
Unveiling Breast Cancer Causes Through Knowledge Graph Analysis
and BioBERT-Based Factuality Prediction
Hasna El Haji
1 a
, Nada Sbihi
1
, Kaoutar El Handri
2,3 b
, Adil Bahaj
1
, Mohammed Elkasri
1
,
Amine Souadka
4 c
and Mounir Ghogho
1 d
1
TiCLab, International University of Rabat, Rabat, Morrocco
2
MedBiotech Laboratory, Faculty of Medicine and Pharmacy (FMPR), University Mohammed V, Rabat, Morocco
3
Aivancity School of AI & Data for Business & Society, Cachan, France
4
Surgical Oncology Department, National Institute of Oncology, University Mohammed V, Rabat, Morocco
Keywords:
Knowledge Graph, Breast Cancer, Bioinformatics, BioBERT, Factuality Prediction.
Abstract:
Worldwide, millions of women are affected by breast cancer, with the impact significantly worsened in under-
served regions. The profound effect of breast cancer on women’s health has driven research into its causes,
with the aim of developing methods for the prevention, diagnosis, and treatment of the disease. The significant
influx of research on this subject is overwhelming and makes manual exploration arduous, which motivates
automated knowledge exploration approaches. Knowledge Graphs (KGs) are one of these approaches that
attracted significant attention in the last few years for their ability to structure and present knowledge, making
it easier to explore and analyze. Current KGs that include causes of breast cancer are deficient in contextual
information, highlighting the uncertainty of these causes (facts). In this work, we present a method for extract-
ing a sub-graph of breast cancer causes and fine-tuning BioBERT to evaluate the uncertainty of these causes.
Our automated approach, which simulates human annotation, computes uncertainty scores based on textual
factuality and assesses cause reliability using a Closeness Score. We also create a web-based application for
easy exploration
a
.
a
https://bckg.datanets.org/
1 INTRODUCTION
Breast cancer is a complex and multifaceted disease,
affecting approximately 2.3 million women world-
wide each year, according to the “WHO” (World
Health Organization, 2024). With about one in eight
women diagnosed during their lifetime (American
Cancer Society, 2024), the disease significantly im-
pacts quality of life, particularly in disadvantaged
countries. This has led to extensive research in var-
ious disciplines to explore prevention (El Haji et al.,
2024), diagnosis, and prognosis (El Haji et al., 2023).
One crucial research focus is identifying the lead-
ing causes of breast cancer, as addressing the root
cause can increase the success of treatments. How-
a
https://orcid.org/0009-0000-0437-2731
b
https://orcid.org/0000-0001-6732-2627
c
https://orcid.org/0000-0002-1091-9446
d
https://orcid.org/0000-0002-0055-7867
ever, abundant research papers on this topic can make
manually exploring findings challenging and require
multiple specialists’ involvement. Automated knowl-
edge extraction methods have been studied to answer
this need.
There has been significant progress in automated
knowledge extraction from scientific literature, par-
ticularly in the biomedical field (Hogan et al., 2021),
(Zheng et al., 2021), (Wang et al., 2023). The aim is
to organize and structure knowledge to facilitate ex-
ploration and discovery (Bahaj et al., 2022). In KGs,
facts are organized into triplets, where each triplet
comprises a head entity and a tail entity linked by a
semantic relation. Mathematically, a KG G can be
presented as a triplet G = (V , R , E), where V is the
set of entities, R is the set of relations and E is the set
of entities such that E = {(h, r,t)|h,t ∈ V , r ∈ R }.
KGs can be utilized for knowledge exploration us-
ing visualization tools, improving search engine re-
sults, question-answering systems, recommendation
El Haji, H., Sbihi, N., El Handri, K., Bahaj, A., Elkasri, M., Souadka, A. and Ghogho, M.
Unveiling Breast Cancer Causes Through Knowledge Graph Analysis and BioBERT-Based Factuality Prediction.
DOI: 10.5220/0013179700003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 2: HEALTHINF, pages 141-148
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
141

systems (El Handri and Idrissi, 2020), and tasks
such as node classification and link prediction (Huang
et al., 2019), (Guo et al., 2020), (Rossi et al., 2021).
In biomedicine, research in biomedical KGs has seen
a surge in recent years. This is due to the influx of new
biomedical publications in multiple disciplines on
various medical concepts. Existing biomedical KGs
in literature are either domain-specific or general.
COVID19KG (Kejriwal, 2020) is a domain-specific
KG with knowledge about COVID19. SemMedDB
(Kilicoglu et al., 2012) is a general-purpose biomedi-
cal KG that contains a wide range of biomedical con-
cepts. However, KGs assume the accuracy and relia-
bility of their facts. Nonetheless, uncertainty can im-
pact them (Sosa and Altman, 2022). This uncertainty
arises from various sources of noise in the source data
or in the extraction process. Uncertain knowledge
graphs (UKGs) have been developed, whereby a con-
fidence score is assigned to each fact, quantifying its
validity (Chen et al., 2019). Formally, these UKGs
assign a confidence score s
l
to each triple l = (h, r,t).
This confidence score reflects the level of veracity of
that fact in the KG.
