
How to Box Your Cells: An Introduction to Box Supervision for 2.5D Cell
Instance Segmentation and a Study of Applications
Fabian Schmeisser
1,2 a
, Maria Caroprese
3,4 b
, Gillian Lovell
3,4 c
, Andreas Dengel
1,2 d
and Sheraz Ahmed
1 e
1
German Research Center for Artificial Intelligence (DFKI) GmbH, Kaiserslautern 67663, Germany
2
RPTU Kaiserslautern-Landau, Kaiserslautern 67663, Germany
3
Sartorius BioAnalytics, Royston, U.K.
4
Sartorius Corporate Research, Royston, U.K.
Keywords:
Cell Segmentation, 2.5D, 3D, Weak Supervision.
Abstract:
Cell segmentation in volumetric microscopic images is a fundamental step towards automating the analysis
of life-like representations of complex specimens. As the performance of current Deep Learning algorithms
is held back by the lack of accurately annotated ground truth, a pipeline is proposed that produces accurate
3D cell instance segmentation masks solely from slice-wise bounding box annotations. In an effort to further
reduce the time requirements for the annotation process, a study is conducted on how to effectively reduce the
size of the training set. To this end, three slice-reduction strategies are suggested and evaluated in combination
with bounding box supervision. We find that as low as 1% of weakly labeled training data suffices to produce
accurate results, and that predictions produced by a 10 times smaller dataset are of equal quality to when the
full dataset is exploited for training.
1 INTRODUCTION
The technological means for rapidly acquiring micro-
scopic images in a life-like 3D representation are ad-
vancing at a tremendous pace. While ever-increasing
image quality and quantity theoretically allow for
greater insight into cell behavior in general, the swift
acquisition of data outpaces the speed at which re-
searchers are capable of manually analyzing its con-
tents. With the surge of capable Deep Learning (DL)
methods in the past decade, many of the most time-
consuming tasks in cell analysis have been success-
fully automated. Among these tasks, cell segmenta-
tion can be considered a fundamental stepping stone
towards further analytic steps. The tracking of mor-
phological changes, spatial movement of single cells,
mitotic behavior and many other aspects hinge on
the accurate estimation of physical space occupied
by single cells. Naturally, an impressive number of
a
https://orcid.org/0000-0001-8222-7900
b
https://orcid.org/0009-0009-2170-1459
c
https://orcid.org/0009-0004-5180-9704
d
https://orcid.org/0000-0002-6100-8255
e
https://orcid.org/0000-0002-4239-6520
sophisticated cell segmentation algorithms were pro-
duced and published (Stringer et al., 2020) (Edlund
et al., 2021) (Schmidt et al., 2018) (Weigert et al.,
2020). Only a fraction of these studies, however,
target the analysis of three-dimensional microscopic
images. The relative lack of 3D-capable machine-
learning based segmentation strategies can be at-
tributed to a number of issues inherent to volumetric
data. Among these issues, the most prolific are the ex-
cessive size of 3D files compared to 2D images, and
the severe lack of accurately annotated ground truth
for supervised learning algorithms. Specifically, the
latter problem, lack of accurate ground truth, can in
large part be attributed to the expensive and complex
process of manual mask creation. With an estimate in
contemporary literature of approximately 5 minutes
for manually annotating a single cell instance in 3D in
a crowded dataset (Jelli et al., 2023), extrapolated an-
notation times for full datasets with several thousands
of cells quickly show the unfeasibility of large-scale
human annotation. While substantial research exists
on how to alleviate this problem in 2D cell segmen-
tation, (Khalid et al., 2023) (Zhao et al., 2018), fewer
approaches have been published to tackle this prob-
Schmeisser, F., Caroprese, M., Lovell, G., Dengel, A. and Ahmed, S.
How to Box Your Cells: An Introduction to Box Supervision for 2.5D Cell Instance Segmentation and a Study of Applications.
DOI: 10.5220/0013189800003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 853-860
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
853

lem in the third dimension. In this study, we focus on
significantly reducing annotation time for 3D micro-
scopic images by introducing bounding box supervi-
sion to 2.5D cell segmentation. The proposed pipeline
uses polygon tracing to estimate segmentation masks
in multi-slice, depth information-preserving pseudo-
2D inputs and reconstructs full 3D instance segmen-
tation masks from these predictions. In addition to
bounding box supervision, a detailed study on the ef-
fects of few-slice training is provided. To preserve
dataset diversity, a limited number of slices are seeded
from the complete dataset to be used as training in-
put to further minimize annotation time. The find-
ings show, that bounding box supervision as well as
few-slice training produce high-quality segmentation
masks, comparable to State-of-the-Art (SOTA) weak
supervision algorithms that require more complex an-
notations and the full dataset.
