
Meta-Ensemble Learning for Multi-Trait Optimization in Maize
Breeding: Combining Gradient Boosting, Random Forests, and Deep
Learning with SVM Integration
Dupuy Rony Charles
1,2 a
, Pascal Pultrini
2
and Andrea Tettamanzi
1 b
1
Universit
´
e C
ˆ
ote d’Azur, I3S, Inria, Sophia Antipolis, France
2
Doriane Research Softare & Consulting, Av. Jean Medecin, Nice, France
Keywords:
Plant Breeding, Multi-Trait Selection Indices (MTSI), Meta-Ensemble Machine Learning.
Abstract:
Plant breeding aims to enhance traits such as yield, drought tolerance, and disease resistance. Traditional
Multi-Trait Selection Indices (MTSI) struggle with high-dimensional genomic data and complex trait inter-
actions. We present a meta-ensemble machine learning framework integrating Gradient Boosting, Random
Forest, and Deep Neural Networks (DNNs) with a Support Vector Machine (SVM) meta-model to address
these challenges. This meta-ensemble approach leverages the strengths of multiple algorithms for improved
predictive accuracy and robustness. Experiments on maize datasets show that our meta-ensemble significantly
outperforms traditional MTSI methods and individual machine learning models. The meta-ensemble achieves
superior predictive accuracy and operational efficiency, with a marked reduction in mean squared error (MSE)
and consistent performance across validation sets. This study advances meta-ensemble machine learning in
plant breeding, providing a robust framework for multi-trait selection. Our approach improves trait prediction
reliability and sets a new standard in maize breeding, with potential applications in other crop species, enhanc-
ing agricultural productivity and resilience.
1 INTRODUCTION
Maize (Zea mays L.), one of the world’s most impor-
tant staple crops, is essential for global food security,
livestock feed, and industrial raw materials. However,
contemporary agricultural challenges, including cli-
mate change, pest infestations, and the rising demand
for higher yields, require breeding strategies that are
both more efficient and innovative. Traditional breed-
ing methods, although effective in the past, often
struggle to optimize multiple agronomic traits simul-
taneously, especially when confronted with the com-
plexities of modern breeding programs.
Recent advancements in machine learning (ML)
techniques have emerged as promising solutions to
these challenges. ML excels in its ability to an-
alyze large datasets and identify intricate patterns,
making it a powerful tool for improving the preci-
sion and efficiency of breeding programs. Numer-
ous studies have demonstrated the applicability of
ML in various agricultural domains, including crop
a
https://orcid.org/0009-0006-4879-9933
b
https://orcid.org/0000-0002-8877-4654
yield prediction and phenotypic trait analysis (Ahmed
et al., 2024), (Reddy and Kumar, 2021), (Westhues
et al., 2021), (Chandana and Parthasarathy, 2022), and
(Crossa et al., 2017).
Multi-trait selection indices (MTSI) are crucial for
modern plant breeding, enabling breeders to improve
multiple agronomic traits simultaneously. Traits such
as yield, drought tolerance, disease resistance, and nu-
tritional quality often exhibit complex genetic corre-
lations. Traditional indices, such as those proposed
by (Smith et al., 1981) and (Hazel et al., 1994), have
been fundamental in this field. However, these meth-
ods face limitations when applied to modern genomic
and phenotypic data, which is characterized by high-
dimensionality and non-linear trait interactions. Their
linear nature often constrains their ability to capture
these complexities.
To address these limitations, advanced ML-based
models, including ensemble methods, deep learning,
and support vector machines (SVM), offer a transfor-
mative approach. These models are capable of captur-
ing non-linear relationships and interactions among
traits, leading to more accurate and efficient selection
876
Charles, D. R., Pultrini, P. and Tettamanzi, A.
Meta-Ensemble Learning for Multi-Trait Optimization in Maize Breeding: Combining Gradient Boosting, Random Forests, and Deep Learning with SVM Integration.
DOI: 10.5220/0013191900003890
In Proceedings of the 17th International Conference on Agents and Artificial Intelligence (ICAART 2025) - Volume 3, pages 876-883
ISBN: 978-989-758-737-5; ISSN: 2184-433X
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

strategies. Meta-ensemble models, which combine
the strengths of multiple base models, provide im-
proved prediction accuracy by capturing diverse pat-
terns and interactions in the data.
This paper investigates the application of a meta-
ensemble ML framework in maize breeding, inte-
grating Gradient Boosting, Random Forest, and Deep
Neural Networks (DNNs), with a Support Vector Ma-
chine (SVM) as a meta-model. We demonstrate how
this approach improves the efficiency and accuracy of
multi-trait selection, outperforming traditional MTSI
methods. Our contributions are as follows: (1) pre-
senting a novel ML-based MTSI that outperforms tra-
ditional indices, (2) showcasing its practical imple-
mentation using real-world genomic and phenotypic
data, and (3) providing a comprehensive analysis of
the algorithm’s performance, emphasizing its robust-
ness and scalability.
The remainder of the paper is structured as fol-
lows: Section 2 reviews related literature, Section 3
outlines the methodology, Section 4 presents the ex-
perimental results, and Section 5 discusses the impli-
cations of the findings and future research directions.
2 RELATED WORK
The complexity of improving multiple agronomic
traits simultaneously has driven significant research
into multi-trait selection indices and their evolution.