In this direction, language modeling has gained
considerable attention recently. The creation of the
transformer-based architecture and self-supervised
pretraining made considerable strides to have more
reliable models. BERT (Devlin et al., 2018) is one
such model. BERT uses a transformer-based architec-
ture relying on a masked language modeling training
process. BERT initiated a research direction, which
extended from general language modeling to more
domain-specific language models in law (Sun, 2023),
biomedicine (Chen et al., 2023), marketing (Li et al.,
2022) and other domains. These variants are gener-
ally fine-tuned versions of BERT, where a domain-
specific corpus is curated and labeled, then used to
retrain BERT. Language model fine-tuning is essen-
tial in modeling small datasets (Zhang et al., 2020)
and low-resource languages (Hangya et al., 2022).
One specific application of these language models
is factuality prediction, which predicts the veracity
of an event expressed in a sentence (Veyseh et al.,
2019). Factuality prediction is generally formulated
as a classification problem, where the model is tasked
to predict the factuality class of a sentence (Kilicoglu
et al., 2017). Other approaches, such as formulat-
ing the problem as a regression problem, where a
factuality value that ranges between -3 and 3 is pro-
vided for each sentence. Negative and positive val-
ues signify negative and positive polarity, respectively
(Stanovsky et al., 2017). The main difficulty in fac-
tuality prediction stems from the nature of factuality
itself. Practically, factuality is expressed using certain
cue words, and corresponding to their dependence on
different sentence elements, the sentence factuality
gets affected (Jiang and de Marneffe, 2021). This syn-
tactic dependency referred to as text paradigmatics is
generally challenging to model since most models fo-
cus on modeling syntax with no regard to long-range
dependencies.
To address this gap, advancements in Natural
Language Processing (NLP) have been significantly
driven by novel pre-training techniques, such as those
presented in XLNet, ERNIE, and ELECTRA. XLNet
(Yang et al., 2019), with its generalized autoregres-
sive pretraining, enhances language understanding by
capturing bidirectional context through permutation-
based training, which surpasses traditional models in
handling complex language tasks. ERNIE (Zhang
et al., 2019) further builds on this foundation by incor-
porating informative entities, offering a more context-
aware representation that is particularly effective for
tasks involving entity-level understanding. Mean-
while, ELECTRA (Clark et al., 2020) introduces a
discriminator-based pre-training approach, improving
the efficiency and effectiveness of language models
by focusing on distinguishing real from generated
tokens. In this evolving landscape, our study con-
tributes by fine-tuning BioBERT (Lee et al., 2020)
specifically for factuality prediction of triplets, lever-
aging its domain-specific pre-training for biomedical
text.
Our work builds on these foundational approaches
by applying BioBERT for factuality prediction. Adi-
tionally, our work proposes a metric to evaluate the
reliability of triplets within our knowledge sub-graph.
This enables visibility and clarity for physicians and
scientists working on breast cancer. Furthermore, we
design a web application that serves as an interactive
exploration platform displaying the entire sub-graph.
Our approach uniquely advances existing method-
ologies by addressing key limitations in handling un-
certainty and factuality prediction within biomedi-
cal knowledge graphs. By integrating BioBERT—a
domain-specific language model—with the construc-
tion of a breast cancer causes knowledge sub-graph,
we introduce a novel framework that not only predicts
factuality but also quantifies the reliability of triplets
through a Closeness Score. This dual-layered ap-
proach significantly enhances the interpretability and
trustworthiness of the extracted causal relationships.
Unlike existing models that assume the veracity of
their facts, our methodology explicitly evaluates and
categorizes factuality, making it particularly impact-
ful for applications in biomedical research where un-
certainty can hinder effective decision-making.
The paper is structured as follows: section 2 de-
HEALTHINF 2025 - 18th International Conference on Health Informatics
142

tails our methodology for constructing the knowledge
sub-graph and evaluating the factuality of triplets.
Section 3 presents our experiments and results, while
sections 4 and 5 provide discussion and concluding
remarks on the added value of our approach.
2 METHODOLOGY
The framework of our method consists of sub-
graph construction (extraction of triplets), fine-tuning
BioBERT (Lee et al., 2020) for factuality prediction
and triplet reliability evaluation based on Closeness
Score. We detail each part in the subsections 2.1, 2.2
and 2.3, respectively. The overall workflow is shown
in Figure 1.
2.1 Knowledge Sub-Graph
Construction
For constructing our knowledge sub-graph of breast
cancer causes, we utilize a generic biomedical litera-
ture database known as SemMedDB (Kilicoglu et al.,
2012). SemMedDB is a valuable resource for inte-
grating and analyzing information from biomedical
literature. It captures relationships between concepts
such as genes, diseases, drugs, and other biomedical
concepts using multiple semantic relations that ex-
press general association, cause, influence and other
relations. SemMedDB provides a rich network of in-
terconnected biomedical knowledge and it includes
contextual details like negation and modality and al-
lows tracing back to PubMed articles via unique iden-
tifiers, enabling researchers to explore relationships
and generate hypotheses grounded in biomedical lit-
erature.
