
Cross-Site Relational Distillation for Enhanced MRI Segmentation
Eddardaa Ben Loussaief
a
, Mohammed Ayad
b
, Hatem A. Rashwan
c
and Domenec Puig
d
Department of Computer Science and Mathematics of Security, University Rovira I Virgili, Tarragona, Spain
Keywords:
MRI Segmentation, Adaptive Affinity Module, Kernel Loss, Unseen Generalization.
Abstract:
The joint use of diverse data sources for medical imaging segmentation has emerged as a crucial area of re-
search, aiming to address challenges such as data heterogeneity, domain shift, and data quality discrepancies.
Integrating information from multiple data domains has shown promise in improving model generalizabil-
ity and adaptability. However, this approach often demands substantial computational resources, hindering
its practicality. In response, knowledge distillation (KD) has garnered attention as a solution. KD involves
training lightweight models to emulate the behavior of more resource-intensive models, thereby mitigating
the computational burden while maintaining performance. This paper addresses the pressing need to de-
velop a lightweight and generalizable model for medical imaging segmentation that can effectively handle
data integration challenges. Our proposed approach introduces a novel relation-based knowledge framework
by seamlessly combining adaptive affinity-based and kernel-based distillation. This methodology empowers
the student model to accurately replicate the feature representations of the teacher model, facilitating robust
performance even in the face of domain shift and data heterogeneity. To validate our approach, we con-
ducted experiments on publicly available multi-source MRI prostate. The results demonstrate a significant
enhancement in segmentation performance using lightweight networks. Notably, our method achieves this
improvement while reducing both inference time and storage usage.
1 INTRODUCTION
Problems with data privacy and sharing have recently
slowed medical progress. This brings us to domain
generalization, a method for protecting data while
also creating robust models that excel across various,
previously unknown data sources. Thus, there is a
growing demand for collaborative data efforts among
multiple medical institutions to enhance the develop-
ment of precise and resilient data-driven deep net-
works for medical image segmentation (Liu et al.,
2021; Liu et al., 2020a; Li et al., 2020). In prac-
tical applications, Deep learning (DL) models often
exhibit decreased performance when tested on data
from a different distribution than that used for train-
ing, which is referred to as domain shift. A major fac-
tor contributing to domain shift in the medical field
is the variation in image acquisition methods such
as imaging modalities, scanning protocols, or device
manufacturers, termed acquisition shift. Hence, ad-
dressing the issue of domain shift has led to investi-
gations into methodologies like unsupervised domain
a
https://orcid.org/0000-0002-6147-642X
b
https://orcid.org/0009-0002-5885-5143
c
https://orcid.org/0000-0001-5421-1637
d
https://orcid.org/0000-0002-0562-4205
adaptation (UDA) (Ganin and Lempitsky, 2015) and
single-source domain generalization (SDG) (Li et al.,
2023; Ouyang et al., 2023). Nevertheless, the effec-
tiveness of these strategies can be hindered by their
dependence on training data from either the target
domain or a single source domain, which frequently
proves inadequate for creating a universally appli-
cable model. A more practical approach is multi-
source domain generalization (MDG) (Muandet et al.,
2013), wherein a DL model is trained to be resilient
to domain shifts using data from various source do-
mains. Since we are dealing with a multi-source do-
main, we adopt The Adaptive Affinity Loss (AAL)
(Ke et al., 2018; Zhang et al., 2021; He et al., 2019)
to minimize the distribution gap across models’ fea-
tures. It’s designed to address challenges related to
domain shift, where the distribution of data in the
training and testing phases differs significantly. The
traditional loss functions used in semantic segmenta-
tion, such as cross-entropy or dice losses, focus on
pixel-wise classification accuracy. However, they of-
ten fail to capture the structural information and rela-
tionships between neighboring pixels, which are cru-
cial for accurate segmentation, especially in medical
images where objects of interest can have complex
shapes and textures. Adaptive Affinity Loss aims to
508
Ben Loussaief, E., Ayad, M., Rashwan, H. A. and Puig, D.
Cross-Site Relational Distillation for Enhanced MRI Segmentation.
DOI: 10.5220/0013181300003912
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 20th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (VISIGRAPP 2025) - Volume 3: VISAPP, pages
508-515
ISBN: 978-989-758-728-3; ISSN: 2184-4321
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

overcome these limitations by incorporating spatial
relationships and structural information into the loss
function. It does so by considering the affinities or
similarities between pixels in addition to their classi-
fications.