2 RELATED WORK
Several works attend to the topic of 3D cell segmen-
tation using semantic segmentation methods such as
modernized variations of U-Net (Chen et al., 2024)
(Arbelle et al., 2022). While these methods are highly
successful in metrics reflecting semantic segmenta-
tion quality, or on images containing easily separa-
ble objects, crowded images still present a significant
challenge. Few methods are specifically developed
for instance segmentation in 3D. As a prominent ex-
ample, Stardist (Weigert et al., 2020) and its improved
version (Jelli et al., 2023) achieve solid performance
on datasets containing star-convex cell shapes, but in
turn have to deal with extraordinarily high computa-
tional resource costs. Meanwhile, 2.5D methods of-
ten trade substantially lower computational require-
ments for lower performance. Examples like (Scherr
et al., 2021) and (Wagner and Rohr, 2022) rank sig-
nificantly below fully 3D methods, and also rely on
post-processing semantic segmentation results to re-
trieve instance segmentation masks. In (Schmeisser
et al., 2024a) and (Schmeisser et al., 2024b) a 2.5D
instance segmentation method is proposed, that pro-
vides a SOTA baseline for instance segmentation and
ranks above the previously mentioned 2.5D seman-
tic segmentation algorithms. The common hindrance
of lacking or inaccurate ground truth for training a
supervised learning algorithm is addressed in various
ways in contemporary literature. Weakly supervised
approaches either employ strategies to learn from par-
tially annotated data, or use more cost-effective anno-
tation strategies to fully label a dataset. In the case of
missing ground truth masks, loss calculation can be
ignored at unannotated image regions (Arbelle et al.,
2022) (Zhao et al., 2018), or artificially generated
training data is used to diversify the sparse training
set (Wu et al., 2023). Weak annotations, on the other
hand, are typically required to cover all instances in
the dataset. Two recent examples of algorithms lever-
aging weak annotations propose two-step approaches,
where either points or lines are seeded inside manu-
ally annotated bounding boxes or 3D boxes respec-
tively (Schmeisser et al., 2024a) (Schmeisser et al.,
2024b). This, however, necessitates a two-step ap-
proach, where annotators have to re-visit instances
already marked with bounding boxes and define if
points or line segments belong to the fore- or back-
ground of the image volume. As a single-step ap-
proach, bounding box supervision has recently been
introduced to 2D cell segmentation (Khalid et al.,
2024), but without an extension to 3D microscopic
images.
3 DATASET
Suitable open-source datasets with natural images and
highly accurate and complete ground truth annota-
tions are exceedingly rare. The dataset chosen for this
study is therefore a synthetic, but sufficiently com-
plex, series of images that come with perfect anno-
tation masks. An additional benefit of this dataset
comes with its usage in previous studies on the sub-
ject of weakly supervised segmentation for 3D images
(Schmeisser et al., 2024a), thus providing the possi-
bility of a direct comparison to the state of the art.
3.1 Dataset Description
The dataset N3DH-SIM+ is provided by the ISBI Cell
Tracking Challenge. It contains two distinct time
series of 150 and 80 volumes, showing simulated
C.elegans cells. The anisotropic volumes with resolu-
tions between 59x639x349 and 59x652x642 are split
into training, test, and validation sets based on their
occurrence in the respective time series. The first
120/50 images of each time series is used for train-
ing, the next 10/10 images are used for validation, and
the final 20/20 images form the test set. This split-
ting strategy is the same as proposed in (Schmeisser
et al., 2024b) to ensure comparability to other SOTA
methods. Next to comparability, this partitioning also
allows for checking the pipeline’s capabilities to ex-
trapolate information learned on images earlier in the
sequence to images situated at later time steps.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
854

3.2 Slice Reduction
The process of slice reduction is used to simulate a
lack of available ground truth. For this, a variable per-
centage p% of annotated slices is extracted from the
fully annotated volume using one of three strategies:
Initial Slice Extraction: Only the initial p% of slices
in the dataset are kept. This slice reduction pro-
cedure emulates having fewer annotated volumes in
the dataset. Choosing fewer volumes to reduce the
dataset size is how set reduction has to be handled for
fully supervised 3D methods. With this procedure,
the diversity of the dataset is significantly diminished.
Regular Slice Extraction: Slices are kept in the train-
ing data based on a regular selection. I.e. if the dataset
is reduced to 10% of its original size, every 10th slice
is kept. This deletion procedure ensures examples
from every volume are present in the training set, even
for very low values for p. Thus, sufficient diversity of
training samples stemming from all available volumes
is guaranteed.
Random Slice Extraction: The slices to be kept are
chosen at random. While this method is statistically
likely to produce a diverse dataset that presents the
initial distribution sufficiently for a larger p, this can-
not be ensured.
4 METHODS
The proposed pipeline is composed of multiple com-
ponents, further discussed in the subsections below.
Multi-slice input, a Swin-Transformer (Liu et al.,
2021) based segmentation algorithm, Box supervision
(Yang et al., 2023), and a multi-view capable 3D re-
construction algorithm (Zhou et al., 2024) are com-
bined for producing high-quality 3D instance seg-
mentation masks from weak box annotations.