Even though an API is available, we download all
the resources to enable offline work and ensure effi-
cient local processing. We extract triplets related to
breast cancer causes for sub-graph construction and
source sentences for factuality prediction. Each triplet
represents a relationship between a cause and breast
cancer. We also retrieve the paper’s PubMed Identi-
fier (PMID) and its citation count to further evaluate
the relevance of the extracted triplets.
We are interested in the causes of breast cancer,
thus we filter the extracted triplets related to breast
cancer by retaining only those with the relationship
pattern:
“Subject” < causes > “Breast cancer”.
Each “Subject” is identified by a Concept Unique
Identifier (CUI), a unique code used to retrieve
information about entities across various medical
databases. In our context, the object “breast can-
cer” refers specifically to female breast cancer, which
can appear under different synonyms in the biomedi-
cal literature. We utilize UMLS to extract these syn-
onyms (Rossi et al., 2021).
2.2 Fine-Tuning BioBERT for
Factuality Prediction
Although the automatic construction of KGs offers
a more practical alternative to manual annotation, it
suffers multiple pitfalls. For example, according to
SemMedDB, secondary Malignant neoplasm of the
skin causes breast cancer. But, upon closer exami-
nation of the sentence: “ We present the rare clinical
entity of a breast cancer which was first diagnosed due
to the skin metastasis away from the breast tumor”. It
is clear that the sentence does not allow us to confirm
this relationship with certainty. Therefore, retrieving
the sentence corresponding to each triplet is essential
as it enriches our knowledge sub-graph.
Given the large number of triplets, it is difficult
to manually assign a factuality score to each one. As
a result, we use a language model to predict the fac-
tuality score of a triplet using the source sentence as
input. To construct the factuality model, we use a fac-
tuality dataset (Kilicoglu et al., 2017). This dataset is
built using 500 PubMed abstracts. Sentences in these
abstracts were annotated to extract seven factuality
values: fact, probable, possible, doubtful, counter-
act, uncommitted, conditional. Although the dataset
contains 500 abstracts, the number of triples (sub-
ject, relation, object) extracted from them amounts
to 3,149 (Kilicoglu et al., 2017). The dataset is un-
balanced, with 87% of the sentences classified as
facts, 4.5% probable, 3.8% uncommitted, 2% pos-
sible, 1.8% counterfact, 0.25% Doubtful, and just 1
case conditional. We removed the last two classes due
to their minimal representation.
Sentences can act as explanatory variables in a
predictive model, with factuality as the target vari-
able. In this context, the objective is to leverage
the information within the sentences to predict the
factuality or outcome of interest. We use BioBERT
(Lee et al., 2020) as the basis for constructing our
predictive model since it is a common and effec-
tive approach, especially for tasks involving natural
language processing and understanding. We fine-
tune the pre-trained BioBERT model to adapt it to
our specific NLP task: factuality classification. We
use the “WeightedRandomSampler” sampling strat-
egy, assigning higher probabilities to samples from
underrepresented classes. This approach helps pre-
vent the model from being biased towards the major-
Unveiling Breast Cancer Causes Through Knowledge Graph Analysis and BioBERT-Based Factuality Prediction
143
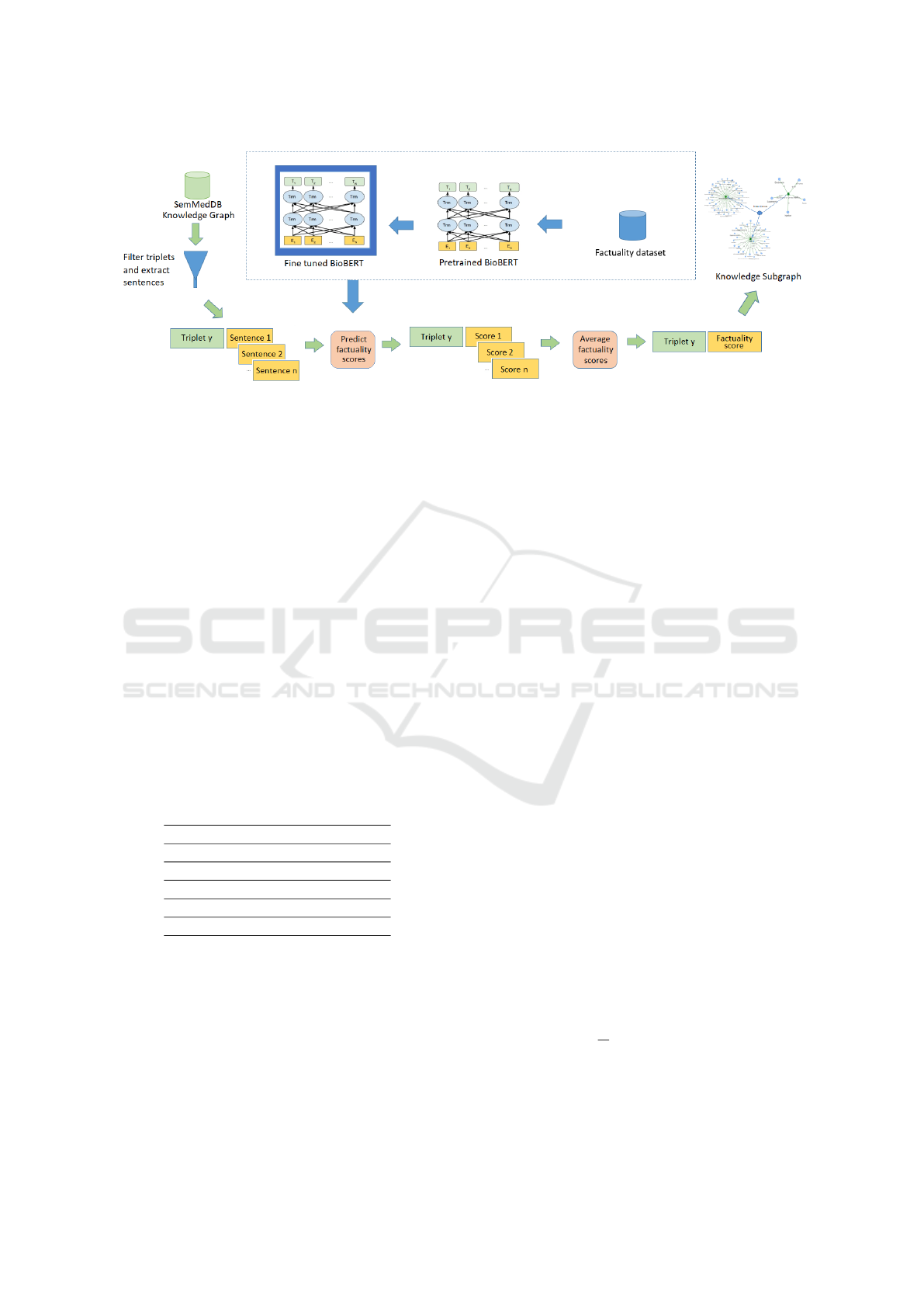
Figure 1: Sub-graph construction and factuality prediction: Triplets are extracted from the SemMedDB knowledge graph,
filtered to include only cases where the “Subject” causes “breast cancer,” followed by the extraction of sentences related to
each triplet. For each triplet y, fine-tuned BioBERT is used to predict the factuality score for each sentence from which the