The affinity loss is calculated based on features
extracted from the deep neural network, which cap-
tures contextual information about the image. The
adaptive aspect of AAL refers to its ability to dynam-
ically adjust the importance of affinity terms based
on the characteristics of the input data. By inte-
grating spatial context and adaptively adjusting the
loss function, AAL helps improve semantic segmen-
tation models’ robustness and generalization capabil-
ity, especially in the presence of domain shift. This
adaptability is particularly useful in scenarios where
data distribution varies across different domains or
imaging modalities. In addition, the gram matrix has
proven its ability to allow the network to capture the
style representation of an input image. In the context
of KD to produce lightweight models, the gram ma-
trix is derived from the feature maps of the teacher
and student models. It serves as a form of representa-
tion of style or correlation between different features.
The difference between the matrices of the student
and teacher feature maps is calculated using a loss
function. This loss encourages the students’ feature
maps to have styles similar to the teacher’s.
In this work, we introduce an innovative segmen-
tation pipeline that leverages a combination of knowl-
edge transfer and unseen generalization techniques.
Our primary objective is to develop a lightweight and
highly generalizable model suitable for real-time clin-
ical applications. Our methodology revolves around
utilizing teacher models, trained on known data, to fa-
cilitate the training of student models with previously
unseen data. Unlike traditional generalization ap-
proaches that focus on minimizing distribution shifts
within the same network across different domains,
our emphasis lies in minimizing the distribution gap
between the domains of teachers and students. To
achieve this, we propose a relation-based KD tech-
nique that incorporates two key modules to tackle
the domain alignment: the Adaptive Affinity Module
(AAM) and the Kernel Matrix Module (KMM). These
modules work in tandem to optimize the discrepancy
across feature maps, thereby enhancing model perfor-
mance. Additionally, we integrate a Logits Module
(LM) that utilizes Kullback-Leibler (KL) divergence
to reduce the distribution shift between the logits of
teachers and students. Thus, our learning structure
comprises three main components: AAM and KMM
for addressing feature map discrepancies, and LM for
handling logits distribution shift. We validate our ap-
proach through segmentation tasks on prostate MRI
imaging (Liu et al., 2020b), demonstrating its superior
effectiveness compared to conventional off-the-shelf
generalization methods. Our segmentation pipeline
offers a robust solution for medical imaging segmen-
tation, with potential applications in real-world clini-
cal settings while preserving data privacy and patient
confidentiality making it suitable for deployment in
sensitive medical environments.
2 METHOD
Existing generalization techniques typically aim to al-
leviate distribution gaps across sources, necessitat-
ing alignment between input domains trained on the
unified network. Consequently, this results in a sce-
nario where data is shared between seen data used
for training and unseen data employed for general-
ization during testing. Nonetheless, we propose an
innovative fusion of knowledge transfer and gener-
alization paradigm for MRI prostate tumor segmen-
tation. This section aims to detail the incorporated
modules in our learning pipeline as shown in Fig.
1. The Holistic pipeline encompasses four distinct
objective functions: AAM and KMM, which are re-
sponsible for transferring intermediate information by
learning pixel-level affinities and gauging pixel sim-
ilarity via a gram matrix, respectively. The logits
module (LM) is designed to narrow the distribution
gap between the logits of teachers and students. Fi-
nally, to fully implement the distillation scheme, it is
imperative to integrate the segmentation loss across
the ground truth and the student’s input domain. We
delve into the relation-based distillation modules ex-
plored to address feature map discrepancies, empow-
ering lightweight models to emulate the capabilities
of powerful teachers.