This section provides context for the need to move
beyond traditional methods and explores the role of
machine learning in addressing these challenges.
Multi-trait selection indices (MTSI) have long
been pivotal in plant breeding, particularly for en-
hancing key agronomic traits such as yield, drought
tolerance, and disease resistance. Classic indices like
the Smith-Hazel index (Smith, 1936), (Hazel, 1943)
and the desired gains index have been instrumental
in selecting for multiple traits concurrently, optimiz-
ing the balance between competing trade-offs. How-
ever, these traditional approaches operate under the
assumption of simple additive correlations between
traits, which limits their capacity to capture the com-
plex genetic and phenotypic relationships inherent in
modern breeding programs (C
´
eron-Rojas and Crossa,
2018).
The advent of high-dimensional genomic data,
generated by next-generation sequencing technolo-
gies, has introduced non-linear relationships and in-
tricate interactions between traits (Mrode, 2014). Tra-
ditional MTSI methods are ill-equipped to manage
these complexities, as they lack the ability to model
interactions across multiple genetic loci and pheno-
typic traits. This limitation is particularly evident in
maize breeding, where traits like yield, moisture con-
tent, and disease resistance are influenced by both ad-
ditive and non-additive genetic factors (Crossa et al.,
2013). Consequently, the limitations of linear meth-
ods necessitate a shift toward more sophisticated ap-
proaches.
Multi-trait selection is integral to modern plant
breeding, allowing breeders to improve several key
traits simultaneously. However, traditional indices of-
ten fail to account for the complexity of genomic and
phenotypic data. The rise of machine learning (ML)
presents a powerful alternative, offering algorithms
that can capture non-linear relationships and complex
trait interactions prevalent in high-dimensional data
(Crossa et al., 2017).
ML techniques have been successfully applied to
genomic selection (GS) and phenotypic trait analy-
sis in various crops, significantly improving upon tra-
ditional methods. Commonly used ML models in-
clude ensemble algorithms such as Random Forests
(Breiman, 2001) and Gradient Boosting Machines
(Friedman, 2001), as well as more advanced mod-
els like Deep Neural Networks (DNNs) (Montesinos-
L
´
opez et al., 2019). These methods excel at uncov-
ering complex interactions between genomic markers
and phenotypic traits, making them highly suitable for
multi-trait selection.
Random Forests and Gradient Boosting are partic-
ularly adept at handling non-linear relationships and
are frequently used in plant breeding to predict phe-
notypic traits (Spindel et al., 2015). These models
combine multiple decision trees to effectively manage
complex trait dependencies and feature importance
estimation. Furthermore, DNNs add value by pro-
viding advanced representation learning, especially in
high-dimensional datasets, enabling the capture of ab-
stract relationships within genomic data (Montesinos-
L
´
opez et al., 2019).
Despite the advantages of individual ML mod-
els, challenges such as overfitting and limited gen-
eralization across environments persist. To mitigate
these issues, meta-ensemble learning approaches have
been developed, combining the strengths of multi-
ple base models to enhance predictive accuracy and
robustness. Meta-ensembles aggregate outputs from
different ML models, improving both the accuracy
of multi-trait selection and the overall efficiency of
breeding programs (Gonz
´
alez-Camacho et al., 2018).
Recent studies in maize breeding have demon-
strated the superiority of meta-ensemble models over
both traditional selection indices and individual ML
models. For example, (Spindel et al., 2015) found
that combining models like Random Forests and
Meta-Ensemble Learning for Multi-Trait Optimization in Maize Breeding: Combining Gradient Boosting, Random Forests, and Deep
Learning with SVM Integration
877

DNNs improved the accuracy of multi-trait predic-
tions in rice, which has parallels in maize breed-
ing. (Gonz
´
alez-Camacho et al., 2018) further empha-
sized that integrating multiple models reduces Mean
Squared Error (MSE) and increases the reliability of
genomic predictions across diverse environments.
Building on this foundation, the current study in-
troduces a novel meta-ensemble framework that com-
bines Gradient Boosting, Random Forests, DNNs, and
an SVM meta-model specifically for maize breed-
ing. Our experiments, conducted on real-world maize
datasets, reveal that this framework outperforms tra-
ditional MTSI methods and individual ML models,
offering significant reductions in MSE and improved
operational efficiency. This research establishes a
new benchmark for genomic selection in maize breed-
ing.
The findings of this study underscore the poten-
tial of meta-ensemble models to optimize breeding
decisions, not only in maize but across a wide range
of crops. The robustness and scalability of this ap-
proach, coupled with its ability to generalize across
environments, position it as a valuable tool for fu-
ture breeding programs aimed at enhancing agricul-
tural productivity and resilience.
Overall, machine learning, and particularly meta-
ensemble approaches, represent a significant ad-
vancement in plant breeding. Traditional methods,
though historically successful, are increasingly un-
able to cope with the complexity of high-dimensional
genomic data and multi-trait interactions. The meta-
ensemble framework proposed in this study offers a
robust and scalable solution to these challenges. Fu-
ture research should explore the integration of envi-
ronmental factors and the application of these meth-
ods to other crop species, maximizing the potential of
ML in agricultural genomics.