triplet was derived. The factuality scores are averaged across sentences corresponding to the same triplet, resulting in a unique
score assigned to each triplet (indicating the degree of factuality of a subject that causes breast cancer).
ity class and improve its ability to learn from the mi-
nority class samples. After testing different parame-
ter values, the following combination gives us the best
validation accuracy: a learning rate of 1e-5, bach size
of 8, and number of epochs of 10.
As proposed by (Kilicoglu et al., 2017), we con-
vert factuality classes to numeric scores as illustrated
in Table 1. Finally, to calculate the factuality of a
breast cancer cause, we compute the average of all
factuality scores predicted from sentences involving
that cause. This approach may be biased, as some
breast cancer causes are cited numerous times while
others are cited only once. Consequently, we propose
further evaluation in the next section to assess cause
reliability.
Table 1: Conversion of factuality classes to numeric scores
(Kilicoglu et al., 2017).
Class Factuality score
Fact 1
Probable 0.75
Possible 0.5
Uncommitted 0.25
Counterfact 0
2.3 Triplet Reliability Evaluation Based
on Closeness Score
The assessment of triplet reliability in knowledge
graphs is a multifaceted challenge that requires evalu-
ating both the impact and contextual relevance of ex-
tracted relations. Previous studies have highlighted
the role of citation counts as a proxy for significance
in research evaluation, where more frequently cited
works are often considered more impactful (Waltman,
2016). On the other hand, contextualized language
models, such as BERT and its biomedical variants
like BioBERT, have been increasingly used to as-
sess the relevance of extracted triplets by capturing
semantic similarity within specific contexts (Soares
et al., 2019). Despite these advances, the integration
of citation-based metrics with contextual relevance
scores for triplet reliability assessment has not been
fully explored.
We evaluate triplet reliability based on Closeness
Score, which quantifies the degree of proximity of the
triplet to the core objective of the publication. By
assessing the closeness between the triplet’s mention
and the publication title, we can determine how rel-
evant the triplet is to the study’s main focus. This
ensures that the triplet is not merely referenced as the
result of other research but is central to the study it-
self.
For each triplet T
k
, we identify the papers i that
have cited it and retrieve the titles Title
i
of these pa-
pers using the PubMed API. Then, using BioBERT
embeddings, we calculate the similarity score be-
tween each title and the sentence S(T
k
) from which
the triplet was extracted in that paper. The overall
triplet closeness score is computed as the mean of the
similarity scores across all citing papers, where N is
the number of citing papers (Equation 1).
Closeness(T
k
) =
1
N
N
∑
i=1
Similarity(Title
i
, S(T
k
)) (1)
For an effective visual representation of our find-
ings, we design a web application using the Python
framework Dash.
HEALTHINF 2025 - 18th International Conference on Health Informatics
144

3 EXPERIMENTS AND RESULTS
As depicted in the framework in Figure 1, we rep-
resent all the potential causes of breast cancer as a
knowledge sub-graph and we assign a degree of fac-
tuality to each cause linked to breast cancer. After fil-
tering the triplets, there are 1,039 subjects identified
as causing breast cancer. In terms of their types, the
most frequently detected are hazardous or poisonous
substances, genes or genomes, amino acids, peptides,
and proteins. Figure 2 illustrates the most commonly
identified types of causes in the literature; however,
this does not necessarily imply that these causes are
the most influential. Conversely, the figure highlights
also types that are less studied and need further re-
search and exploration by the scientific community.