2.1 Adaptive Affinity Module
Our Module implementation is derived from the idea
of (Ke et al., 2018), where instead of incorporating an
additional network, it utilizes affinity learning for net-
work predictions. We take advantage of affinity loss
in (Zhang et al., 2021), introducing an adaptive affin-
ity loss that encourages the network to learn inter-
and inner-class pixel relationships within the feature
maps. For this purpose, we employ the labeled seg-
mentation predictions of the teacher, which contain
precise delineations of each semantic class, to ex-
tract region information based on classes from fea-
ture maps. The pairwise pixel affinity is based on the
teacher’s prediction label map, where for each pixel
Cross-Site Relational Distillation for Enhanced MRI Segmentation
509
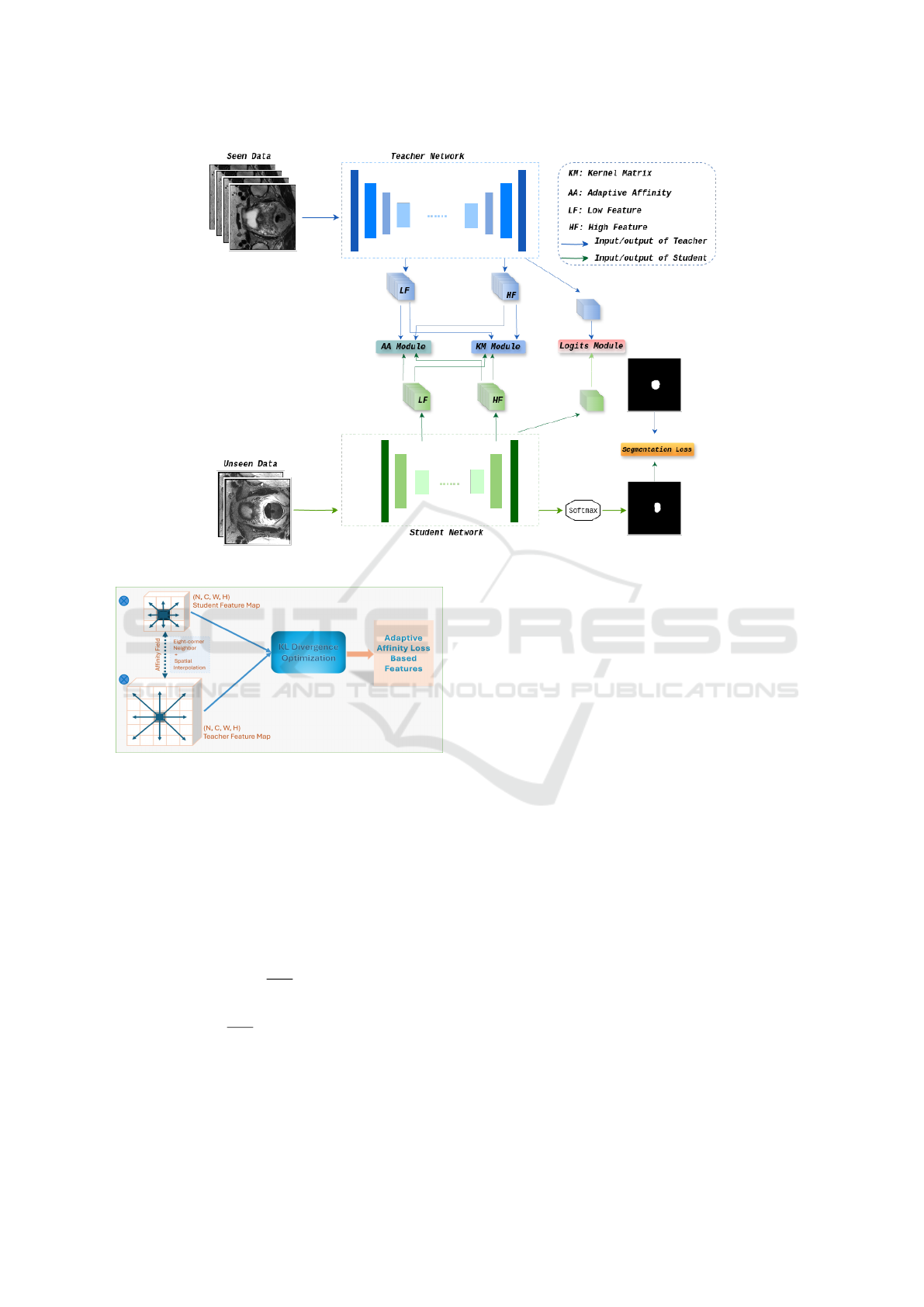
Figure 1: Overall framework of the proposed generalization method across teacher’s and student’s networks.