3 METHODOLOGY
3.1 Dataset Description
The dataset utilized in this study was sourced from
the Genome to Fields (G2F) initiative,
1
covering the
period from 2018 to 2021. It encompasses a com-
prehensive collection of 4,372 maize lines charac-
terized by 98,027 single nucleotide polymorphism
(SNP) markers. The dataset was collected from 38
diverse locations across the United States, represent-
1
https://datacommons.cyverse.org/browse/iplant/home/
shared/commons repo/curated/GenomesToFields G2F
2016 Data Mar 2018 Accessed on December 27
th
, 2023.
ing a broad spectrum of environmental conditions.
These locations span 27 U.S. cities, contributing to
the robustness and generalizability of the study’s find-
ings.
The primary phenotypic traits analyzed in this
study are Grain yield (RDT), Grain moisture (HUM),
Plant stand (PS), Date of anthesis (ANT), Date of silk-
ing (SILK), and Anthesis-silking interval (ASI).
3.1.1 Data Cleaning and Preprocessing
To ensure the integrity and reliability of the dataset,
the following steps were performed:
• Genotypic Data Cleaning. SNP markers with
null, missing (NA), or inconsistent values were re-
moved, ensuring that only high-quality genotypic
data were used for analysis.
• Phenotypic Data Imputation. Missing values
in phenotypic traits were imputed using the mean
values corresponding to the relevant environmen-
tal conditions (year, location, block, replication).
This step helped mitigate missing data issues
while maintaining data accuracy.
• Outlier Detection. A Subspace Outlier Detection
method was applied to both genomic and pheno-
typic data to identify and remove outliers, enhanc-
ing the dataset’s robustness and ensuring reliable
model performance.
3.1.2 Feature Selection and Engineering
To further refine the dataset for model training,
several feature selection and engineering techniques
were employed:
• SNP Matrix Centering. The SNP matrix was
centered to reduce the influence of rare variants,
allowing for a more balanced contribution of ge-
netic markers to the models.
• Genetic Relationship Matrix (GRM). A genetic
relationship matrix was constructed to capture
population structure and relatedness among the
maize lines. This matrix was integrated across all
analytical methods to account for population-level
correlations.
• Principal Component Analysis (PCA). To ad-
dress the high-dimensional nature of the genomic
data, PCA was applied, retaining components
that explained over 95% of the total variance.
This dimensionality reduction step helped in im-
proving computational efficiency while preserv-
ing critical information.
• Normalization. Both genotypic and phenotypic
data were normalized to ensure equal contribu-
tion from different traits during model training,
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
878
preventing any single feature from dominating the
predictions.
This curated dataset underpins the meta-ensemble
learning framework in this study, facilitating the ex-
ploration of multi-trait optimization in maize breed-
ing. Its rich genomic and phenotypic data enable ad-
vanced model training and provide key insights into
optimizing agronomic traits.
3.2 Multi-Trait Selection Methods &
Models
Optimizing multi-trait selection requires a thorough
evaluation of both conventional and modern ap-
proaches to identify the most effective methods. The
first subsection, Traditional Multi-Trait Selection In-
dex (MTSI) Methods, reviews established techniques
for combining multiple phenotypic traits into a sin-
gle index. These traditional methods provide a bench-
mark for assessing the performance of more advanced
approaches.
Next, Advanced Meta-Ensemble Learning Frame-
work, presents a cutting-edge methodology that com-
bines multiple machine learning models—Gradient
Boosting, Random Forests, Deep Learning, and Sup-
port Vector Machines (SVM). This meta-ensemble
framework leverages the strengths of each model to
potentially improve predictive accuracy and selection
performance.
A comparison of these two approaches will deter-
mine whether the meta-ensemble framework outper-
forms traditional MTSI methods in multi-trait opti-
mization.
3.2.1 Multi-Trait Selection Index Methods
(MTSI)
The Smith-Hazel Index (SHI), also known as the Eco-
nomic Selection Index, and the Genomic Selection
Index, which employs techniques such as Ridge Re-
gression BLUP (rrBLUP), are among the most widely
utilized methods in plant breeding.
3.2.2 Meta-Ensemble Learning Framework
The proposed meta-ensemble machine learning
framework is designed to enhance multi-trait selec-
tion in plant breeding by harnessing the complemen-
tary strengths of various machine learning models.