Figure 2: Top 20 breast cancer cause types.
As for factuality prediction, Table 2 summarizes
the performance metrics of the fine-tuned BioBERT
model for factuality prediction across various classes.
The model demonstrates strong overall accuracy
(0.927). It also showed very good performance in the
“Fact” class, with a precision of 0.970, recall of 0.918,
and an F1-score of 0.944, reflecting its high accuracy
in identifying factual statements.
For the “Probable” class, the model shows a pre-
cision of 0.526 and a recall of 0.833, resulting in an
F1-score of 0.645. This indicates that while the model
is fairly effective at recognizing probable statements,
there is room for improvement in its precision.
The “Possible” class has a precision of 0.600
and recall of 0.500, leading to an F1-score of 0.545.
Despite achieving an overall accuracy of 0.927, the
model’s performance in predicting possible state-
ments is less robust compared to the ”Fact” class, sug-
gesting that further refinement is needed.
The “Uncommitted” class presents a precision of
0.412 and recall of 0.778, with an F1-score of 0.538.
This reflects moderate performance, indicating that
while the model can identify uncommitted statements
to some extent, its precision remains relatively low.
Finally, for the “Counterfact” class, the model ex-
hibits a precision of 0.286 and recall of 0.667, result-
ing in an F1-score of 0.400. This performance sug-
gests that the model has challenges in accurately iden-
tifying counterfactual statements, highlighting an area
for potential improvement.
Table 2: Performance metrics of the fine-tuned BioBERT
for factuality prediction.
Class Precision Recall F1-Score Accuracy
Fact 0.970 0.918 0.944
Probable 0.526 0.833 0.645
Possible 0.600 0.500 0.545 0.927
Uncommitted 0.412 0.778 0.538
Counterfact 0.286 0.667 0.400
Our web interface serves as an interactive explo-
ration platform and consists of a dropdown list where
the user can choose “All” to display the entire knowl-
edge sub-graph or select a specific segment of the sub-
graph (Figure 3). If the user is interested in viewing
only the causes related to hormones (Figure 3a), they
should select the “Hormones” option. This will dis-
play the causes categorized as “Hormones” each with
the predicted factuality score. Similarly, Figure 3b
highlights factors related to hazardous or poisonous
substances that contribute to breast cancer. These
visualizations incorporate factuality scores, offering
valuable insights into the confidence levels associated
with various causal relationships. These are just two
sub-graphs of breast cancer causes, with the remain-
ing causes accessible through our web application.
Finally, we notice that some breast cancer causes
are cited numerous times while others are cited only
once. Figure 4 depicts the most frequently cited
causes. As shown, some causes have been cited over
200 times. However, among the 1,039 causes we ex-
tracted, over 90% are cited fewer than five times.
Figure 5 presents the calculated Closeness Scores
for the extracted triplets. The x-axis shows the in-
dex of each triplet after sorting to provide a clear vi-
sual representation of how the scores change across
the most to least relevant or popular triplets. Figure
5 shows that the closeness score starts high, around
0.90, and gradually decreases as the triplet index in-
creases. This indicates that the first few triplets are
more closely aligned with the main objective of the
publication. As the index increases, the triplets be-
come less relevant. Plotting all 1039 triplets could
lead to a dense or unreadable chart. Limiting the view
to the top 80 keeps the visualization clean and inter-
pretable.