4.1 Depth Context Preserving
Multi-Slice Input
Several studies have shown the benefit of using multi-
slice input for 2.5D learning algorithms (Bouyssoux
et al., 2022). Providing depth context through the ad-
dition of neighboring slices greatly improves model
performance and is especially beneficial for 3D re-
constructions. Specifically for anisotropic images
with low depth resolution, the usage of more than
two neighboring slices has been shown to deliver
diminishing returns, however (Zhang et al., 2022)
(Schmeisser et al., 2024b). For these reasons, a
consistent 3-slice input is chosen for the proposed
pipeline. For each 3-slice input, only the segmenta-
tion of the middle slice is predicted.
4.2 Deep Learning Architecture
Figure 1 provides an overview of the employed deep
learning architecture. As an upgrade over previous
approaches mostly relying on traditional CNN archi-
tectures, the method used in this study is based on a
more effective variation of the standard Vision Trans-
former (ViT) architecture (Dosovitskiy, 2020). The
Shifted Window (Swin) Transformer (Liu et al., 2021)
maintains computational efficiency while capturing
long-range dependencies in images with high accu-
racy. By integrating the image features hierarchi-
cally extracted by the Swin Feature Pyramid Network
(FPN) backbone into an instance segmentation frame-
work, in this case a Cascade Mask RCNN-like (Cai
and Vasconcelos, 2018) structure, fine-grained details
and contextual information help to successfully gen-
erate accurate segmentation masks. The three main
stages of the segmentation pipeline are:
Swin FPN. Responsible for the extraction of feature
maps. These maps are directly generated from the
input image and come at varying scales to capture de-
tails of arbitrary sizes.
RPN. Feature maps are passed to a Region Proposal
Network (RPN) which generates Regions of Interest
(RoIs), that are likely to contain objects.
Prediction Head. The final stage of the pipeline
has the purpose of generating bounding boxes, object
classes, and instance segmentation masks. This stage
traditionally is trained by directly relating ground
truth and prediction using a similarity metric like
Cross Entropy or DICE. In the case of the proposed
weak box supervision approach, however, polygons
are fitted around object boundaries and refined us-
ing a combination of local and global pairwise loss-
functions, as further explained in 4.3.
4.3 Box Supervision
To predict segmentation masks inside the bound-
ing boxes proposed by the RPN, a polygon-based
approach employing point-based unary loss and
distance-aware pairwise loss is employed (Yang et al.,
2023). The value computed with these loss functions
is used to tighten a predicted polygon around object
boundaries. The point-based unary loss function en-
sures that the predicted polygon vertices are fully en-
closed within the respective ground-truth bounding
box. By computing a bounding box b
p
that tightly
fits the polygon using corresponding minimum and
maximum values in both image dimensions, the dif-
How to Box Your Cells: An Introduction to Box Supervision for 2.5D Cell Instance Segmentation and a Study of Applications
855
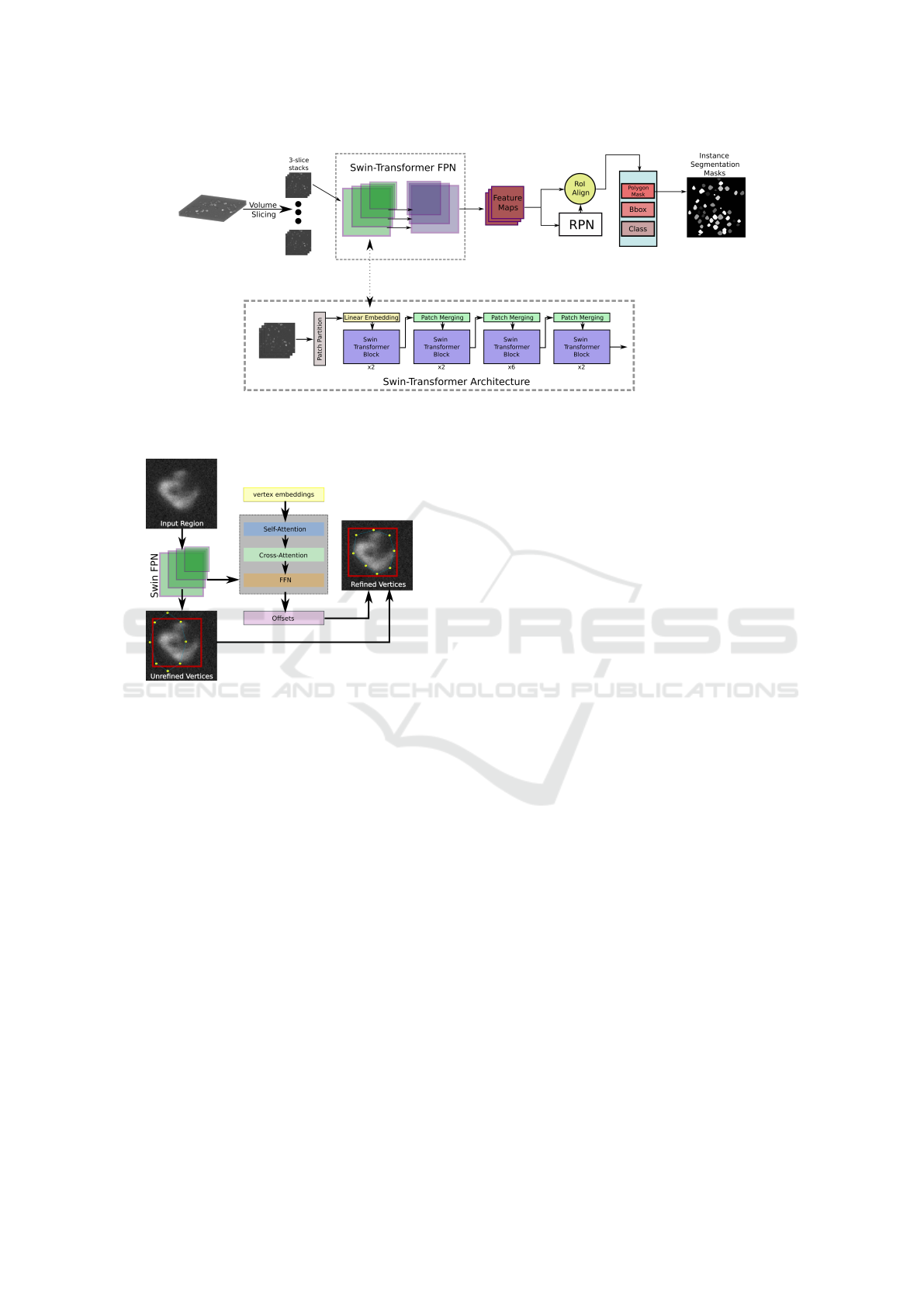
