
BOVNet: Cervical Cells Classifications Using a Custom-Based Neural
Network with Autoencoders
Diogen Babuc
a
and Darian Onchis¸
b
Computer Science Department, West University of Timis¸oara,
Blvd. Vasile P
ˆ
arvan 4, Timis¸oara, Romania
Keywords:
Neural Network, Machine Learning Techniques, Expert Rules, Cervical Cells, Autoencoders.
Abstract:
Cervical cancer is a major global health challenge being the fourth-most common type of cancer. This em-
phasizes the need for accurate and efficient diagnostic tools that work well for small clinical datasets. This
paper introduces an approach to computer-aided cervical scanning by integrating a custom-based neural net-
work with autoencoders. The proposed architecture, Baby-On-Vision neural network (BOVNet), is tailored
to extract intricate features from cervical images, while the autoencoders mitigate noise and enhance image
quality. State-of-the-art architectures and the BOVNet architecture are trained on three comprehensive data
sets (496, 484, and 1050 samples) that include Pap smear scans and histopathological findings. We demon-
strate the effectiveness of our approach in accurately predicting cervical cancer risk and stratifying patients
into appropriate risk categories. A comparative analysis with existing screening methods indicates the superior
performance of BOVNet in terms of sensitivity (between 90.9% and 98.81% for three data sets), general pre-
dictive accuracy (between 92% and 94.86%), and time efficiency in identifying the increased risk of cervical
abnormalities.
1 INTRODUCTION
One of the biggest threats to world health is cervical
cancer, especially in areas with poor access to med-
ical treatment (Tsikouras et al., 2016). The morbid-
ity and mortality rates related to cervical cancer are
still alarmingly high, despite improvements in screen-
ing programs and diagnostic methods (Bedell et al.,
2020). This highlights the need for more reliable and
easily available diagnostic tools.
Computer-aided diagnostic (CAD) technologies
have become a viable addition to conventional screen-
ing techniques in recent years, with the potential to
increase the efficiency and accuracy of cervical ab-
normality detection (Tekchandani et al., 2022). Exist-
ing CAD systems have certain drawbacks. Many of
them rely on oversimplified algorithms or do not han-
dle issues such as image noise, fluctuation in tissue
appearance, and the subtlety of abnormalities in the
early stages (Athinarayanan et al., 2016). Anomalies
or outliers in the input data often result in poor re-
constructions compared to normal instances (Lehman
et al., 2015).
To overcome these limitations, we suggest creat-
ing the Baby-On-Vision neural network (BOVNet), a
a
https://orcid.org/0009-0000-5126-6480
b
https://orcid.org/0000-0003-4846-3752
unique CAD system designed especially for cervical
diagnosis. The goal of this system is to improve the
accuracy and reliability of cervical scans by combin-
ing autoencoders with cutting-edge machine-learning
approaches. BOVNet aims to provide a reliable and
adaptable tool for medical professionals.
The three major objectives of this study are:
1. A summary of the reasoning for the creation of the
suggested model, offering insights into the diffi-
culties in diagnosing cervical cancer and the ways
CAD systems may be able to help with these dif-
ficulties;
2. Thorough rundown of BOVNet’s features and
components, emphasizing its novel methodol-
ogy and potential benefits for enhancing cervical
health outcomes;
3. Proof of BOVNet’s efficacy and dependability as
a useful addition to the toolkit for diagnosing cer-
vical cancer, which will ultimately help with early
identification, individualized care, and better pa-
tient outcomes.
These targets act as a road map for developing,
implementing, and evaluating the recommended CAD
cervical analysis principle (Figure 1).
Babuc, D. and Onchi¸s, D.
BOVNet: Cervical Cells Classifications Using a Custom-Based Neural Network with Autoencoders.
DOI: 10.5220/0013201500003938
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 11th International Conference on Information and Communication Technologies for Ageing Well and e-Health (ICT4AWE 2025), pages 173-180
ISBN: 978-989-758-743-6; ISSN: 2184-4984
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.
173
Reasoning for the creation of an adjusted model
Proof of BOVNet’s efficacy
Emphasizing the novel methodology
Figure 1: Objectives’ diagram for the cervical cells classifi-
cations.
