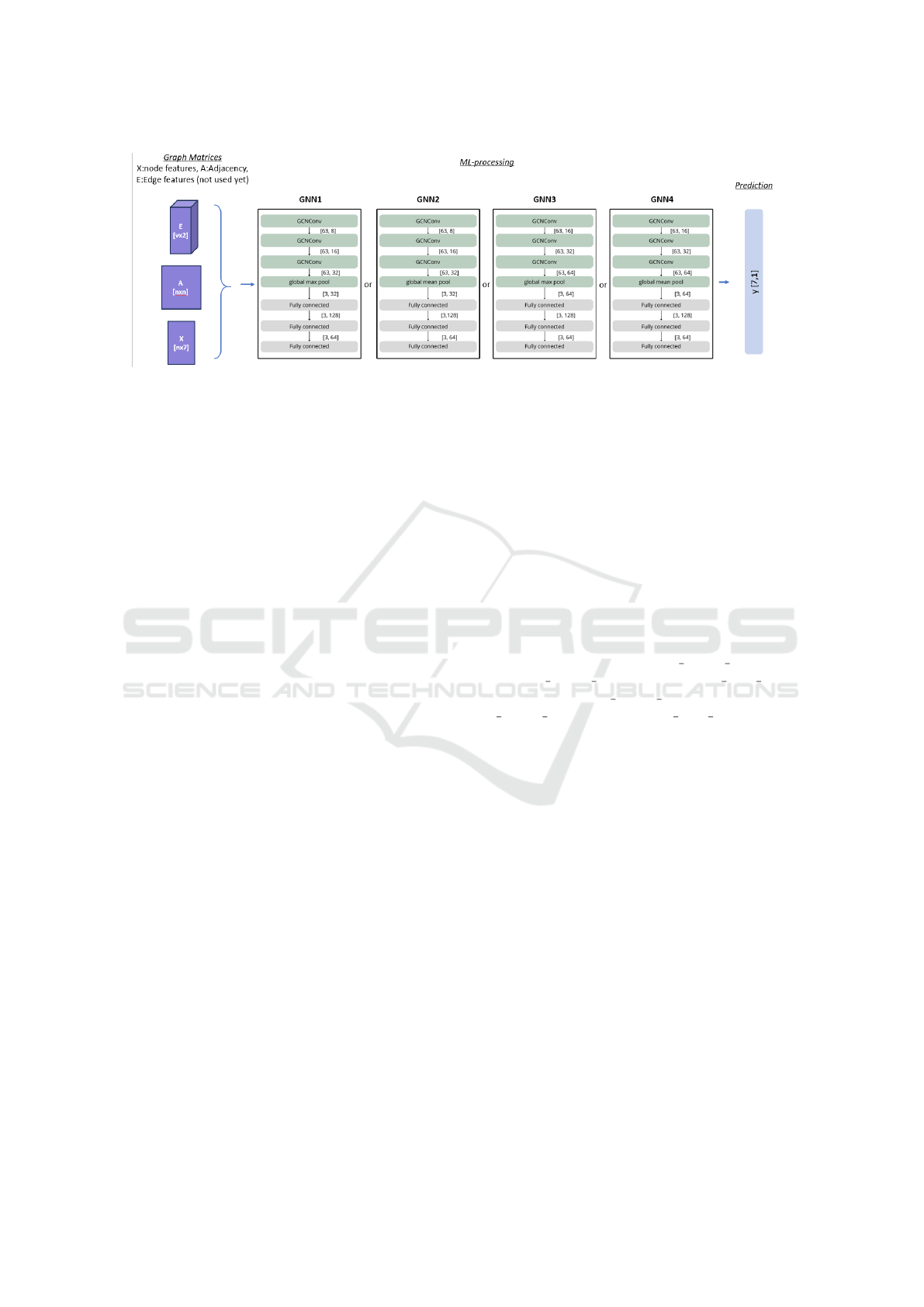
Figure 2: different GNN architectures according to the number of convolutions and the type of readout pooling have been
evaluated for the different types of generated graphs presented in Figure 1.
layer to a 64-dimensional space and the third FC layer
reduces this to a 7-dimensional space, to match with
the (K)=7 age classes of the sheep brain.
In total, we have tested 4 different architectures
by combining the 2 sets of convolution parameters
and the 2 global poolings (GNN1, GNN2, GNN3 and
GNN4, (Figure 2)).