Figure 1: Schematic representation of the employed DL Instance Segmentation Architecture. The Swin-Transformer FPN
Backbone extracts detailed features that are passed on to the Region Proposal Network (RPN) and finally turned into Segmen-
tation Masks by the Box Head which is further described in section 4.3.
Figure 2: System overview of the box head, predicting poly-
gon masks from a set of initial vertices refined by features
extracted using the Swin FPN backbone.
ference between b
p
and ground truth box b
gt
can be
minimized with
L
b
= 1 −CIoU(b
p
, b
gt
) (1)
where L
b
is the point-based unary loss, i.e. the dis-
crepancy between predicted and actual bounding box,
and CIoU is the complete intersection over union.
The distance-aware pairwise lose is composed of two
major components, a local pairwise loss and a global
pairwise loss. The local pairwise loss is based on the
hypothesis that objects boundaries are typically de-
fined by local color variations in an image (Gonzalez,
2009). This idea is expressed in the equation:
L
l p
=
∑
(p,q)∈
ˆ
Ω(i, j)
w
[(i, j),(p,q)]
|U
′
C
(i, j) −U
′
C
(p, q)| (2)
where U
′
C
(·, ·) is a sigmoid-normalized mapping func-
tion expressing the minimal distance from a poly-
gon to a pixel. This function is further explained in
the original proposal of the local pairwise loss (Yang
et al., 2023).
The global pairwise loss is used to reduce the ef-
fects of noise that might introduce unwanted segmen-
tation boundaries due to color changes in the vicin-
ity of noisy pixels. Assuming internal regions of ob-
jects should be nearly homogeneous (Chan and Vese,
2001), the global pairwise loss is formulated as:
L
gp
=
∑
(x,y)∈Ω
||I(x, y) −U
in
||
2
·U
′
C
(x, y)
+
∑
(x,y)∈Ω
||I(x, y) − u
out
||
2
· (1 −U
′
C
(x, y))
(3)
where u
in
and u
out
represent the average image color
inside and outside the polygon, respectively.
The full polygon loss function is then calculated as
a sum of the partial losses, modified with modulated
weight parameters α, β, and γ:
L
polygon
= αL
u
+ βL
l p
+ γL
gp
(4)
In short, L
u
carries the responsibility for enclosing the
polygon into the ground truth box, and L
l p
and L
gp
ensure a proper fit of the polygon along the object
boundary. A schematic representation of the polygon
refinement process is shown in figure 2.
4.4 3D Reconstruction
Following the approach published in (Zhou et al.,
2024) the reconstruction of slice-wise predicted 2D
segmentation masks is handled by a multistep pro-
cess, based on the gradients calculated from 2D seg-
mentations via distance transform. In the specific
case of single-view segmentation masks (i.e. seg-
mentations on slices along one dimension only), the
matching of object instances is conceptually similar to
stitching predictions slice-wise along the depth-axis.
After reconstructing the 3D semantic masks, they are
combined with reconstructed 3D gradients to retrieve
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
856

3D instance segmentation masks using 3D gradient
tracking. As this method is not reliant on complex and
computational expensive deep learning architectures,
the computational resource requirements are entirely
manageable even by lower-end hardware.