Figure 2: The architecture of Adaptive Affinity module
(AAM).
pair we have two categories of label relationships:
whether their labels are identical or distinct. We de-
note set a pixel pair P, segregated into two distinct
subsets based on whether a pixel i and its neighbor
j share the same label or belong to different regions:
P
+
and P
−
represents pairs with the same object label
and with different labels respectively. Specifically, we
define the pairwise affinities losses as follows:
Loss
P
+
=
1
|P
+
|
∑
i, j∈P
+
W
i j
, (1)
Loss
P
−
=
1
|P
−
|
∑
i, j∈P
−
max(0, m −W
i j
), (2)
where, pixel i and its neighbor j, belong to the same
class c in the feature map F. W
i j
is the KL divergence
between the classification probabilities, m is the mar-
gin of the separating force, and W
i j
can be defined as:
W
i j
= D
KL
(F
c
i
||F
c
j
). (3)
The total Adaptive Affinity Loss can be defined:
Loss
AA
= L oss
P
+
+ Loss
P
−
. (4)
2.2 Kernel Matrix Module
The similarity between two images, x
i
and x
j
, can be
assessed using a kernel function denoted as k(x
i
, x
j
) =
⟨φ(x
i
), φ(x
j
)⟩. Here, φ(x
i
) represents a projection
function that transforms examples from their original
space into a more suitable space for the target task.
We regard a specific layer within a neural network as
this projection function to generate the intermediate
features by f
S
(x
i
) = x
i
S
and f
T
(x
i
) = x
i
T
from the stu-
dent network S and teacher network T, respectively.
Subsequently, the similarity between x
i
and x
j
in the
gram matrix for S and T networks can be computed
as:
K
S
(x
i
, x
j
) = ⟨ f
s
(x
i
), f
s
(x
j
)⟩ = x
iT
S
x
j
S
,
K
T
(x
i
, x
j
) = ⟨ f
T
(x
i
), f
T
(x
j
)⟩ = x
iT
T
x
j
T
,
(5)
where x
i
and x
iT
refer to the feature map and its corre-
sponding transpose, while K
S
and K
T
denote the n × n
gram matrix derived from the S and T networks, re-
spectively. n is the total input samples for each net-
work separately, which represents the total input sam-
ples for each network individually. To gauge the sim-
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
510

ilarity between the teacher’s feature maps and the stu-
dent’s feature map, we adopt a depth-wise layer ap-
proach by incorporating a convolutional layer with a
1 × 1 kernel. This adjustment ensures that there is
alignment in the spatial resolution of the features de-
rived from both S and T networks. Thereby, we tend
to transfer the full kernel matrix from the T model to
the S model to enable this latter to mimic the teacher’s
performance. Then, the distillation loss of this mod-
ule can be defined by:
Loss
KM
=
1
n
n
∑
i=0
(K
S
− K
T
)
2
+ ||K
S
− K
T
||
L
1
, (6)
where K
S
and K
T
are defined in (5). ||.||
L
1
represents
the L
1
normalization, which assigns a weight to adjust
the loss.
2.3 Logits Module
Logits-based distillation stands as a foundational
technique in knowledge distillation, first introduced
by Hinton et al. (Hinton et al., 2015). It involves
the student network learning from the teacher net-
work by replicating its output probability distribu-
tion. This is crucial because these output probabili-
ties contain valuable insights into inter-class similari-
ties that might not be entirely reflected in the ground
truth labels. By mimicking these probabilities, the
student network can develop a better grasp of the un-
derlying knowledge embedded in the teacher’s predic-
tions. The traditional knowledge-transferring method
applies a softmax function on the logit layer to soften
the output and then measure the loss using the teacher
and student outputs. However, in our case, due to that
we aim to build not just a lightweight student model
but a generalizable one as well. and due to that the
student has no prior knowledge of the input domain,
so we adopt to minimize the distribution probabili-
ties of the logit layers. Thus the loss in our method is
calculated using the KL divergence between the prob-
abilities p
s
i
and p
t
i
of the i
th
class derived from the S
and the T networks, respectively.
Loss
Logits
=
1
N
N
∑
i
KL(p
s
i
||p
t
i
), (7)
where N is the total pixel number derived from the
logits’ output.
As illustrated in Fig. 1, we adopt a global function
loss, given here, to train the student in an end to end
manner with unseen data.
Loss
Total
= Loss
Seg
+ λ
1
Loss
Logits
+ λ
2
Loss
KM
+ λ
3
Loss
AA
,
(8)
where L oss
Seg
is the general segmentation loss that
can either dice loss (Hinton et al., 2015) and focal
Table 1: The rank of the models in ascending order of the
number of parameters.
Method Params(M) Flops(G)
ESPNet 0.183 1.27
ENet 0.353 2.2
ERFNet 2.06 16.56
Unet++ 36.6 621.38
DeepLabV3+ 56.8 273.94
loss (Sudre et al., 2017). In this work, we empirically
set the weighted parameters λ
1
, λ
2
, and λ
3
to 0.2, 0.9,
and 0.9, respectively.