This framework is tailored to address the complexities
of high-dimensional genomic data and intricate trait
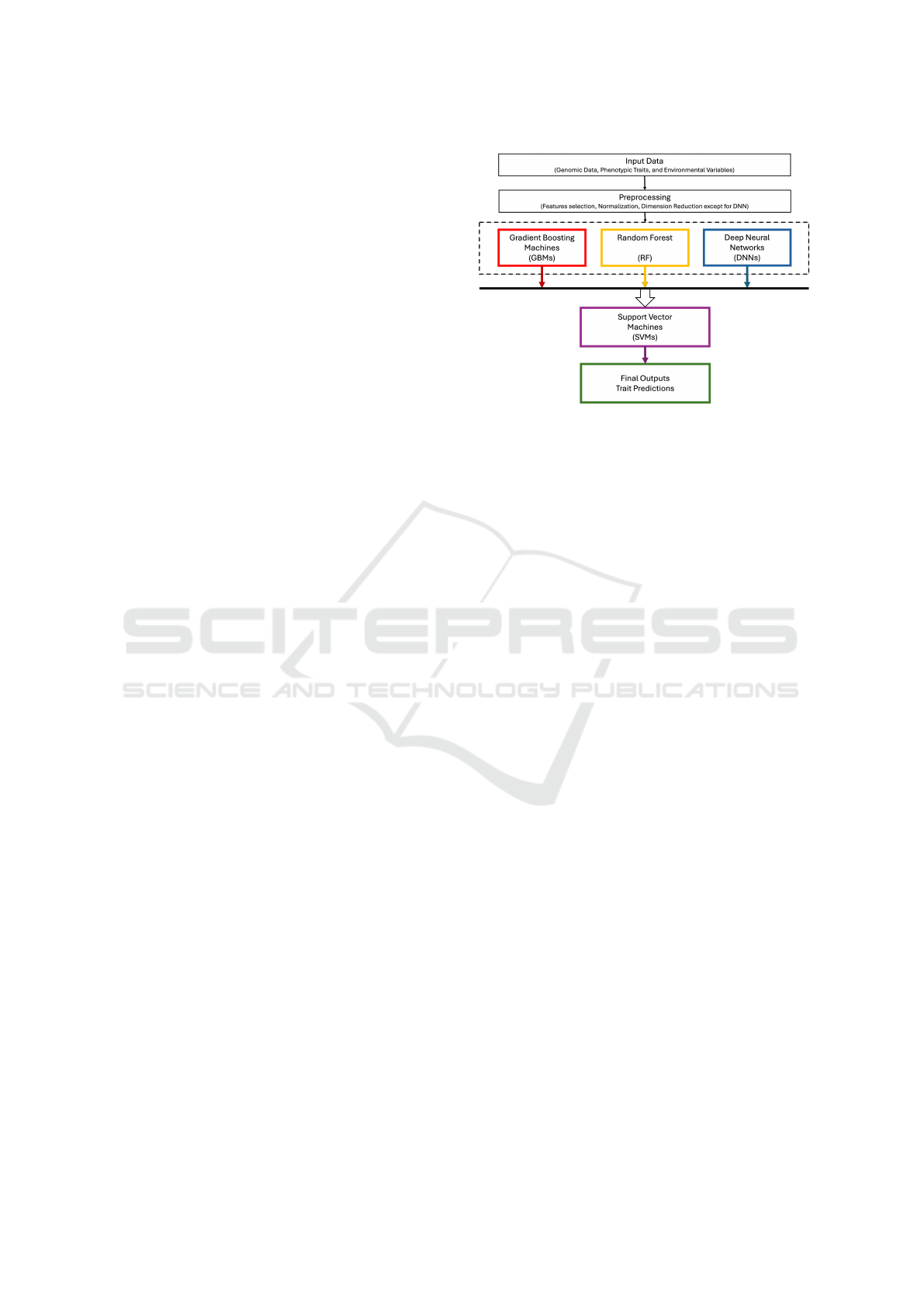
interactions. It consists of two main layers, as shown
in Figure-1: base models and a meta-model. These
layers work in tandem to optimize predictive perfor-
Figure 1: A two-layer Meta-Ensemble Framework for
multi-trait selection.
mance, accuracy, and robustness. The key compo-
nents of the proposed framework are outlined below:
• Base Models
The framework begins with three diverse base
models that capture different aspects of the data
and provide multiple perspectives on the predic-
tion task:
– Gradient Boosting Machines (GBM): are
a powerful ensemble learning method de-
signed to iteratively improve predictive accu-
racy by minimizing residual errors at each step
(Natekin and Knoll, 2013).
– Random Forests (RF): is an ensemble learn-
ing method that constructs multiple decision
trees and combines their predictions to enhance
accuracy and robustness (Genuer et al., 2020).
– Deep Neural Networks (DNN): are machine
learning models that learn hierarchical repre-
sentations of data through multiple layers of in-
terconnected neurons. DNN consist of an in-
put layer, several hidden layers, and an out-
put layer, with each layer containing multiple
neurons. These networks model complex, non-
linear relationships by learning features at vari-
ous levels of abstraction (LeCun et al., 2015).
• Meta-Model
In this ensemble learning framework, the meta-
model is a Support Vector Machines (SVMs) that
acts as a second-level learner.
Support Vector Machines (SVMs): are a pow-
erful classification and regression technique that
seeks to find the optimal hyperplane to maximize
the margin between classes. In ensemble frame-
works, SVMs serve as the second layer, enhancing
model performance through their ability to handle
Meta-Ensemble Learning for Multi-Trait Optimization in Maize Breeding: Combining Gradient Boosting, Random Forests, and Deep
Learning with SVM Integration
879

complex decision boundaries and improve gener-
alization (Sch
¨
olkopf and Smola, 2002).
3.2.3 Overall Architecture of the Framework
The architecture of the meta-ensemble machine learn-
ing framework is depicted in Figure-1, illustrating its
overall structure and component interactions.
1. The input data is first processed by the Pre-
processing Unit to ensure compatibility with the
base models. Dimensionality reduction tech-
niques are applied to optimize the performance of
GBMs and RF models; however, for Deep Neu-
ral Networks (DNNs), no reduction is performed.
This decision is based on the observation that
DNNs achieve superior outcomes when operating
on the dataset’s full dimensionality.
2. The preprocessed data is then fed into the
Base Model Layer, where predictions are gener-
ated independently by GBMs, RF, and DNNs.
3. The predictions from each base model are
aggregated and fed into the Meta-Model Layer
(SVM), produces the final set of predictions for
each trait. These predictions are optimized to bal-
ance the strengths of each base model, resulting
in enhanced overall accuracy and reduced error
rates.