Unveiling Breast Cancer Causes Through Knowledge Graph Analysis and BioBERT-Based Factuality Prediction
145
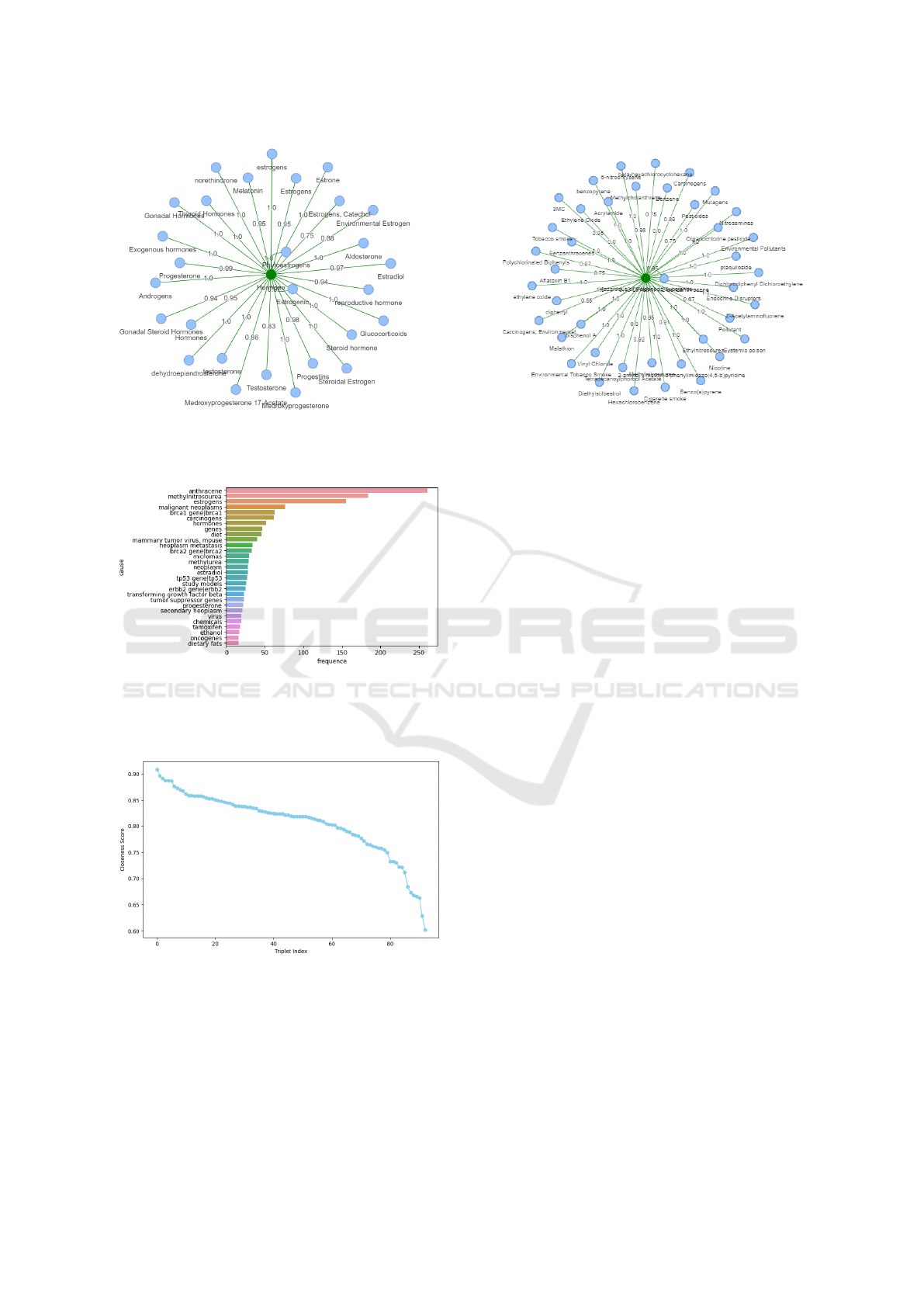
(a) Hormones (b) Hazardous or poisonous substances
Figure 3: Segments of the knowledge sub-graph of breast cancer causes with corresponding factuality scores, displayed as
screenshots from our web application.
Figure 4: Causes of breast cancer with the highest citation
frequency (frequency corresponds to the number of research
articles from which the cause and its associated sentence
were extracted).
Figure 5: Closeness Scores: Measurement of the alignment
between triplets and the primary focus of the scientific arti-
cle in which they are mentioned.
4 DISCUSSION
Our framework (Figure 1) aids in summarizing the
literature on the causes of breast cancer by provid-
ing a factuality score automatically computed using
BioBERT. This factuality is subsequently categorized
as reliable or unreliable based on the number of ci-
tations of the relationship and the alignment of sen-
tences revealing the triplets with those papers. Here,
we discuss the significance of our findings and their
implications.
Figure 2 highlights the most frequently identified
types of breast cancer causes in the biomedical liter-
ature, such as hazardous substances and genetic fac-
tors. This distribution not only reflects existing re-
search priorities but also uncovers underexplored ar-
eas, such as less-studied molecular factors. These
findings emphasize the need for targeted investiga-
tions in less-documented categories, guiding future
research toward filling critical gaps in the literature.
The visual segments in Figure 3, focusing on hor-
mones and hazardous substances, demonstrate the
breadth of our knowledge sub-graph and its capacity
to categorize causal relationships with associated fac-
tuality scores. This categorization enables researchers
to focus on specific domains while maintaining a
comprehensive understanding of the underlying re-
lationships. The integration of factuality scores into
these segments provides clarity on the reliability of
each causal link, enhancing trust in automated knowl-
edge extraction systems.
Figure 4 underscores the uneven citation distribu-
tion among the identified breast cancer causes. While
some causes are supported by extensive literature,
others are based on minimal citations, indicating vari-
ability in research focus. This variability emphasizes
the importance of combining citation counts with con-
textual evaluation, such as the Closeness Score, to en-
sure a balanced representation of causes in the knowl-
HEALTHINF 2025 - 18th International Conference on Health Informatics
146

edge graph.
As depicted in Figure 5, the Closeness Score pro-
vides a quantitative measure of triplet relevance to the
core objective of each publication. Higher scores cor-
respond to triplets directly aligned with the study’s
primary focus, while lower scores represent periph-
eral relationships. This metric demonstrates the ef-
fectiveness of integrating contextual relevance with
citation-based significance, addressing a key limita-
tion in existing knowledge graph methodologies.
As for BioBERT, while it performs well in identi-
fying factual statements, its performance varies across
different classes, particularly concerning precision
and F1-score. The results highlight areas where the
model’s predictive capabilities could be improved, es-
pecially for less frequently occurring classes.