2 BACKGROUND INFORMATION
AND RELATED WORKS
2.1 Important Image Features
The degree of detail, obtained in an image, is re-
ferred to as its resolution, and it is commonly ex-
pressed in pixels per unit area, such as pixels per
inch or millimeter (Sabottke and Spieler, 2020). More
specific information is provided by higher-resolution
photographs, which is advantageous for identifying
minute anomalies. The way colors are portrayed in
a picture depends on its color space, which can affect
how the properties of the cervical tissue are analyzed
(Wang et al., 2020).
The contrast of an image is the variation in bright-
ness between its various components. Different tissue
types and anomalies are easier to discern from one
another on high-contrast images (Zhang et al., 2020).
The clarity of edges and details in a picture is referred
to as sharpness, sometimes called image sharpness.
Sharper images enable clearer visibility of the fea-
tures of cervical tissue, which is necessary for precise
analysis (Li et al., 2021). Image noise can obfuscate
crucial information and compromise the precision of
diagnostic algorithms. Improving image quality re-
quires evaluating and lowering noise levels using pre-
processing methods such as denoising filters.
The spatial arrangement of the pixel intensities
in an image is called texture, and it tells us some-
thing about the surface properties of the cervical tis-
sue (Chen et al., 2022). Finding unusual patterns or
anomalies might be aided by analyzing textural prop-
erties. The size of an object within a picture on a ref-
erence scale is referred to as its scale. Comprehending
the magnitude of the characteristics of the cervical tis-
sue is crucial to measuring irregularities and contrast-
ing images of various patients or imaging techniques
(Rahaman et al., 2021).
The uniformity of pixel intensities inside an im-
age is measured by homogeneity. Whereas regions of
poor homogeneity may indicate the presence of ab-
normalities or lesions, areas of high homogeneity may
indicate normal tissue. The geometric properties of
the objects in the image, such as size, symmetry, and
irregularity, are described by shape features (Attallah,
2023). The directionality or alignment of texture pat-
terns within an image is referred to as texture orienta-
tion. Evaluating texture orientation can help identify
abnormalities and reveal information about how cer-
vical tissue structures are organized.
2.2 Autoencoders
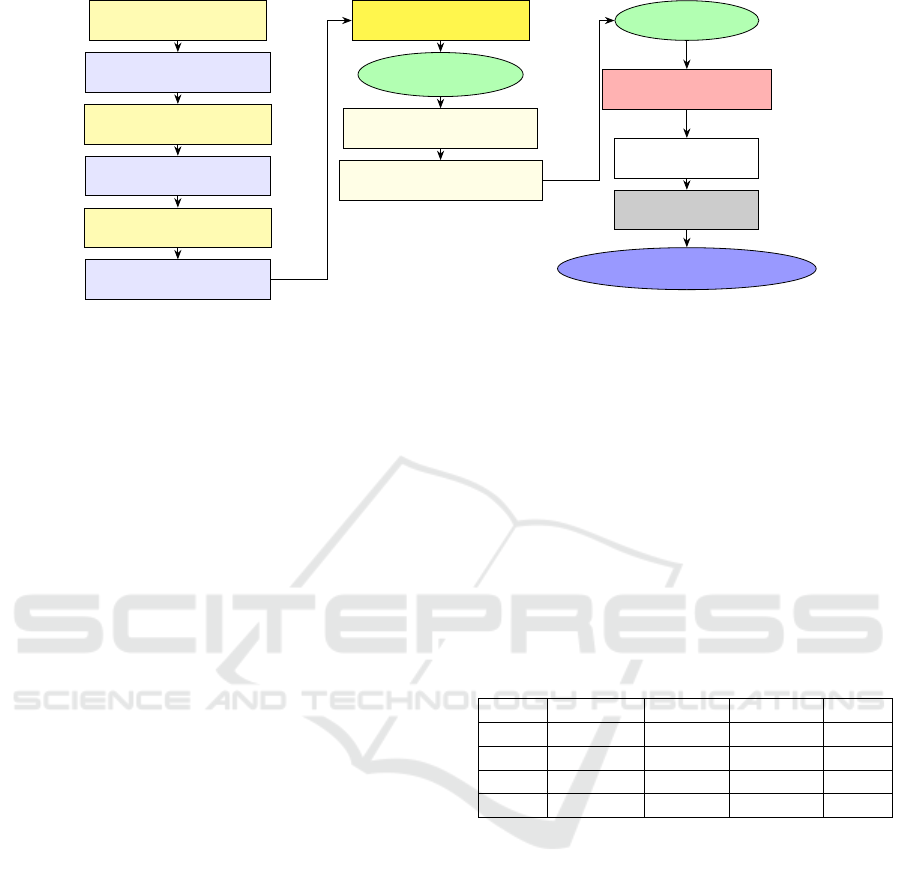
Figure 2: Abnormal carcinoma in situ for cervical cell num-
ber 5749-001.