3 EXPERIMENTS AND RESULTS
3.1 Datasets
We assess the effectiveness of our generalization
pipeline using prostate MRI segmentation data gath-
ered from six distinct sites sourced from three public
datasets: NCI-ISBI2013 (Bloch N, 2015), I2CVB
(Lema
ˆ
ıtre et al., 2015), and Promise12 (Litjens et al.,
2014). Specifically, sites 1, 2, and 3 correspond to
data from ISBI2013 and I2CVB respectively, while
sites 4, 5, and 6 are obtained from Promise12. We
adopt a dice similarity score to evaluate the models’
performance and to conduct a comparison with the
state-of-the-art (SOTA) generalization methods.
3.2 Implementation Setup
To evaluate our structured generalization-distillation
framework, we meticulously select NestedUnet
(Unet++) (Zhou et al., 2018) and DeepLabv+3 (Chen
et al., 2016) as the teacher networks. In the scope
of lightweight student networks, we employed ENet
(Paszke et al., 2016), ESPNet (Mehta et al., 2018),
and ERFNet (Romera et al., 2018). All networks
were trained using the Adam optimizer, initialized
with a learning rate of 0.01. We employed a Cycli-
cLR to schedule the learning rate with a step size of
2000, gradually decreasing it until reaching a min-
imum of 1e
−6
. We conducted the experiments to
converge within 100 epochs with a batch size of 16
on an NVIDIA RTX 3050 ti with 16 GB of mem-
ory. The computational complexity of the aforemen-
tioned models in terms of the number of parameters
and Flops is listed in Table 1.
Cross-Site Relational Distillation for Enhanced MRI Segmentation
511

Table 2: Cross experiments results between a single teacher and student on prostate. “w/o” denotes the baseline performance,
and “w/” stands for the performance with our distillation. We denote by S1, S2, S3, S4, S5, and S6 the input domains and
target sites for the teacher and student networks, respectively.
Models
Prostate
S1 S2 S3 S4 S5 S6
T1:
DeepLabV3+
0.895 0.886 0.889 0.874 0.878 0.885
T2: Unet++ 0.901 0.893 0.883 0.880 0.869 0.871
Generalization’s performances distilled from different teachers
ENet: w/o 0.714 0.604 0.539 0.782 0.701 0.772
T1: w/ 0.810 0.642 0.734 0.881 0.796 0.876
T2: w/ 0.808 0.637 0.684 0.858 0.894 0.869
ESPNet: w/o 0.703 0.482 0.573 0.738 0.745 0.715
T1: w/ 0.819 0.563 0.604 0.826 0.819 0.832
T2: w/ 0.837 0.594 0.643 0.842 0.836 0.808
ERFNet: w/o 0.788 0.720 0.736 0.760 0.725 0.694
T1: w/ 0.823 0.751 0.799 0.869 0.801 0.861
T2: w/ 0.843 0.746 0.752 0.892 0.877 0.852
3.3 Experimental Evaluation
We intend in this section to demonstrate the effi-
ciency of the generalization ability of our proposed
framework. As mentioned earlier, we adopted various
combinations of the off the shelf teacher and student
models, specifically, (Unet++) (Zhou et al., 2018) and
DeepLabv+3 (Chen et al., 2016) as the teachers due to
their high performance in medical imaging segmenta-
tion. Meanwhile, we opted for ENet (Paszke et al.,
2016), ESPNet (Mehta et al., 2018), and ERFNet
(Romera et al., 2018) as lightweight student models.
Table 2 presents the results of applying our distil-
lation framework including the aforementioned mod-
ules, i.e. affinity module AAM, the kernel module
KMM, and the logits module LM. All student mod-
els show significant improvements in Prostate MRI
segmentation with the unseen data compared to the
baseline. We listed in the table 2 the primary results
after applying the three modules. The students ex-
hibit a notable improvement depending on the input
domains. There are obvious difficulties in enabling
the students to enhance their performance when they
have been trained with site 3, this latter has differ-
ent institution source and device settings compared to
the remaining sites. Our pipeline enables Enet to sur-
pass the performance of the large teachers in terms of
segmentation dice, i.e. for the target S4 and S5 Enet
yielded dice scores of 0.881 and 0.894, which were
distilled from Deeplabv3+ and Unet++ respectively.