Several aspects of our work are still under de-
velopment. For instance, the model struggles with
detecting negation. For example, in the sentence,
“These findings do not support a role for HAAs from
meat or NAT2 in the etiology of breast cancer,” the
algorithm incorrectly classifies the factuality of meat
causing breast cancer as probable, disregarding the
negation.
Another challenge is illustrated in the relationship
between tea and breast cancer. In the sentence, “The
role of tea in the aetiology of breast cancer is contro-
versial,” the algorithm fails to interpret the term “con-
troversial” correctly. Although the relationship is a
fact, the algorithm misclassifies it as probable due to
its misunderstanding of the term.
Incorporating crowd-sourcing could improve the
reliability of this knowledge sub-graph. Volunteers
could assist in classifying relationships and validating
the classes detected by the model, with these correc-
tions serving as training examples for further refine-
ment.
5 CONCLUSION
Ultimately, this study showcases the potential of ar-
tificial intelligence to revolutionize our understand-
ing of breast cancer. We introduce a knowledge sub-
graph to illustrate the causes contributing to breast
cancer. By mapping potential causal relationships
and assigning factuality scores to these links, the sub-
graph provides a valuable resource for researchers and
clinicians. The interactive web application enhances
usability, allowing for customized data exploration.
While offering a comprehensive overview, the sub-
graph also highlights areas for further investigation,
including expanding the representation of viral fac-
tors and elucidating the underlying biological mech-
anisms. Furthermore, we plan to include gene inter-
actions and causal antecedents in our knowledge sub-
graph to better understand the causal pathways lead-
ing to breast cancer.
The results underscore the importance of collabo-
rative efforts across disciplines to address the chal-
lenges posed by breast cancer. By combining this
knowledge sub-graph with clinical data, researchers
can develop more accurate risk prediction models and
improve patient care. Future research could expand
the sub-graph’s scope, validate causal relationships
through experimental studies, and explore integrating
additional data from domain-specific sources.
Limitations and Future Directions
As a limitation, this study lacks a comprehensive
comparison with state-of-the-art models. To address
this, we are adapting ERNIE and XLNet to our fac-
tuality dataset for prediction, and we will compare
their performance with BioBERT to assess potential
improvements in factuality classification.
Another limitation of the current study is the
reliance on a single biomedical knowledge graph
(SemMedDB) for triplet extraction. Future work will
involve integrating additional datasets from domain-
specific sources, such as clinical trial repositories, ge-
netic databases, and epidemiological studies. These
diverse datasets will enrich the knowledge sub-graph,
capturing a wider range of causal relationships and
improving the comprehensiveness of our approach.
Finally, the inclusion of real-world clinical data,
such as electronic health records (EHRs) and patient
registries, will provide context-specific insights into
breast cancer causes and their interactions. This inte-
gration will allow for personalized causal inference,
aligning our findings more closely with clinical prac-
tice.
REFERENCES
American Cancer Society (2024). American cancer society.
https://www.cancer.org/. Accessed: 2024-06-30.
Bahaj, A., Lhazmir, S., Ghogho, M., and Benbrahim, H.
(2022). Covid-19-related scientific literature explo-
ration: Short survey and comparative study. Biology,
11(8):1221.
Chen, Q., Du, J., Hu, Y., Keloth, V. K., Peng, X., Raja,
K., Zhang, R., Lu, Z., and Xu, H. (2023). Large lan-
guage models in biomedical natural language process-
ing: benchmarks, baselines, and recommendations.
arXiv preprint arXiv:2305.16326.
Chen, X., Chen, M., Shi, W., Sun, Y., and Zaniolo, C.
(2019). Embedding uncertain knowledge graphs. In
Unveiling Breast Cancer Causes Through Knowledge Graph Analysis and BioBERT-Based Factuality Prediction
147

Proceedings of the AAAI conference on artificial intel-
ligence, volume 33, pages 3363–3370.
Clark, K., Luong, M.-T., Le, Q. V., and Manning, C. D.
(2020). Electra: Pre-training text encoders as dis-
criminators rather than generators. arXiv preprint
arXiv:2003.10555.
Devlin, J., Chang, M.-W., Lee, K., and Toutanova, K.
(2018). Bert: Pre-training of deep bidirectional trans-
formers for language understanding. arXiv preprint
arXiv:1810.04805.
El Haji, H., Sbihi, N., Guermah, B., Souadka, A., and
Ghogho, M. (2024). Epidemiological breast cancer
prediction by country: A novel machine learning ap-
proach. PLOS ONE, 19(8):e0308905.
El Haji, H., Souadka, A., Patel, B. N., Sbihi, N., Ra-
masamy, G., Patel, B. K., Ghogho, M., and Banerjee,
I. (2023). Evolution of breast cancer recurrence risk
prediction: a systematic review of statistical and ma-
chine learning–based models. JCO Clinical Cancer
Informatics, 7:e2300049.
El Handri, K. and Idrissi, A. (2020). Parallelization of topk
algorithm through a new hybrid recommendation sys-
tem for big data in spark cloud computing framework.