4 EXPERIMENTS AND RESULTS
We conducted experiments with different CNN and
GNN architectures to analyse which ones are the best
suited to predict the age of brains of non-conventional
animal models such as sheep. The experiments were
conducted on a machine equipped with an Intel Core
i7-11850H CPU running at 2.50 GHz, 32 GB of
RAM, and an NVIDIA GeForce RTX A3000 Laptop
GPU. All computations were performed using Python
3.11.8 with Pytorch 2.2.2+cu121 frameworks. This
section describes the used datasets and protocols.
4.1 Datasets, Protocol and Metric
Description
4.1.1 Datasets
The dataset used is composed of 195 3D-T1w-MRI
from different projects which were all conduct in
”Ile de France” growing and adult sheep from the
“Unit
´
e Exp
´
erimentale de Physiologie Animale de
l’Orfrasi
`
ere” (UEPAO, INRAE Val de Loire, France;
https://doi.org/10.15454/1.5573896321728955E12).
MRI were acquired in vivo with a 3 Teslas Siemens
Magnetom Verio® scanner (Erlangen, Germany)
located at the imaging platform PIXANIM. A 4-
channel Siemens FLEX coil was used. T1-weighted
images were acquired with the 3D-MPRAGE se-
quence with 2 or 4 numbers of excitations according
to the project. The plane resolution of the T1-w
MRI was between 0.4x0.4mm and 0.5x0.5mm. We
created 5 segmentations for each MRI in total : 3
from our histogram-based algorithm and 2 from
our split and merge algorithm. The parameter of
histogram-based algorithm is set to N=6, N=20 and
N=30. N=6 gives a segmentation similar to a tissue
segmentation (White matter, gray matter and CSF).
The other parameters produced visually interesting
results, maximizing the expressiveness of the image
content while ensuring that the segmentation remains
clear and not overly noisy. After testing different set
of parameters we chose 2 sets of parameters for the
split and merge : {homogeneity during split=10, ho-
mogeneity during merge=40, minimal cube size=1}
and {homogeneity during split=20, homogene-
ity during merge=60, minimal cube size=1}. Each
segmentation are then transformed into an adjacency
graph (distance threshold d=0) with every attribute
described previously.
4.1.2 Protocol and Metrics
During our experiments, we try to learn and predict
the age of the subjects. Age prediction can be a hard
task regarding our data. That’s why we decided to
learn a classification task instead of a regression task,
the latter considered as more difficult to perform. We
decided to distribute the images into K=7 classes of
ages (Table 1) in order to obtain balanced with enough
representative images into each class. The ranges as-
sociated with each class have been defined based on
available data, corresponding to 10 to 20 days (with
no data available for the range between 70 to 120
days).
The proposed pipelines based on GNN architec-
tures were compared with a classical CNN architec-
ture working with low level image features (Figure
3). In the following we compare our pipeline with the
CNN architecture that presented the best results after
Comparison Between CNN and GNN Pipelines for Analysing the Brain in Development
479

Table 1: Organization of the 7 classes according to the range of age (days) and number of subjects per class.
Class 1 2 3 4 5 6 7
Age (days) 0-10 11-20 21-30 31-40 41-50 51-70 123-139
Numbers 33 16 22 24 61 24 15
Figure 3: Optimized CCN pipeline. The architecture con-
tains 2 convolution layers (Conv3d), a flattening layer and
3 linear layers.
learning with different layer numbers and sizes. The
CNN consists of a 3D convolution layer with a kernel
size of 3x3x3, followed by a 3D max-pooling layer of
size 2x2. This sequence of convolution and pooling
layers is repeated once. The output is then flattened
into a vector, which is passed through three fully con-
nected layers that progressively reduce its size to 128,
64, and finally 7 dimensions.
The 195 images (see dataset section) were used to
create the training (50% of the MR-images), the vali-
dation (25% of the MR-images) and the test datasets
(25% of the MR-images). We carefully design the
test dataset to be representative of all the data by bal-
ancing the classes and projects to remove any project
bias. We performed a shuffle split validation which is
well suited for small datasets. The training lasts 300
epochs. The loss function is the cross-entropy loss
with a learning rate of 0.001.
4.2 Results
In terms of accuracy, the GNN pipeline remains bet-
ter than a classical CNN pipeline with an average ac-
curacy of 63.22% compared to 59.77% for the CNN
(table 2). These results show that our pipeline could
be convenient for a classical task like age prediction.