In Addition, ERFNet outperforms the teacher Unet++
achieving scores of 0.892 and 0.877 with the target
domains S4 and S5. ESPNetV2, on the other hand,
demonstrated comparable performance to the teacher
networks, showcasing an increase of up 13% (0.703 to
0.837) when paired with Unet++ and S1 as the target
domain. To further illustrate the effectiveness of our
approach, we present the segmentation performance
on MRI prostate in Fig 3. To assess the impact of our
introduced modules, we conducted an ablation study,
the results of which are summarized in the table 3. We
selected Deeplabv3+ and Enet as teacher and student
models respectively. As shown in table 3, the highest
dice score for all the target sites is obtained when we
integrated our three modules simultaneously.
To exhibit the superiority of our structured frame-
work, we compare it with various advanced SOTA
generalizations and KD. Specifically, we adhered to
the same generalization scenario as developed in our
work for distillation. Table 4 provides a summary of
the comparison outcomes.
In (Zhao et al., 2023), the generalization results
exceeded our own, albeit employing a traditional ap-
proach with a complex model. It’s worth noting
that (Zhao et al., 2023) utilized a different method
from ours. While their approach yielded superior re-
sults, it relied on a complex model. In contrast, our
method offers a significant advantage: we achieved
comparable performance with a lightweight model
that requires minimal memory storage and computa-
tional resources with trainable parameters of 353k and
FLOPS of 2.2G. To the best of our knowledge, no ex-
isting work has explored similar methods to ours, un-
derscoring our approach’s novelty and potential im-
pact.
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
512

Table 3: The effectiveness of the components of our generalization method. We select Enet and Deeplabv3+ as the student
and teacher models respectively.
Model
Prostate
S1 S2 S3 S4 S5 S6
Teacher:
Deeplabv3+
0.895 0.886 0.889 0.874 0.878 0.885
Student:Enet 0.714 0.604 0.539 0.782 0.701 0.772
+LM 0.691 0.571 0.505 0.741 0.619 0.721
+AAM 0.700 0.599 0.648 0.780 0.691 0.740
+KMM 0.698 0.583 0.630 0.767 0.690 0.738
+KMM + LM 0.763 0.679 0.659 0.807 0.769 0.775
+AAM + LM 0.772 0.691 0.688 0.810 0.762 0.798
+AAM + KMM 0.781 0.611 0.709 0.849 0.781 0.816
+AAM + KMM
+ LM
0.810 0.642 0.734 0.881 0.796 0.876
Figure 3: Segmentation Performance of our method. The first row refers to the ESPNet’s prediction and the second row
presents results derived from ERFNet Student. The red and green contours denote the GT and predicted mask of the student
after our KD, respectively.
Table 4: Comparative results of our method with two advanced distillation methods and two generalization methods.
Method
Prostate
S1 S2 S3 S4 S5 S6
Teacher:
DeepLabV3+
0.895 0.886 0.889 0.874 0.878 0.885
Student (Ours):
Enet
0.810 0.642 0.734 0.881 0.796 0.876
Distillation methods
KD (Hinton
et al., 2014)
0.778 0.765 0.761 0.786 0.699 0.745
AT (Zagoruyko
and Komodakis,
2017)
0.789 0.641 0.585 0.816 0.802 0.774
MSKD (Zhao
et al., 2023)
0.762 0.643 0.532 0.809 0.790 0.658
EMKD (Qin
et al., 2021)
0.635 0.524 0.681 0.696 0.544 0.711
Generalization methods
SAML (Liu
et al., 2020a)
0.896 0.875 0.843 0.886 0.873 0.883
DRDG (Lu
et al., 2021)
0.709 0.758 0.665 0.739 0.857 0.826
Cross-Site Relational Distillation for Enhanced MRI Segmentation
513

4 CONCLUSION
We have outlined three key distillation modules:
Adaptive Affinity Module (AAM), Kernel Matrix
Module (KMM), and Logits Module (LM) to develop
a lightweight, generalizable model for medical imag-
ing segmentation. A unique aspect of our approach
is its ability to enhance the student model by leverag-
ing detailed contextual information from feature maps
through the integration of AAM and KMM. Experi-
mental results on MRI prostate data demonstrate that
our method significantly outperforms related state-of-
the-art (SOTA) techniques, improving both segmen-
tation accuracy and the generalization capabilities of
lightweight networks. Future work will focus on ex-
panding our ablation study by incorporating deeper
teacher models and refining the proposed method to
further improve segmentation outcomes. Addition-
ally, we will explore the model’s applicability to dif-
ferent medical imaging tasks, addressing potential
limitations to achieve the highest possible segmenta-
tion accuracy.