4.5 Evaluation Metrics
A large variety of metrics exist in the space of instance
segmentation. While formal, mathematical defini-
tions might differ and as a result numerical disparities
are implied, fundamentally all metrics aim to mea-
sure the relationship between the prediction of a sin-
gle object and the matching ground truth object. Due
to this fundamental similarity, the commonly used
metrics DICE, mean Average Precision, Accuracy,
F1, etc. are highly correlated and reporting multi-
ple can be seen as redundant. To focus on the two
aspects of comparability and expressiveness, the two
metrics SEG (et al., 2017) and Accuracy@X are cho-
sen. Both metrics are based on the Intersection over
Union (IoU) of ground truth (GT) and Predicted (P)
instances, defined as:
IoU (GT, P) =
GT ∩ P
GT ∪ P
(5)
The SEG metric as defined in (et al., 2017) formulates
an instance segmentation metric from this semantic
metric by introducing the condition
|GT ∩ P| >
|GT |
2
(6)
which functions as a matching function. Using this
formulation, each GT object can at most be assigned
one matching predicted object. If a GT object has
a matching predicted object, it is assigned the corre-
sponding IoU value, if it has none, it is assigned a
value of 0. The final SEG score is then calculated
as the mean of all matching values for all GT objects.
While this metric fulfills the requirement of compara-
bility, as it is the official metric employed by the ISBI
Cell Tracking Challenge and comes with an official
implementation that is used for this study, it lacks in
expressiveness. Due to only matching GT objects to
predictions, the SEG value does not cover False Posi-
tive (FP) predictions. Here, Acc@X gives a more ac-
curate estimate. Accuracy for instance segmentation
tasks is defined as:
Acc(GT, P) =
T P
T P + FP + FN
(7)
where T P and FN indicate True Positives and False
Negatives, respectively. Objects are considered TPs
if their IoU score relative to a GT object exceeds a
pre-defined threshold X. Similarly for objects consid-
ered FPs or FNs. The thresholds X are set to val-
ues in the range of [0.1, 0.2, ·· · , 0.9] and correspond-
ing accuracy values are reported with the abbrevia-
tion Acc@X. Next to Acc@X, Precision@X and Re-
call@X are reported as:
Prec(GT, P) =
T P
T P + FP
(8)
Rec(GT, P))
T P
T P + FN
(9)
5 RESULTS
The results reported for this study are split into two
sections, performance achieved on the full dataset and
performance achieved on reduced datasets. For the
full dataset evaluation, the proposed pipeline is com-
pared against two SOTA weakly supervised 2.5D cell
instance segmentation methods, as well as two fully
supervised 2.5D methods. All comparisons are con-
ducted on the same train/test/validation split to en-
sure consistency and comparability of metrics. More
specifically, in the case of few-slice training, only the
train set is modified and test and validation splits are
unchanged.
5.1 Full Dataset Training
The pipeline is trained and evaluated on the full
dataset, split as described in 3. For this, all 170 train-
ing volumes containing a total of 10030 2D image
slices have to be fully annotated with ground truth
bounding boxes.
Table 1: Comparison of metrics for fully supervised
(2.5DCMRCNN (Schmeisser et al., 2024a), KIT-SCHE