3.3 Performance Metrics
The performance of the meta-ensemble machine
learning framework is evaluated using a combination
of well-established metrics that assess both predictive
accuracy and model robustness: Mean Squared Error
(MSE), R-squared (R²), and Predictive/Selection Ac-
curacy (SA).
To ensure the robustness and generalizability of
the model, 5-fold cross-validation is used, where the
data is divided into 5 subsets.
These metrics, along with cross-validation, pro-
vide a comprehensive evaluation framework, ensur-
ing that the proposed model not only performs well
on the training data but also generalizes effectively to
new, unseen data.
4 EXPERIMENTAL RESULTS
This section presents the evaluation of the base mod-
els, the meta-ensemble framework, and traditional
multi-trait selection indices (MTSI), including the
Smith-Hazel Index (SHI) and Genomic Selection In-
dex (GSI). Two key metrics, MSE and R
2
, are used
Figure 2: Comparison of MSE and R
2
for Grain Yield
(RDT) across SHI, GSI, GBM, RF, DNN, and SVM mod-
els.
Figure 3: Comparison of MSE and R
2
for Grain Moisture
(HUM) across SHI, GSI, GBM, RF, DNN, and SVM mod-
els.
to assess performance in the six primary phenotypic
traits.
The analysis of Grain Yield (RDT) reveals that
GSI achieved the lowest MSE (7.5706), while Gradi-
ent Boosting Machine (GBM) closely followed with
an MSE of 7.6016. In terms of R
2
, the Support Vector
Machine (SVM) outperformed other methods with a
value of 3.0522, demonstrating its superior predictive
accuracy for this trait.
For Grain Moisture (HUM), meta-ensemble mod-
els, particularly Random Forest (RF) and GBM,
showed similar performance with MSE values around
11.52. The SVM model, however, achieved the best
R
2
(0.5101), significantly outperforming the tradi-
tional indices and other machine learning models,
while SHI displayed the lowest predictive accuracy.
For Plant Stand (PS), GSI had the lowest MSE
(3.7087), while SVM achieved the highest R
2
(0.0616), surpassing both traditional indices and base
models in predictive accuracy.
GSI produced the lowest MSE (23.7178) for Date
of Anthesis (ANT), while SVM demonstrated the best
R
2
(11.9788), showcasing its capacity for improved
predictive accuracy despite a higher MSE than tradi-
tional methods.
The SHI model provided the lowest MSE
(24.5171), while the SVM model achieved the high-
est R
2
(0.1708), demonstrating its effectiveness in ex-
plaining trait variance for Date of Silking (SILK).
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
880
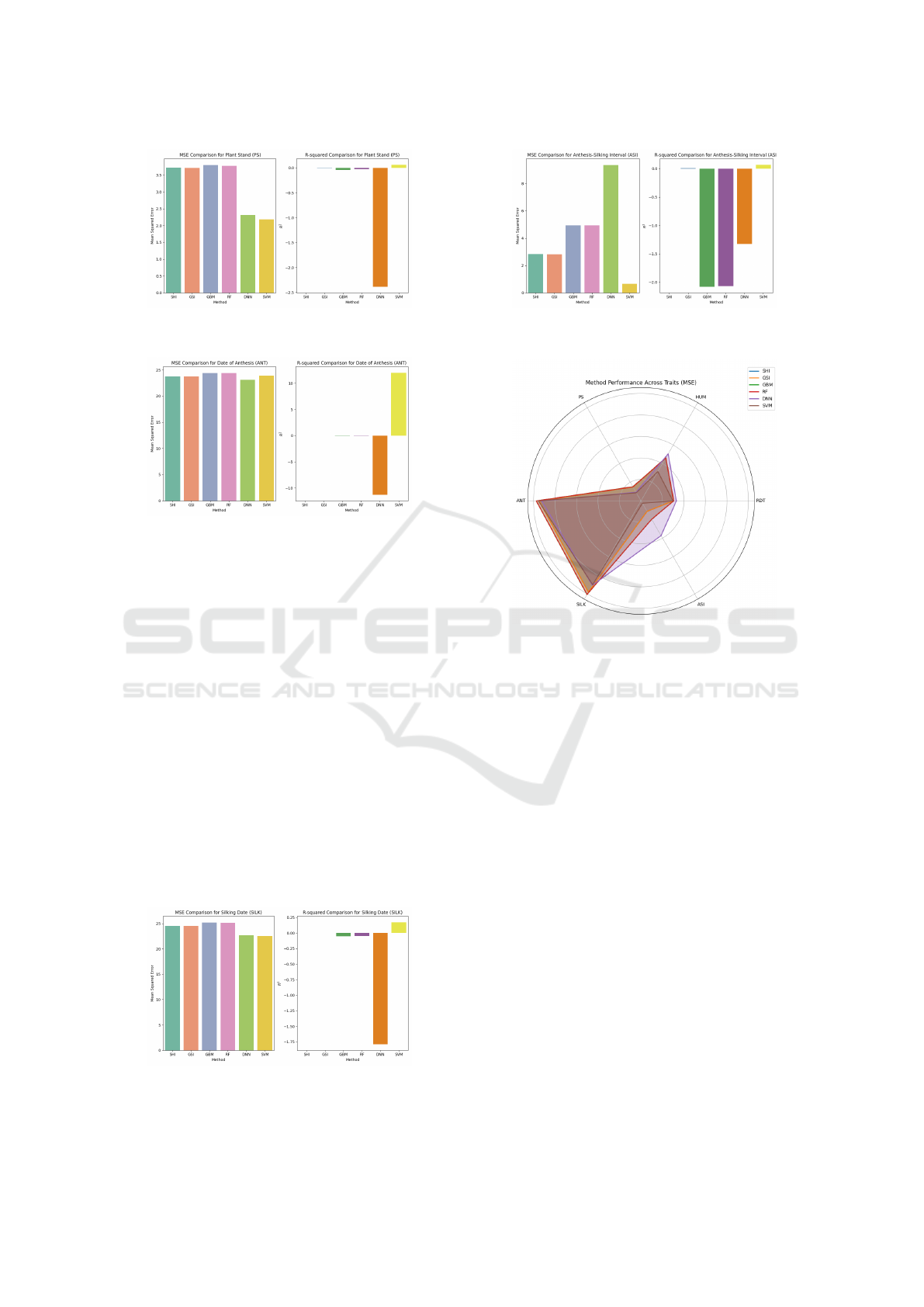
Figure 4: Comparison of MSE and R
2
for Plant Stand (PS)
across SHI, GSI, GBM, RF, DNN, and SVM models.