IEEE Systems Journal, 15(4):4876–4886.
Guo, Q., Zhuang, F., Qin, C., Zhu, H., Xie, X., Xiong, H.,
and He, Q. (2020). A survey on knowledge graph-
based recommender systems. IEEE Transactions on
Knowledge and Data Engineering, 34(8):3549–3568.
Hangya, V., Saadi, H. S., and Fraser, A. (2022). Improv-
ing low-resource languages in pre-trained multilingual
language models. In Proceedings of the 2022 Confer-
ence on Empirical Methods in Natural Language Pro-
cessing, pages 11993–12006.
Hogan, A., Blomqvist, E., Cochez, M., d’Amato, C., Melo,
G. D., Gutierrez, C., Kirrane, S., Gayo, J. E. L.,
Navigli, R., Neumaier, S., et al. (2021). Knowledge
graphs. ACM Computing Surveys (Csur), 54(4):1–37.
Huang, X., Zhang, J., Li, D., and Li, P. (2019). Knowledge
graph embedding based question answering. In Pro-
ceedings of the twelfth ACM international conference
on web search and data mining, pages 105–113.
Jiang, N. and de Marneffe, M.-C. (2021). He thinks he
knows better than the doctors: Bert for event factu-
ality fails on pragmatics. Transactions of the Associa-
tion for Computational Linguistics, 9:1081–1097.
Kejriwal, M. (2020). Knowledge graphs and covid-19: op-
portunities, challenges, and implementation. Harv.
Data Sci. Rev, 11:300.
Kilicoglu, H., Rosemblat, G., and Rindflesch, T. C. (2017).
Assigning factuality values to semantic relations ex-
tracted from biomedical research literature. PloS one,
12(7):e0179926.
Kilicoglu, H., Shin, D., Fiszman, M., Rosemblat, G., and
Rindflesch, T. C. (2012). Semmeddb: a pubmed-
scale repository of biomedical semantic predications.
Bioinformatics, 28(23):3158–3160.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H.,
and Kang, J. (2020). Biobert: a pre-trained biomedi-
cal language representation model for biomedical text
mining. Bioinformatics, 36(4):1234–1240.
Li, P., Castelo, N., Katona, Z., and Sarvary, M. (2022). Lan-
guage models for automated market research: A new
way to generate perceptual maps. Available at SSRN
4241291.
Rossi, A., Barbosa, D., Firmani, D., Matinata, A., and Meri-
aldo, P. (2021). Knowledge graph embedding for link
prediction: A comparative analysis. ACM Transac-
tions on Knowledge Discovery from Data (TKDD),
15(2):1–49.
Soares, L. B., FitzGerald, N., Ling, J., and Kwiatkowski,
T. (2019). Matching the blanks: Distributional
similarity for relation learning. arXiv preprint
arXiv:1906.03158.
Sosa, D. N. and Altman, R. B. (2022). Contexts and contra-
dictions: a roadmap for computational drug repurpos-
ing with knowledge inference. Briefings in Bioinfor-
matics, 23(4):bbac268.
Stanovsky, G., Eckle-Kohler, J., Puzikov, Y., Dagan, I., and
Gurevych, I. (2017). Integrating deep linguistic fea-
tures in factuality prediction over unified datasets. In
Proceedings of the 55th Annual Meeting of the Associ-
ation for Computational Linguistics (Volume 2: Short
Papers), pages 352–357.
Sun, Z. (2023). A short survey of viewing large lan-
guage models in legal aspect. arXiv preprint
arXiv:2303.09136.
Veyseh, A. P. B., Nguyen, T. H., and Dou, D. (2019). Graph
based neural networks for event factuality prediction
using syntactic and semantic structures. In Proceed-
ings of the 57th Annual Meeting of the Association for
Computational Linguistics, pages 4393–4399.
Waltman, L. (2016). A review of the literature on citation
impact indicators. Journal of informetrics, 10(2):365–
391.
Wang, Z., Nie, H., Zheng, W., Wang, Y., and Li, X. (2023).
A novel tensor learning model for joint relational
triplet extraction. IEEE Transactions on Cybernetics,
54(4):2483–2494.
World Health Organization (2024). World health
organization. Accessed: 01:08:2024, url =
https://www.who.int/en,.
Yang, Z., Dai, Z., Yang, Y., Carbonell, J., Salakhutdinov,
R. R., and Le, Q. V. (2019). Xlnet: Generalized au-
toregressive pretraining for language understanding.
Advances in neural information processing systems,
32.
Zhang, T., Wu, F., Katiyar, A., Weinberger, K. Q., and Artzi,
Y. (2020). Revisiting few-sample bert fine-tuning.
arXiv preprint arXiv:2006.05987.
Zhang, Z., Han, X., Liu, Z., Jiang, X., Sun, M., and
Liu, Q. (2019). Ernie: Enhanced language repre-
sentation with informative entities. arXiv preprint
arXiv:1905.07129.
Zheng, W., Wang, Z., Yao, Q., and Li, X. (2021). Wrtre:
Weighted relative position transformer for joint entity
and relation extraction. Neurocomputing, 459:315–
326.
HEALTHINF 2025 - 18th International Conference on Health Informatics
148