(Scherr et al., 2021)) and weakly supervised (Point
(Schmeisser et al., 2024a), Line (Schmeisser et al., 2024b))
2.5D Cell Segmentation algorithms.
Method SEG Superv. Type
2.5DCMRCNN 0.732 full
KIT-SCHE 0.639 full
Line 0.721 weak
Point 0.738 weak
Box (Ours) 0.738 weak
Table 1 shows a comparison between four SOTA
approaches, two weakly and two fully supervised. Al-
though bounding box-only annotation is far more effi-
cient than the two-step weakly supervised approaches
Point and Line, there is no significant reduction in
segmentation performance. This can partly be at-
tributed to the more effective pipeline architecture
How to Box Your Cells: An Introduction to Box Supervision for 2.5D Cell Instance Segmentation and a Study of Applications
857
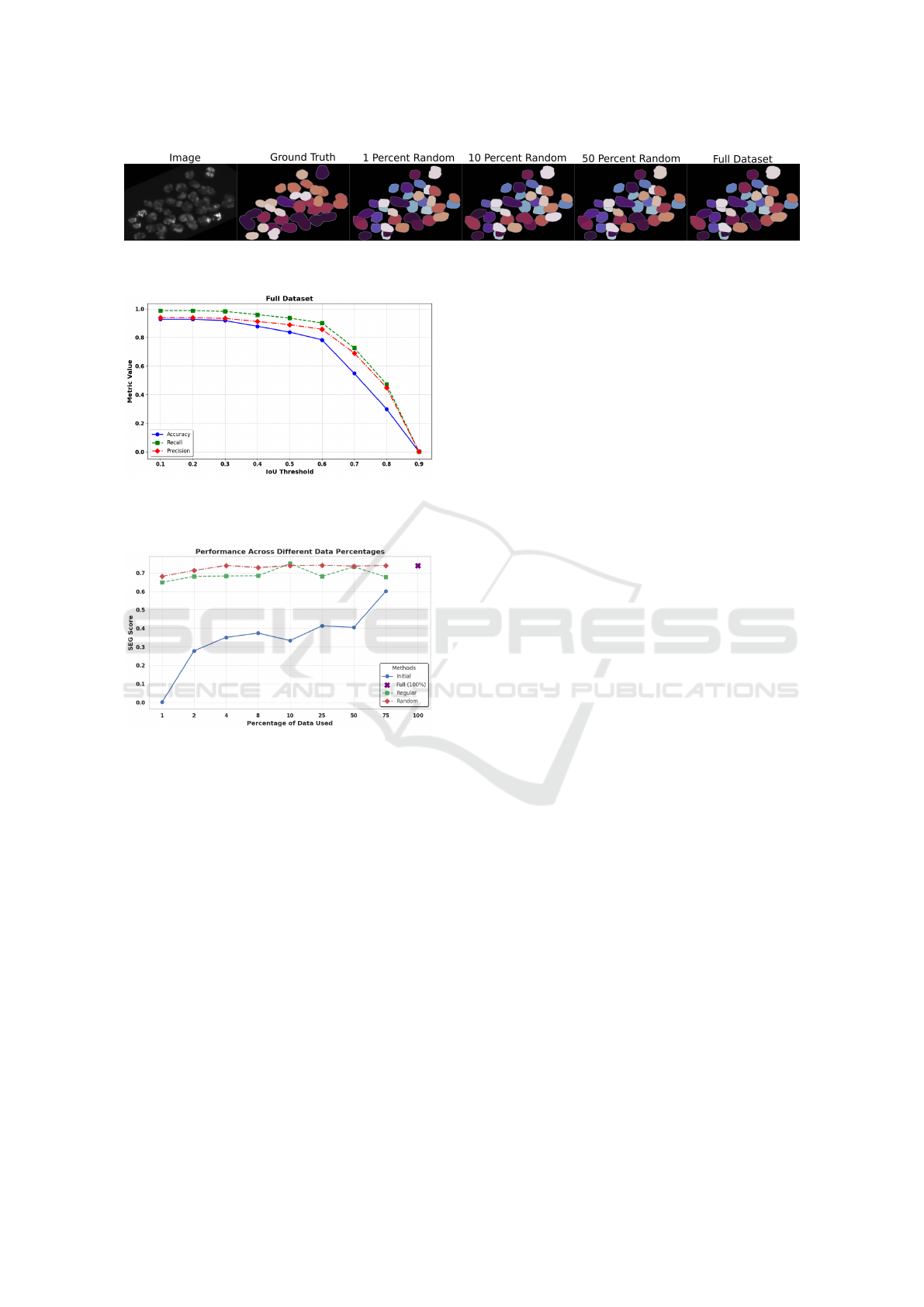
Figure 3: Example 3D segmentation for different slice reduction percentages, compared to ground truth and full dataset
training. Even with 1% of training data, visual differences between segmentation results are minimal.
Figure 4: Precision, Recall, and Accuracy for the bounding-
box supervised 2.5D instance segmentation pipeline trained
on the full dataset.
Figure 5: Performance of the pipeline w.r.t. the SEG metric,
with different dataset percentages kept and different reduc-
tion strategies.
employed in this approach, as well as the extremely
unclear boundaries of cell instances in volumetric mi-
croscopic images which inherently require border ap-
proximation, even when full segmentation masks are
provided during training.
Figure 4 presents an overview of the accuracy, pre-
cision, and recall achieved at different IoU thresholds.
The sharp drop in all three metrics at the IoU thresh-
old of 0.7 is especially noticeable. As even human ex-
pert annotators rarely exceed an IoU of 0.8 for single
cell instance masks (Jelli et al., 2023), this decline is
expected and can be attributed to the high ambiguity
of cell boundaries.
5.2 Few-Slice Training
For few-slice training, the dataset is reduced as de-
scribed in 3.2. With 10, 030 slices in the origi-
nal dataset, this implies that for 1 percent training,
only 100 slices are available as training data. Fig-
ure 5 shows the SEG values achieved by the proposed
method. Using only the initial slices of the dataset,
i.e. directly reducing the number of volumes, yields
the lowest scores for any subset percentage. Conse-
quently, exploiting the capabilities of a 2.5D segmen-
tation algorithm turns out to be highly valuable. In
both cases, regular slice reduction and random slice
reduction, the method is capable of learning complex
features and produces accurate segmentation masks
from very limited data. Even with as few as 100
training images, the proposed pipeline is capable of
achieving SEG scores of 0.649 and 0.680 for regular
and random slice reduction, respectively. Addition-
ally, results for the random slice reduction strategy
start to converge as early as when 4%, or 400 images
are used for training. Higher percentages yield sig-
nificantly diminishing returns, meaning with the ran-
dom slice reduction strategy the training set can be
reduced radically without noticeable loss in segmen-
tation accuracy. Random slice reduction provides the
overall best results, while regular slice extraction only
achieves a higher SEG score at the 10 percent level.