Figure 5: Comparison of MSE and R
2
for Date of Anthesis
(ANT) across SHI, GSI, GBM, RF, DNN, and SVM mod-
els.
For Anthesis-Silking Interval (ASI), GSI had the
lowest MSE (2.8077) and a slightly positive R
2
, while
SVM demonstrated strong performance with the low-
est MSE among machine learning models (0.6559)
and a positive R
2
(0.0673).
Across all traits, GSI consistently achieved competi-
tive MSE values, while SVM consistently produced
the highest R
2
values, particularly for Grain Yield,
Plant Stand, Date of Anthesis, and Anthesis-Silking
Interval. Machine learning models such as GBM and
RF displayed stable performance but generally did not
outperform the SVM model in predictive accuracy.
The results summarized in Table-1 indicate that
traditional indices like SHI and GSI remain effective
for minimizing error, but SVM offers superior per-
formance in capturing trait variability. These results
suggest that incorporating SVM into multi-trait selec-
Figure 6: Comparison of MSE and R
2
for Date of Silking
(SILK) across SHI, GSI, GBM, RF, DNN, and SVM mod-
els.
Figure 7: Comparison of MSE and R
2
for Anthesis-Silking
Interval (ASI) across SHI, GSI, GBM, RF, DNN, and SVM
models.
Figure 8: Summary of MSE and R
2
across traits.
tion frameworks could significantly improve predic-
tion accuracy.
4.1 Comparison of Methods
• Gradient Boosting Machines (GBM) and Ran-
dom Forest (RF). Both GBM and RF mod-
els demonstrate stable performance across traits.
However, they consistently underperform relative
to SVM, producing higher MSE values and lower
R
2
scores, indicating less effective prediction ac-
curacy.
• Deep Neural Networks (DNN). While DNNs
achieve competitive MSE values for certain traits,
their R
2
scores are significantly lower compared to
SVM, reflecting challenges in capturing complex
trait variability and interactions as effectively.
• Traditional Indices (SHI and GSI). Although
SHI and GSI remain competitive benchmarks in
multi-trait selection, their predictive performance
lags behind SVM. They generally produce higher
MSE values and lower R
2
scores, suggesting that
these indices could be enhanced through integra-
tion with advanced machine learning techniques.
Meta-Ensemble Learning for Multi-Trait Optimization in Maize Breeding: Combining Gradient Boosting, Random Forests, and Deep
Learning with SVM Integration
881
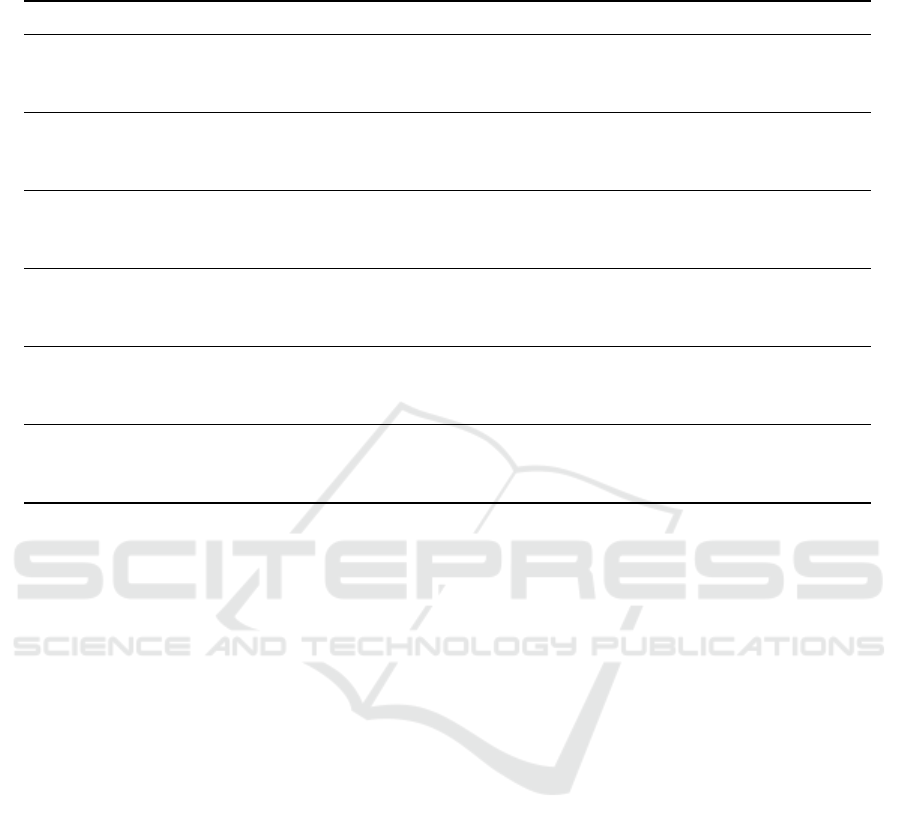
Table 1: Evaluation of Base Models, Meta-ensemble Framework, and Traditional Multi-Trait Selection Indices Using Mean
Squared Error, R-squared, and Selection Accuracy (SA).