In cervical diagnosis, autoencoders can be quite
useful, especially when used in conjunction with
computer-aided diagnostic systems such as BOVNet.
From cervical images, autoencoders may effec-
tively extract meaningful features that capture perti-
nent information necessary for a precise diagnosis.
Autoencoders allow for the identification of subtle
patterns and anomalies that would not be visible with
typical image-analysis approaches (Khamparia et al.,
2021).
Due to their high pixel count and intricate spa-
tial information, cervical images are frequently high
dimensional. By mapping the high-dimensional in-
put space to a lower-dimensional latent space, autoen-
coders can achieve dimensionality reduction, keeping
the most important information while removing un-
necessary or noisy features (Adem et al., 2019).
The resolution, contrast, and look of cervical im-
ages obtained in various clinical settings might vary
significantly. The ability of autoencoders to acquire
a uniform representation of cervical images from a
variety of data sets allows diagnostic systems to gen-
eralize and adapt to a variety of heterogeneous data
sources (Nandy et al., 2020) (see Figure 2). This im-
proves the resilience and suitability of the system for
use in actual clinical settings.
The authors of (Adem et al., 2019) investigate two
primary categories of autoencoder-based methods for
cervical diagnosis utilizing Pap smear images: vari-
ational autoencoders (VAE) and denoising autoen-
coders. By reconstructing clear images from noisy
ICT4AWE 2025 - 11th International Conference on Information and Communication Technologies for Ageing Well and e-Health
174

input, denoising autoencoders improve image qual-
ity and enable more precise feature extraction (Bodin
et al., 2017). Conversely, VAE generates images more
reliably and flexibly by learning a probabilistic latent
space representation of the input images.
The intricate and multidimensional nature of med-
ical imaging data can provide overfitting problems
(owing to irrelevant patterns) for autoencoders used
for cervical cell categorization (Xue et al., 2021).
Moreover, class imbalances in medical data and a lack
of labeled data sets might increase the likelihood of
overfitting and impair the model’s capacity for suc-
cessful generalization (Corlan et al., 2023). Careful
regularization methods, data augmentation plans, and
model validation methodologies adapted to the unique
properties of cervical cell images are needed to ad-
dress these problems (Adem et al., 2019).
2.3 Related Works
In the paper (Hussain et al., 2020), authors obtained
a ResNet-50 accuracy value of 91.78% for the test-
ing data, a VGG-16 accuracy value of 87.16%, and
an AlexNet accuracy of 82% for liquid-based cytol-
ogy (AN, 2004) data set. For the conventional data
set (AN, 2004), the authors obtained 92% for ResNet-
50, 87% for VGG-16, and 82% for AlexNet. For the
complete Herlev’s benchmark (pap, 2024), they ob-
tained 89.37% for ResNet-50, an accuracy of 83.37%
for VGG-16, and 80% for AlexNet. The sensitivity,
precision, and average accuracy of all models were
between 79% and 97% on Herlev’s data sets. But
they did not approach the individual data sets and their
outcomes, for a more performant prediction and, af-
terwards, classification.
In the article (Park et al., 2021), authors deter-
mined an area under the ROC curve (ROC-AUC) of
97% for ResNet-50, precision was around 93%, sen-
sitivity around 89%, and accuracy of 91% for data set
described in this paper but not listed. The authors
used a 5-fold cross-validation for calculating all the
evaluation metrics. This paper made a comparison
between ResNet-50 and some shallow models, such
as Extreme Gradient Boost (Chen et al., 2015), Sup-
port Vector Machine (Hearst et al., 1998), and Ran-
dom Forest (Breiman, 2001) but did not analyze other
potentially performant deep learning models such as
AlexNet or VGG-16.
The authors of (Kudva et al., 2020) received a hy-
brid model architecture between AlexNet and VGG-
16 with an accuracy value of 91.46% using the data
set described in (Ribeiro et al., 2016). Accuracy
for AlexNet was 84.31%, sensitivity of 93.50%, and
specificity of 75%. For the VGG-16 individually, they
achieved 84.15% accuracy, a sensitivity of 83.13%,
and specificity of 85.18%. Their hybrid model outper-
formed both deep learning architectures also in terms
of sensitivity and specificity, with 89.16% sensitivity
and 93.83% specificity.