Specifically, in the case of random slice extraction,
further experiments have to be conducted to compute
an average score for multiple runs with differing ran-
dom slice choices.
Accuracy, Precision, and Recall as shown in figure
6 show a similar pattern of the dominating random
slice reduction strategy. Interestingly, lower subset
percentages tend to show better results in the case of
low IoU thresholds, indicating that cells are more ac-
curately detected and located, but less accurately seg-
mented, when fewer training examples are used. The
much steeper decline of Accuracy, Precision, and Re-
call at higher IoU values for training with 1 percent
of the dataset additionally shows the improvement in
robustness as more data becomes available.
5.3 Estimated Time Savings
Few resources in contemporary literature exist that
provide comprehensible studies on the time effort of
annotating 3D microscopic images, due to the time-
and resource intensive nature of the task. The esti-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
858

Figure 6: Precision, Recall, and Accuracy at IoU thresholds in the range of [0.1, 0.2, ··· , 0.9].
mates provided in (Khalid et al., 2022), (Khalid et al.,
2024), and (Schmeisser et al., 2024a), however, pro-
vide a starting point for gauging the time savings pro-
vided by bounding box supervision. With an approx-
imate speed-up of 11x to annotate a cell with bound-
ing boxes instead of a full voxel mask as stated in
(Schmeisser et al., 2024a), this value can be linearly
scaled to time savings when only annotating a fraction
of the dataset. We therefore expect a speed-up of an-
notation time of over 100 times when only 10 percent
of the slices have to be annotated with boxes. This
enormous acceleration of annotation does not come
with any significant reduction in segmentation perfor-
mance, as shown in 5.2.
6 CONCLUSION
This study introduces box supervision to the realm
of 2.5D Cell Segmentation. In contrast to previous
weak supervision approaches for volumetric micro-
scopic images which require a two-step annotation
approach, bounding box annotations can be generated
in a single step. Additionally, bounding box annota-
tion is extremely cost-efficient, reducing annotation
time for a dataset by an estimated 11 times. With the
proposed cell instance segmentation pipeline, bound-
ing box annotations suffice to produce segmentation
masks of comparable quality to other weak supervi-
sion and fully supervised SOTA approaches while be-
ing more resource effective. Next to the introduction
of box supervision, a study was conducted to reduce
the dataset size by up to 100 times, with impressive
results. Using only 1% of the dataset and weak box
annotations, the pipeline produces 3D instance seg-
mentation masks with 92.1% of the SEG score of a
fully supervised SOTA method. With 10% of the an-
notated slices of a full dataset, the proposed segmen-
tation algorithm performs on a level comparable to a
fully supervised method trained on the full dataset.
The combination of efficient bounding box-based an-
notation and slice reduction for training enables re-
searchers to generate ground truth for complex 3D
dataset 100 times faster and reduces the probability
of error occurrences during the annotation process.
Without the need for complex, voxel-wise mask an-
notations for each cell instance and by significantly
reducing the amount of data that has to be labeled,
this approach describes a first step towards collect-
ing, labelling, and analyzing 3D microscopic data on
a much larger scale than previously possible.
ACKNOWLEDGEMENTS
This work is partially funded by SAIL (Sartorius AI
Lab), a collaboration between the German Research
Center for Artificial Intelligence (DFKI) and Sarto-
rius AG.
How to Box Your Cells: An Introduction to Box Supervision for 2.5D Cell Instance Segmentation and a Study of Applications
859

REFERENCES
Arbelle, A., Cohen, S., and Raviv, T. R. (2022). Dual-
task ConvLSTM-UNet for instance segmentation of
weakly annotated microscopy videos. IEEE Transac-
tions on Medical Imaging, 41(8):1948–1960.
Bouyssoux, A., Fezzani, R., and Olivo-Marin, J.-C. (2022).
Cell instance segmentation using z-stacks in digital
cytology. In 2022 IEEE 19th International Sympo-
sium on Biomedical Imaging (ISBI). IEEE.
Cai, Z. and Vasconcelos, N. (2018). Cascade r-cnn: Delving
into high quality object detection. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 6154–6162.
Chan, T. F. and Vese, L. A. (2001). Active contours with-
out edges. IEEE Transactions on image processing,
10(2):266–277.
Chen, T., Ding, C., Zhu, L., Xu, T., Ji, D., Zang, Y., and
Li, Z. (2024). xlstm-unet can be an effective 2d\&
3d medical image segmentation backbone with vision-
lstm (vil) better than its mamba counterpart. arXiv
preprint arXiv:2407.01530.
Dosovitskiy, A. (2020). An image is worth 16x16 words:
Transformers for image recognition at scale. arXiv
preprint arXiv:2010.11929.
Edlund, C., Jackson, T. R., Khalid, N., Bevan, N., Dale,
T., Dengel, A., Ahmed, S., Trygg, J., and Sj
¨
ogren, R.