Phenotypic Trait Metric SHI GSI GBM RF DNN SVM
Grain Yield MSE 7.6122 7.5706 7.6016 7.6061 8.2437 7.4256
R
2
-0.0007 0.0102 0.0021 0.0009 1.4571 3.0522
SA 0.85 0.87 0.88 0.89 0.81 0.93
Grain Moisture MSE 11.5851 11.5709 11.5241 11.5232 12.6322 7.8811
R
2
0.0000 0.0025 0.0105 0.0107 -2.0417 0.5101
SA 0.80 0.82 0.85 0.86 0.78 0.90
Plant Stand MSE 3.7189 3.7087 3.7952 3.7730 2.3155 2.1829
R
2
0.0000 0.0055 -0.0415 -0.0293 -2.3854 0.0616
SA 0.83 0.84 0.88 0.87 0.75 0.89
Anthesis Date MSE 23.7234 23.7178 24.3741 24.3707 23.1299 23.8810
R
2
0.0000 0.0004 -0.0556 -0.0553 -11.2793 11.9788
SA 0.79 0.81 0.83 0.82 0.70 0.92
Silking Date MSE 24.5171 24.5113 25.1629 25.1266 22.6813 22.5233
R
2
0.0000 0.0004 -0.0534 -0.0504 -1.7887 0.1708
SA 0.82 0.84 0.85 0.86 0.79 0.94
Anthesis-Silking Interval
MSE 2.8212 2.8077 4.9477 4.9407 9.3398 0.6559
R
2
-0.0016 0.0080 -2.0806 -2.0720 -1.3290 0.0673
SA 0.78 0.79 0.83 0.82 0.74 0.90
In summary, SVM consistently outperforms other
methods in capturing trait variability and improving
prediction accuracy across multiple traits. This indi-
cates that incorporating SVM into multi-trait genomic
selection frameworks offers significant potential for
enhancing breeding efficiency. Traditional indices,
though still valuable, may benefit from complement-
ing them with advanced machine learning approaches
to achieve optimal performance in multi-trait selec-
tion.
5 CONCLUSION
The evaluation of base models, meta-ensemble frame-
works, and traditional multi-trait selection indices has
provided key insights into their effectiveness in pre-
dicting agronomic traits. Several important conclu-
sions can be drawn from this analysis:
1. Superior Performance of SVM. The Sup-
port Vector Machine (SVM) consistently outper-
formed all other methods, demonstrating its capa-
bility to achieve the lowest Mean Squared Error
(MSE) and highest R-squared (R
2
) values, partic-
ularly for traits such as Grain Yield (RDT), Plant
Stand (PS), and Anthesis-Silking Interval (ASI).
This highlights its strength in capturing complex
trait variability and delivering accurate predic-
tions.
2. Competitiveness of Traditional Indices. The
Smith-Hazel Index (SHI) and Genomic Selec-
tion Index (GSI) remain reliable benchmarks for
multi-trait selection. However, while competitive,
they fall short of SVM’s performance in minimiz-
ing MSE and maximizing R
2
. Integrating these
traditional indices with advanced machine learn-
ing techniques could further improve their effec-
tiveness.
3. Performance of Other Methods. Gradient
Boosting Machines (GBM) and Random Forest
(RF) provided stable and reliable predictions but
were generally outperformed by SVM. Similarly,
Deep Neural Networks (DNN) showed potential
but exhibited lower R
2
scores, reflecting limita-
tions in capturing trait variability. While useful,
these methods do not surpass the predictive accu-
racy of SVM.
4. Implications for Multi-Trait Selection. The
superior performance of the SVM model sug-
gests that incorporating advanced machine learn-
ing techniques into multi-trait genomic selection
frameworks can significantly enhance prediction
accuracy. These results underscore the poten-
tial for continued exploration of meta-ensemble
frameworks to improve trait selection and breed-
ing strategies.
In summary, the meta-ensemble framework
presents a notable advancement in multi-trait selec-
ICAART 2025 - 17th International Conference on Agents and Artificial Intelligence
882

tion by leveraging the strengths of diverse machine
learning models to significantly enhance prediction
accuracy. The exceptional performance of SVM
within this framework highlights its substantial poten-
tial for future applications in trait prediction.