ACKNOWLEDGEMENTS
PID2023-146925OB-I00
REFERENCES
Bloch N, Madabhushi A, H. H. F. J. K. J. G. M. E. A. J.
C. C. L. F. K. (2015). Nci-isbi 2013 challenge: Auto-
mated segmentation of prostate structures. the Cancer
Imaging Archive.
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K.,
and Yuille, A. L. (2016). Deeplab: Semantic
image segmentation with deep convolutional nets,
atrous convolution, and fully connected crfs. CoRR,
abs/1606.00915.
Ganin, Y. and Lempitsky, V. (2015). Unsupervised domain
adaptation by backpropagation. In Proceedings of the
32nd International Conference on International Con-
ference on Machine Learning - Volume 37, ICML’15,
page 1180–1189. JMLR.org.
He, T., Shen, C., Tian, Z., Gong, D., Sun, C., and Yan,
Y. (2019). Knowledge adaptation for efficient se-
mantic segmentation. In 2019 IEEE/CVF Conference
on Computer Vision and Pattern Recognition (CVPR),
pages 578–587.
Hinton, G., Dean, J., and Vinyals, O. (2014). Distilling the
knowledge in a neural network. pages 1–9.
Hinton, G. E., Vinyals, O., and Dean, J. (2015). Dis-
tilling the knowledge in a neural network. CoRR,
abs/1503.02531.
Ke, T.-W., Hwang, J.-J., Liu, Z., and Yu, S. X. (2018).
Adaptive affinity fields for semantic segmentation. In
Ferrari, V., Hebert, M., Sminchisescu, C., and Weiss,
Y., editors, Computer Vision – ECCV 2018, pages
605–621, Cham. Springer International Publishing.
Lema
ˆ
ıtre, G., Mart
´
ı, R., Freixenet, J., Vilanova, J. C.,
Walker, P. M., and Meriaudeau, F. (2015). Computer-
aided detection and diagnosis for prostate cancer
based on mono and multi-parametric mri. Comput.
Biol. Med., 60(C):8–31.
Li, H., Li, H., Zhao, W., Fu, H., Su, X., Hu, Y., and Liu, J.
(2023). Frequency-mixed single-source domain gen-
eralization for medical image segmentation. In Med-
ical Image Computing and Computer Assisted Inter-
vention – MICCAI 2023: 26th International Confer-
ence, Vancouver, BC, Canada, October 8–12, 2023,
Proceedings, Part VI, page 127–136, Berlin, Heidel-
berg. Springer-Verlag.
Li, H., Wang, Y., Wan, R., Wang, S., Li, T.-Q., and Kot,
A. C. (2020). Domain generalization for medical
imaging classification with linear-dependency regu-
larization. In Proceedings of the 34th International
Conference on Neural Information Processing Sys-
tems, NIPS ’20, Red Hook, NY, USA. Curran Asso-
ciates Inc.
Litjens, G., Toth, R., van de Ven, W., Hoeks, C., Kerkstra,
S., van Ginneken, B., Vincent, G., Guillard, G., Bir-
beck, N., Zhang, J., Strand, R., Malmberg, F., Ou,
Y., Davatzikos, C., Kirschner, M., Jung, F., Yuan, J.,
Qiu, W., Gao, Q., Edwards, P.E, Maan, B., van der
Heijden, F., Ghose, S., Mitra, J., Dowling, J., Barratt,
D., Huisman, H., and Madabhushi, A. (2014). Eval-
uation of prostate segmentation algorithms for mri:
The promise12 challenge. Medical Image Analysis,
18(2):359–373.
Liu, Q., Chen, C., Qin, J., Dou, Q., and Heng, P.-A. (2021).
Feddg: Federated domain generalization on medical
image segmentation via episodic learning in contin-
uous frequency space. In CVPR, pages 1013–1023.
Computer Vision Foundation / IEEE.
Liu, Q., Dou, Q., and Heng, P.-A. (2020a). Shape-aware
meta-learning for generalizing prostate mri segmenta-
tion to unseen domains. In Martel, A. L., Abolmae-
sumi, P., Stoyanov, D., Mateus, D., Zuluaga, M. A.,
Zhou, S. K., Racoceanu, D., and Joskowicz, L., ed-
itors, Medical Image Computing and Computer As-
sisted Intervention – MICCAI 2020. Springer Interna-
tional Publishing.