The split and merge pipelines seem to perform bet-
ter than the histogram-based pipelines achieving an
average accuracy of 51.43% compared to 37.73%.
This could be explained by the fact that the number
of nodes is not fixed and could be discriminative be-
tween the classes. For example, we found that class 6
has an average of 1,110 nodes, compared to 2,205 for
class 7, making it very easy to discriminate between
them.
Concerning the GNN architecture, the GNN with
global better results was the GNN4 with a conv1=16,
conv2=32, conv3=64 and a global mean pooling with
an average accuracy of 45.08%. The other architec-
tures have the following average accuracies : GNN1
= 43.49%, GNN2 = 40.48% and GNN3 = 43.81%.
These results suggest that the global max pooling is a
little bit better in average. Other pooling methods and
architectures need to be experimented.
We calculated the confusion matrix of each training
and the table 3 is one of them. The mean accuracy
of the 3 trainings is 63.22% and this matrix is one of
the 3 trainings and has a score of 63.79% of accuracy.
The rows denote the true classes and the columns are
the predicted classes. The results described in this
confusion matrix are quite representative of this GNN
model and graph creation method (which is the best
configuration). We can see that 86.2% of the predic-
tions are either correct or off by only one age class.
5 CONCLUSION AND
PERSPECTIVES
In this study, we propose a novel pipeline to
predict the brain age of nonconventional animal
models without relying on neuro-anatomical pri-
ors to not bias the analysis. We provide an
open access generic graph generation tool from
3D images available at this URL: https://scm.univ-
tours.fr/projetspublics/lifat/3dbrainminer. Our pro-
posed GNN pipeline provides better results in terms
of accuracy than a traditional CNN pipeline. The
process starts with automatic MRI segmentation, fol-
lowed by graph transformation and analysed using
a GNN model. We compared 2 segmentation algo-
rithms with different parameters and GNN architec-
tures.
The uniqueness of our pipeline lies in its ability to
work without anatomical priors, enabling an unbiased
analysis of morphofunctional features. It would be
very interesting to try other parameters and especially
for the split and merge algorithm, which showed
promising results. Adjusting the homogeneity crite-
rion may reveal different brain information. Improv-
ing graph transformation, particularly edge creation,
could enhance learning. Using a higher distance
threshold or other strategies might improve message
passing in the GNN. One approach is to create all pos-
sible edges and let the GNN decide which are impor-
tant during training. Edge features, not used in this
study, could also provide valuable insights, such as
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
480
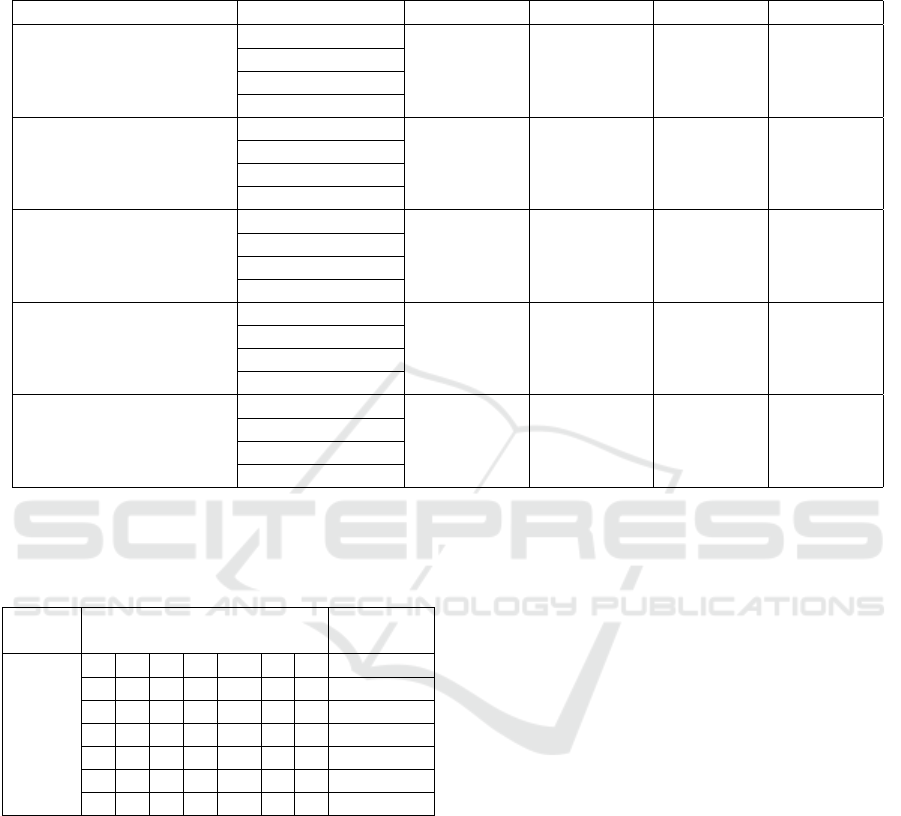
Table 2: Classification performances of the proposed pipelines using different segmentation methods and GNN architectures.