3 OUR APPROACH
In this section, we will discuss the preprocessing stage
of selected data sets, the model’s construction, and
executed experiments.
3.1 Analysis of the Constructed Data
Sets
Carcinoma in situ and normal columnar refer to dif-
ferent types of cervical cell samples in the first data
set (DS1) with 496 observations provided (300 in situ
and 196 normal columnar cells). We modified the cost
function to penalize misclassifications of the minority
class more heavily. The same procedure was applied
on other two data sets. This encouraged the model to
focus more on learning the minority class. We com-
bined these two cell types because we want to pro-
vide the obvious difference between cells such that
our model can classify properly the observations from
the first constructed data set.
There are 484 observations in this second
constructed data set (DS2), which contain nor-
mal/intermediate and superficial cells. There are 148
superficial cancerous cell instances. Cells from this
data set can provide important information about the
condition of the cervix and are often evaluated during
cervical screenings. Using these types of cervical cell
data in the data set, we can train classification models
to differentiate between normal/intermediate and su-
perficial cell types based on their morphological and
pathological features. This enables the development
of automated systems for cervical cell classification,
aiding in the early detection and diagnosis of cervical
abnormalities and diseases.
In the third updated data set (DS3) with 1050 ob-
servations, abnormal dysplastic cells are categorized
into two subtypes: light/moderate (364 + 292 sam-
ples) and severe (394 samples). By categorizing dys-
plastic cells into these subtypes, the data set provides
a more nuanced understanding of the severity of cel-
lular abnormalities present in cervical samples. Re-
searchers can use this information to train classifica-
tion models to differentiate between different grades
of dysplasia.
BOVNet: Cervical Cells Classifications Using a Custom-Based Neural Network with Autoencoders
175

3.2 Model Construction
BOVNet’s architecture, with its series of convolu-
tional and pooling layers, is well-suited for extracting
hierarchical features from images. This hierarchical
feature extraction capability is crucial for distinguish-
ing between different types of cervical cells, which
may exhibit subtle variations in appearance. The
ReLU activation function used throughout BOVNet
introduces non-linearity into the model, enabling it
to learn complex decision boundaries between dif-
ferent cell types. This is important for handling the
potentially non-linear relationships present in cervi-
cal cell images (Corlan et al., 2024). The inclusion
of a rule-based layer in BOVNet provides the flex-
ibility to incorporate domain-specific knowledge or
constraints into the classification process. This can
be particularly valuable in medical diagnosis tasks
where certain rules or guidelines are established by
experts (Babuc et al., 2024). The loss of Tversky focal
points emphasizes the importance of correctly classi-
fying difficult or misclassified examples by introduc-
ing a focal parameter that controls the weight of hard
examples (Abraham and Khan, 2019).
BOVNet process begins with the input shape spec-
ification and continues through several convolutional
layers to improve the network’s capacity to extract
hierarchical information from input pictures. Sub-
sequently, a convolutional autoencoder component is
shown, which consists of an encoder for input data
compression into a latent space and a decoder for data
reconstruction. This addition highlights the model’s
complex feature learning. Through this process, au-
toencoders play a crucial role in dimensionality re-
duction, facilitating the extraction of meaningful rep-
resentations from the data. This procedure will cap-
ture the essential features of the images in a lower-
dimensional space. Modifications to the latent space
involve altering its distribution to enhance the gener-
ative capabilities of the model through regularization
methods. The network’s interpretability and general-
ization skills are further enhanced by the integration
of a layer normalization process.
The first layer of this architecture is convolutional
and applies 64 filters to the input image and uses the
ReLU activation function. Let I be the input image, W
be the filter weights, b be the bias, and σ be the ReLU
activation function (see (1)). The output feature map
O is computed as:
O = σ(W · I + b) (1)
This layer extracts 64 different features from the input
image using convolution. ReLU is chosen as the acti-
vation function to introduce non-linearity and sparsity
to the network. The next layer performs max pooling
with a pool size of 2 × 2 and a stride of 2 × 2. Max
pooling operation selects the maximum value within
each 2 × 2 region of the input feature map. Max
pooling reduces the spatial dimensions of the feature
maps, leading to translation invariance and compu-
tational efficiency. Similar to the first convolutional
layer, this layer applies 128 filters with ReLU activa-
tion. Increasing the number of filters allows the net-
work to learn more complex features from the input.