Liu, Q., Dou, Q., Yu, L., and Heng, P. A. (2020b). Ms-net:
Multi-site network for improving prostate segmenta-
tion with heterogeneous mri data. IEEE Transactions
on Medical Imaging, 39(9):2713–2724.
Lu, Y., Xing, X., and Meng, M. Q.-H. (2021). Unseen do-
main generalization for prostate mri segmentation via
disentangled representations. In 2021 IEEE Interna-
tional Conference on Robotics and Biomimetics (RO-
BIO), pages 1986–1991.
Mehta, S., Rastegari, M., Caspi, A., Shapiro, L., and Ha-
jishirzi, H. (2018). Espnet: Efficient spatial pyra-
mid of dilated convolutions for semantic segmenta-
tion. In Proceedings of the European Conference on
Computer Vision (ECCV).
Muandet, K., Balduzzi, D., and Sch
¨
olkopf, B. (2013).
Domain generalization via invariant feature repre-
VISAPP 2025 - 20th International Conference on Computer Vision Theory and Applications
514

sentation. In Proceedings of the 30th International
Conference on International Conference on Machine
Learning - Volume 28, ICML’13, page I–10–I–18.
JMLR.org.
Ouyang, C., Chen, C., Li, S., Li, Z., Qin, C., Bai, W.,
and Rueckert, D. (2023). Causality-inspired single-
source domain generalization for medical image seg-
mentation. IEEE Transactions on Medical Imaging,
42(4):1095–1106. Publisher Copyright: © 1982-2012
IEEE.
Paszke, A., Chaurasia, A., Kim, S., and Culurciello, E.
(2016). Enet: A deep neural network architec-
ture for real-time semantic segmentation. ArXiv,
abs/1606.02147.
Qin, D., Bu, J.-J., Liu, Z., Shen, X., Zhou, S., Gu, J.-J.,
Wang, Z.-H., Wu, L., and Dai, H.-F. (2021). Efficient
medical image segmentation based on knowledge dis-
tillation. IEEE Transactions on Medical Imaging,
40(12):3820–3831.
Romera, E.,
´
Alvarez, J. M., Bergasa, L. M., and Ar-
royo, R. (2018). Erfnet: Efficient residual factorized
convnet for real-time semantic segmentation. IEEE
Transactions on Intelligent Transportation Systems,
19(1):263–272.
Sudre, C. H., Li, W., Vercauteren, T., Ourselin, S., and
Jorge Cardoso, M. (2017). Generalised dice overlap
as a deep learning loss function for highly unbalanced
segmentations. In Cardoso, M. J., Arbel, T., Carneiro,
G., Syeda-Mahmood, T., Tavares, J. M. R., Moradi,
M., Bradley, A., Greenspan, H., Papa, J. P., Madab-
hushi, A., Nascimento, J. C., Cardoso, J. S., Belagian-
nis, V., and Lu, Z., editors, Deep Learning in Medical
Image Analysis and Multimodal Learning for Clinical
Decision Support, pages 240–248, Cham. Springer In-
ternational Publishing.
Zagoruyko, S. and Komodakis, N. (2017). Paying more
attention to attention: Improving the performance of
convolutional neural networks via attention transfer.
Zhang, X., Peng, Z., Zhu, P., Zhang, T., Li, C., Zhou, H.,
and Jiao, L. (2021). Adaptive affinity loss and erro-
neous pseudo-label refinement for weakly supervised
semantic segmentation. In Proceedings of the 29th
ACM International Conference on Multimedia, MM
’21, page 5463–5472, New York, NY, USA. Associa-
tion for Computing Machinery.
Zhao, L., Qian, X., Guo, Y., Song, J., Hou, J., and Gong, J.
(2023). Mskd: Structured knowledge distillation for
efficient medical image segmentation. Computers in
Biology and Medicine, 164:107284.
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N., and
Liang, J. (2018). Unet++: A nested u-net architecture
for medical image segmentation. In Deep Learning in
Medical Image Analysis and Multimodal Learning for
Clinical Decision Support: 4th International Work-
shop, DLMIA 2018, and 8th International Workshop,
ML-CDS 2018, Held in Conjunction with MICCAI
2018, Granada, Spain, September 20, 2018, Proceed-
ings, page 3–11, Berlin, Heidelberg. Springer-Verlag.
Cross-Site Relational Distillation for Enhanced MRI Segmentation
515