Results are averages (with standard deviation) of performance measured over 3 shuffle splits validation. Layers and sizes of
each GNN architecture : {GNN1 : conv1 = 8, conv2 = 16, conv3 = 32, maxpool; GNN2 : conv1 = 8, conv2 = 16, conv3 = 32,
meanpool; GNN4 : conv1 = 16, conv2 = 32, conv3 = 64, maxpool; GNN4 : conv1 = 16, conv2 = 32, conv3 = 64, meanpool;}.
GNN architecture
Accuracy Recall Precision F1
GNN1 36.78 ± 4.34 0.34 ± 0.03 0.32 ± 0.03 0.33 ± 0.03
GNN2 35.63 ± 8.68 0.35 ± 0.05 0.32 ± 0.06 0.33 ± 0.05
GNN3 43.10 ± 8.62 0.37 ± 0.06 0.35 ± 0.02 0.37 ± 0.04
Histogram-based
n = 6
GNN4 41.95 ± 1.99 0.37 ± 0.03 0.38 ± 0.02 0.37 ± 0.01
GNN1 31.61 ± 4.34 0.33 ± 0.06 0.31 ± 0.03 0.32 ± 0.04
GNN2 33.33 ± 8.84 9.78 ± 16.32 0.30 ± 0.07 0.32 ± 0.08
GNN3 30.46 ± 2.63 0.30 ± 0.01 0.29 ± 0.03 0.29 ± 0.01
Histogram-based
n = 20
GNN4 37.92 ± 3.47 0.39 ± 0.04 0.35 ± 0.06 0.35 ± 0.04
GNN1 40.81 ± 7.78 0.42 ± 0.07 0.41 ± 0.08 0.42 ± 0.08
GNN2 37.36 ± 6.97 0.38 ± 0.07 0.35 ± 0.03 0.37 ± 0.03
GNN3 43.10 ± 6.90 0.47 ± 0.03 0.43 ± 0.04 0.42 ± 0.05
Histogram-based
n = 30
GNN4 40.81 ± 6.53 0.38 ± 0.08 0.39 ± 0.07 0.39 ± 0.08
GNN1 59.20 ± 8.15 0.60 ± 0.08 0.59 ± 0.10 0.59 ± 0.09
GNN2 48.28 ± 2.98 0.53 ± 0.05 0.53 ± 0.04 0.53 ± 0.04
GNN3 54.60 ± 7.18 0.57 ± 0.08 0.60 ± 0.11 0.58 ± 0.10
Split and merge
Split homogeneity = 10
Merge homogeneity = 40
GNN4 63.22 ± 5.27 0.62 ± 0.03 0.65 ± 0.07 0.62 ± 0.05
GNN1 49.05 ± 5.00 0.50 ± 0.04 0.51 ± 0.05 0.51 ± 0.04
GNN2 47.80 ± 4.36 0.49 ± 0.09 0.53 ± 0.11 0.51 ± 0.10
GNN3 47.80 ± 2.88 0.54 ± 0.06 0.53 ± 0.06 0.54 ± 0.06
Split and merge
Split homogeneity = 20
Merge homogeneity = 60
GNN4 41.51 ± 5.33 0.43 ± 0.08 0.48 ± 0.04 0.42 ± 0.07
Table 3: Confusion matrix of the first “shuffle split” training
of the best configuration which has a score of 63.79% of
accuracy.