By integrating a wide range of models, the meta-
ensemble approach not only improves predictive per-
formance but also offers a robust solution for ad-
dressing the complexities of multi-trait genomic se-
lection. This sophisticated methodology promises to
refine breeding strategies and achieve more accurate
trait predictions, advancing precision breeding.
Future research should prioritize integrating en-
vironmental clustering insights and addressing pre-
diction uncertainty to further optimize the meta-
ensemble framework. This involves developing meth-
ods to dynamically adjust predictions based on en-
vironmental conditions and conducting comprehen-
sive uncertainty analyses. Such advancements will
enhance prediction robustness and reliability, leading
to more effective and resilient breeding strategies, ul-
timately boosting agricultural productivity and preci-
sion.
REFERENCES
Ahmed, F. U., Das, A., and Zubair, M. (2024). A ma-
chine learning approach for crop yield and disease
prediction integrating soil nutrition and weather fac-
tors. CoRR, abs/2403.19273.
Breiman, L. (2001). Random forests. Machine learning,
45:5–32.
C
´
eron-Rojas, J. J. and Crossa, J. (2018). Linear selection
indices in modern plant breeding. Springer Nature.
Chandana, C. and Parthasarathy, G. (2022). Efficient ma-
chine learning regression algorithm using na
¨
ıve bayes
classifier for crop yield prediction and optimal utiliza-
tion of fertilizer. Int. J. Perform. Eng., 18(1):47.
Crossa, J., Beyene, Y., Kassa, S., P
´
erez, P., Hickey,
J. M., Chen, C., De Los Campos, G., Burgue
˜
no, J.,
Windhausen, V. S., Buckler, E., et al. (2013). Ge-
nomic prediction in maize breeding populations with
genotyping-by-sequencing. G3: Genes, genomes, ge-
netics, 3(11):1903–1926.
Crossa, J., P
´
erez-Rodr
´
ıguez, P., Cuevas, J., Montesinos-
L
´
opez, O., Jarqu
´
ın, D., De Los Campos, G., Bur-
gue
˜
no, J., Gonz
´
alez-Camacho, J. M., P
´
erez-Elizalde,
S., Beyene, Y., et al. (2017). Genomic selection in
plant breeding: methods, models, and perspectives.
Trends in plant science, 22(11):961–975.
Friedman, J. H. (2001). Greedy function approximation: a
gradient boosting machine. Annals of statistics, pages
1189–1232.
Genuer, R., Poggi, J.-M., Genuer, R., and Poggi, J.-M.
(2020). Random forests. Springer.
Gonz
´
alez-Camacho, J. M., Ornella, L., P
´
erez-Rodr
´
ıguez, P.,
Gianola, D., Dreisigacker, S., and Crossa, J. (2018).
Applications of machine learning methods to genomic
selection in breeding wheat for rust resistance. The
plant genome, 11(2):170104.
Hazel, L., Dickerson, G., and Freeman, A. (1994). The se-
lection index—then, now, and for the future. Journal
of dairy science, 77(10):3236–3251.
Hazel, L. N. (1943). The genetic basis for constructing se-
lection indexes. Genetics, 28(6):476–490.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learn-
ing. nature, 521(7553):436–444.
Montesinos-L
´
opez, O. A., Montesinos-L
´
opez, A.,
Tuberosa, R., Maccaferri, M., Sciara, G., Ammar, K.,
and Crossa, J. (2019). Multi-trait, multi-environment
genomic prediction of durum wheat with genomic
best linear unbiased predictor and deep learning
methods. Frontiers in Plant Science, 10:1311.
Mrode, R. (2014). Linear models for the prediction of ani-
mal breeding values. Cabi.
Natekin, A. and Knoll, A. (2013). Gradient boosting ma-
chines, a tutorial. Frontiers in neurorobotics, 7:21.
Reddy, D. J. and Kumar, M. R. (2021). Crop yield predic-
tion using machine learning algorithm. In 2021 5th In-
ternational Conference on Intelligent Computing and
Control Systems (ICICCS), pages 1466–1470. IEEE.
Sch
¨
olkopf, B. and Smola, A. J. (2002). Learning with ker-
nels: support vector machines, regularization, opti-
mization, and beyond. MIT press.
Smith, H. F. (1936). A discriminant function for plant se-
lection. Annals of eugenics, 7(3):240–250.
Smith, O., Hallauer, A., and Russell, W. (1981). Use of in-
dex selection in recurrent selection programs in maize.
Euphytica, 30:611–618.
Spindel, J., Begum, H., Akdemir, D., Virk, P., Collard, B.,
Redona, E., Atlin, G., Jannink, J.-L., and McCouch,
S. R. (2015). Genomic selection and association map-
ping in rice (oryza sativa): effect of trait genetic archi-
tecture, training population composition, marker num-
ber and statistical model on accuracy of rice genomic
selection in elite, tropical rice breeding lines. PLoS
genetics, 11(2):e1004982.
Westhues, C. C., Mahone, G. S., da Silva, S., Thorwarth,
P., Schmidt, M., Richter, J.-C., Simianer, H., and
Beissinger, T. M. (2021). Prediction of maize pheno-
typic traits with genomic and environmental predic-
tors using gradient boosting frameworks. Frontiers in
plant science, 12:699589.
Meta-Ensemble Learning for Multi-Trait Optimization in Maize Breeding: Combining Gradient Boosting, Random Forests, and Deep
Learning with SVM Integration
883