(2021). LIVECell—a large-scale dataset for label-free
live cell segmentation. Nature Methods, 18(9):1038–
1045.
et al., V. U. (2017). An objective comparison of cell-
tracking algorithms. Nature Methods, 14(12):1141–
1152.
Gonzalez, R. C. (2009). Digital image processing. Pearson
education india.
Jelli, E., Ohmura, T., Netter, N., Abt, M., Jim
´
enez-Siebert,
E., Neuhaus, K., Rode, D. K. H., Nadell, C. D.,
and Drescher, K. (2023). Single-cell segmentation
in bacterial biofilms with an optimized deep learn-
ing method enables tracking of cell lineages and mea-
surements of growth rates. Molecular Microbiology,
119(6):659–676.
Khalid, N., Caroprese, M., Lovell, G., Porto, D. A., Trygg,
J., Dengel, A., and Ahmed, S. (2024). Bounding box
is all you need: Learning to segment cells in 2d micro-
scopic images via box annotations. In Annual Confer-
ence on Medical Image Understanding and Analysis,
pages 314–328. Springer.
Khalid, N., Froes, T. C., Caroprese, M., Lovell, G., Trygg,
J., Dengel, A., and Ahmed, S. (2023). Pace: Point
annotation-based cell segmentation for efficient mi-
croscopic image analysis. In International Confer-
ence on Artificial Neural Networks, pages 545–557.
Springer.
Khalid, N., Schmeisser, F., Koochali, M., Munir, M., Ed-
lund, C., Jackson, T. R., Trygg, J., Sj
¨
ogren, R., Den-
gel, A., and Ahmed, S. (2022). Point2mask: A weakly
supervised approach for cell segmentation using point
annotation. In Medical Image Understanding and
Analysis, pages 139–153. Springer International Pub-
lishing.
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin,
S., and Guo, B. (2021). Swin transformer: Hierar-
chical vision transformer using shifted windows. In
Proceedings of the IEEE/CVF international confer-
ence on computer vision, pages 10012–10022.
Scherr, T., L
¨
offler, K., Neumann, O., and Mikut, R.
(2021). On improving an already competitive segmen-
tation algorithm for the cell tracking challenge-lessons
learned. bioRxiv, pages 2021–06.
Schmeisser, F., Dengel, A., and Ahmed, S. (2024a). Point-
based weakly supervised 2.5 d cell segmentation. In
International Conference on Artificial Neural Net-
works, pages 343–358. Springer.
Schmeisser, F., Thomann, C., Petiot, E., Lovell, G., Carop-
rese, M., Dengel, A., and Ahmed, S. (2024b). A line is
all you need: Weak supervision for 2.5 d cell segmen-
tation. In Annual Conference on Medical Image Un-
derstanding and Analysis, pages 402–416. Springer.
Schmidt, U., Weigert, M., Broaddus, C., and Myers, G.
(2018). Cell detection with star-convex polygons.
Stringer, C., Wang, T., Michaelos, M., and Pachitariu, M.
(2020). Cellpose: a generalist algorithm for cellular
segmentation. Nature Methods, 18(1):100–106.
Wagner, R. and Rohr, K. (2022). Efficientcellseg: Ef-
ficient volumetric cell segmentation using context
aware pseudocoloring.
Weigert, M., Schmidt, U., Haase, R., Sugawara, K., and
Myers, G. (2020). Star-convex polyhedra for 3d ob-
ject detection and segmentation in microscopy. In Pro-
ceedings of the IEEE/CVF winter conference on appli-
cations of computer vision, pages 3666–3673.
Wu, L., Chen, A., Salama, P., Dunn, K., and Delp, E.
(2023). Nisnet3d: Three-dimensional nuclear syn-
thesis and instance segmentation for fluorescence mi-
croscopy images.
Yang, R., Song, L., Ge, Y., and Li, X. (2023). Boxsnake:
Polygonal instance segmentation with box supervi-
sion. In Proceedings of the IEEE/CVF International
Conference on Computer Vision, pages 766–776.
Zhang, Y., Liao, Q., Ding, L., and Zhang, J. (2022).
Bridging 2d and 3d segmentation networks for
computation-efficient volumetric medical image seg-
mentation: An empirical study of 2.5d solu-
tions. Computerized Medical Imaging and Graphics,
99:102088.
Zhao, Z., Yang, L., Zheng, H., Guldner, I. H., Zhang, S.,
and Chen, D. Z. (2018). Deep Learning Based In-
stance Segmentation in 3D Biomedical Images Using
Weak Annotation, pages 352–360. Springer Interna-
tional Publishing.
Zhou, F. Y., Yapp, C., Shang, Z., Daetwyler, S., Marin, Z.,
Islam, M. T., Nanes, B. A., Jenkins, E., Gihana, G. M.,
Chang, B.-J., et al. (2024). A general algorithm for
consensus 3d cell segmentation from 2d segmented
stacks. bioRxiv, pages 2024–05.
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
860