Predicted classes
Prediction
rate
True
classes
6 1 2 1 0 0 0 6/10
1 3 0 0 0 0 0 3/4
0 0 3 2 1 0 0 3/6
0 0 1 4 1 0 0 4/6
0 0 2 2 12 3 0 12/19
0 0 1 1 2 4 0 4/8
0 0 0 0 0 0 5 5/5
distances and surface contact areas.
Future research will focus on exploring various con-
figurations and architectures to optimize our ap-
proach. More complex models, like NNConv or graph
attention layers, could be incorporated to better utilize
edge features. This approach enhances interpretabil-
ity, often lacking in CNN methods, and offers flexibil-
ity in selecting scales or conducting multi-scale anal-
yses for deeper insights into data structures. GNNs
capture complex relationships, leading to more robust
models, making them superior for tasks requiring an
understanding of element relationships. Thus, we be-
lieve automatic segmentation for graph building and
GNN analysis is a promising solution.
ACKNOWLEDGEMENTS
We thank Scott Love for scientific and enthusiastic
discussions on brain MRI segmentations. We ac-
knowledge the financial support the Institut National
de Recherche pour l’Agriculture, l’Alimentation et
l’Environnement D
´
epartement Physiologie Animale
(INRAE-PHASE) et Syst
`
emes d’
´
Elevage and the
R
´
egion Centre Val de Loire for the PhD grant for
Antoine Bourlier. The MR-images were acquired
in projects supported by INRAE - PHASE (Neu-
roimagery, PhenoMatHyp, Prebiostress) and by Val
de Loire regional council (Ovin2A, Convention 2013
00083140, coordinator R. Nowak, France; Neuro2Co,
Convention 2017 00117257, coordinator E. Chaillou,
France). This research was funded, in whole or in
part, by the French National Research Agency (ANR)
under the project ”ANR-23-SSAI-0008-01.” In line
with the objective of open-access publication, the au-
thor holder applies an open-access CC-BY license to
any accepted article (AAM) resulting from this sub-
mission.
Comparison Between CNN and GNN Pipelines for Analysing the Brain in Development
481

REFERENCES
Bullmore, E. T. and Bassett, D. S. (2011). Brain graphs:
Graphical models of the human brain connectome.
7:113–140.
Cai, H., Gao, Y., and Liu, M. (2023). Graph Trans-
former Geometric Learning of Brain Networks Us-
ing Multimodal MR Images for Brain Age Estimation.
42(2):456–466.
Cheng, J., Liu, Z., Guan, H., Wu, Z., Zhu, H., Jiang, J.,
Wen, W., Tao, D., and Liu, T. (2021). Brain Age
Estimation From MRI Using Cascade Networks With
Ranking Loss. 40(12):3400–3412.
Cole, J. H., Poudel, R. P. K., Tsagkrasoulis, D., Caan, M.
W. A., Steves, C., Spector, T. D., and Montana, G.
(2017). Predicting brain age with deep learning from
raw imaging data results in a reliable and heritable
biomarker. 163:115–124.
Coupeau, P., Fasquel, J. B., Mazerand, E., Menei, P.,
Montero-Menei, C. N., and Dinomais, M. (2022).
Patch-based 3D U-Net and transfer learning for
longitudinal piglet brain segmentation on MRI.
214:106563.
Cui, H., Dai, W., Zhu, Y., Li, X., He, L., and Yang, C.
(2021). BrainNNExplainer: An Interpretable Graph
Neural Network Framework for Brain Network based
Disease Analysis.
De Vico Fallani, F., Latora, V., and Chavez, M. (2017).
A Topological Criterion for Filtering Information in
Complex Brain Networks. 13(1):e1005305.
Ella, A., Delgadillo, J. A., Chemineau, P., and Keller, M.
(2017). Computation of a high-resolution MRI 3D
stereotaxic atlas of the sheep brain. 525(3):676–692.
Fedorov, A., Beichel, R., Kalpathy-Cramer, J., Finet, J.,
Fillion-Robin, J.-C., Pujol, S., Bauer, C., Jennings,
D., Fennessy, F., Sonka, M., Buatti, J., Aylward, S.,
Miller, J. V., Pieper, S., and Kikinis, R. (2012). 3D
Slicer as an Image Computing Platform for the Quan-
titative Imaging Network. 30(9):1323–1341.
Fil, J. E., Joung, S., Zimmerman, B. J., Sutton, B. P., and
Dilger, R. N. (2021). High-resolution magnetic res-
onance imaging-based atlases for the young and ado-
lescent domesticated pig (Sus scrofa). 354:109107.
Galisot, G., Ramel, J.-Y., Brouard, T., Chaillou, E., and Ser-
res, B. (2022). Visual and structural feature combi-
nation in an interactive machine learning system for
medical image segmentation. 8:100294.
Gonzalez, R. C. and Woods, R. E. (2017). Digital Image
Processing. Pearson, fourth edition, global edition
edition.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep Resid-
ual Learning for Image Recognition. In 2016 IEEE
Conference on Computer Vision and Pattern Recogni-
tion (CVPR), pages 770–778.
Jiang, H., Lu, N., Chen, K., Yao, L., Li, K., Zhang, J.,
and Guo, X. (2020). Predicting Brain Age of Healthy
Adults Based on Structural MRI Parcellation Using
Convolutional Neural Networks. 10.
Kaur, P. and Gaba, G. S. (2021). Computational Neuro-
science Models and Tools: A Review. In Bhoi, A. K.,
Mallick, P. K., Liu, C.-M., and Balas, V. E., editors,
Bio-Inspired Neurocomputing, Studies in Computa-
tional Intelligence, pages 403–417. Springer.
Krizhevsky, A., Sutskever, I., and Hinton, G. E. (2012). Im-
ageNet Classification with Deep Convolutional Neu-
ral Networks. In Advances in Neural Information Pro-
cessing Systems, volume 25. Curran Associates, Inc.
Lam, P., Zhu, A. H., Gari, I. B., Jahanshad, N., and Thomp-
son, P. M. (2020). 3D Grid-Attention Networks for
Interpretable Age and Alzheimer’s Disease Prediction
from Structural MRI.
Li, G., Wang, L., Yap, P.-T., Wang, F., Wu, Z., Meng, Y.,
Dong, P., Kim, J., Shi, F., Rekik, I., Lin, W., and
Shen, D. (2019). Computational neuroanatomy of
baby brains: A review. 185:906–925.
Li, X., Zhou, Y., Dvornek, N., Zhang, M., Gao, S., Zhuang,
J., Scheinost, D., Staib, L. H., Ventola, P., and Duncan,
J. S. (2021). BrainGNN: Interpretable Brain Graph
Neural Network for fMRI Analysis. 74:102233.
Lim, H., Joo, Y., Ha, E., Song, Y., Yoon, S., and Shin, T.
(2024). Brain Age Prediction Using Multi-Hop Graph
Attention Combined with Convolutional Neural Net-
work. 11(3):265.
Nitzsche, B., Frey, S., Collins, L. D., Seeger, J., Lobsien,
D., Dreyer, A., Kirsten, H., Stoffel, M. H., Fonov,
V. S., and Boltze, J. (2015). A stereotaxic, population-
averaged T1w ovine brain atlas including cerebral
morphology and tissue volumes. 9.
Park, H.-J. and Friston, K. (2013). Structural and Func-
tional Brain Networks: From Connections to Cogni-
tion. 342(6158):1238411.
Poriya, V. (2023). Brain Tumor Classification And Segmen-
tation Using Machine Learning For Magnetic Reso-
nance Images.
Ravinder, M., Saluja, G., Allabun, S., Alqahtani, M. S., Ab-
bas, M., Othman, M., and Soufiene, B. O. (2023). En-
hanced brain tumor classification using graph convo-
lutional neural network architecture. 13(1):14938.
Schmid, S. (2023). Image Intensity Normalization in Med-
ical Imaging.
Sporns, O. (2007). Brain connectivity. 2(10):4695.
Sporns, O. (2018). Graph theory methods: Applications in
brain networks. 20(2):111–121.
Srinivasan, S., Francis, D., Mathivanan, S. K., Rajadurai,
H., Shivahare, B. D., and Shah, M. A. (2024). A hy-
brid deep CNN model for brain tumor image multi-
classification. 24(1):21.
Van Essen, D. C. and Drury, H. A. (1997). Structural and
Functional Analyses of Human Cerebral Cortex Using
a Surface-Based Atlas. 17(18):7079–7102.
Yang, G., Zhou, S., Bozek, J., Dong, H.-M., Han, M., Zuo,
X.-N., Liu, H., and Gao, J.-H. (2020). Sample sizes
and population differences in brain template construc-
tion. 206:116318.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
482