The same principle is applied for following convolu-
tional and max pooling layers. The Flatten layer flat-
tens the 3D feature maps into a 1D vector, preparing
them for input into the fully connected layers. This is
a necessary step in transitioning from convolutional
layers to fully connected layers.
Fully connected layer with 128 neurons and ReLU
activation follows after the Flatten layer (see Fig-
ure 3). This layer introduces non-linearity and learns
high-level representations of the features extracted by
convolutional layers. After the output of the dense
layer is obtained, it is concatenated with directly fed
into the rule-based layer. The rule-based layer eval-
uates the input data based on predefined rules and
makes a decision: malignant or benign (with RB
Layer M/B). This decision is combined with the dense
layer (through weighted combination) to produce the
final classification outcome. Focal Tversky loss can
lead to better performance, especially in tasks where
class imbalance and minimizing false negatives are
important considerations (Abraham and Khan, 2019).
Before all convolutional layers of BOVNet, we in-
troduced an encoder module consisting of convolu-
tional layers followed by max-pooling layers. The
encoder compresses the input cervical cell images
into a lower-dimensional latent space representation.
The output of the encoder serves as the input to both
BOVNet and the decoder module. After the fully con-
nected layers of BOVNet, we added a decoder mod-
ule comprising convolutional transpose layers. The
decoder aims to reconstruct the original input images
from the latent space representation learned by the en-
coder. Reconstruction loss guides the learning pro-
cess, encouraging the autoencoder to capture mean-
ingful features in the latent space. We prefer autoen-
coders (AE) instead of VAE because the primary goal
is to learn a compact and dense representation of the
input data without explicitly modeling its probabil-
ity distribution. AE’s architecture consists of an en-
coder network that compresses the input data into a
latent representation and a decoder network that re-
constructs the original input from this representation.
This simplicity in architecture and training procedure
makes AE suitable for tasks such as dimensional-
ity reduction, feature learning, and data denoising.
ICT4AWE 2025 - 11th International Conference on Information and Communication Technologies for Ageing Well and e-Health
176
Conv2D (64, ReLU)
MaxPooling2D (2, 2)
Conv2D (128, ReLU)
MaxPooling2D (2, 2)
Conv2D (192, ReLU)
MaxPooling2D (2, 2)
Conv2D (32, ReLU)
Latent Space
Conv2D T (64, ReLU)
Conv2D T (3, Sigmoid)
Flatten
Dense (128, ReLU)
RB Layer M/B
Layer Norm.
Focal loss, 0.7, 0.3, 1
Figure 3: Components of the adjusted neural network model, BOVNet.
For the implementation we used five epochs. Using
five epochs for autoencoders is a practical choice to
balance capturing essential features while mitigating
overfitting, particularly for smaller datasets or sim-
pler deep models. Our main principle functions like a
teachable machine that categorizes cervical cells and
distinguishes classes for the training part. This pro-
cess continues with testing the model on the intro-
duced images. The results showed a well-constructed
model that surpasses other deep learning architec-
tures.
However, this architecture has some limitations.
Interpretability of the taught representations may suf-
fer while the model performs better in terms of clas-
sification accuracy. It can be challenging to compre-
hend the precise characteristics that the autoencoder
learned and how they apply to the classification. Cer-
vical cell classification may benefit from consider-
ing global contextual information within the image.
BOVNet’s architecture may not effectively capture
long-range dependencies in the images, potentially
limiting its performance.
4 RESULTS AND DISCUSSION
The data shown displays the performance metrics
of several deep learning models (BOVNet, ResNet,
VGG-16, and AlexNet) on three separate data sets
with various cervical cell types. As performance eval-
uation metrics, we selected accuracy, sensitivity, pre-
cision, and ROC-AUC (Pal et al., 2021). We cal-
culated the average for each performance evaluation
metrics after running the application for 10 times.
The percentage of correctly determined instances
among all instances is known as accuracy. It is a
key performance indicator for assessing a classifi-
cation model’s overall effectiveness. However, the
accuracy alone could not give a clear view of the
performance of the model in unbalanced data sets
when one class predominates over the others (nor-
mal samples outnumber abnormal samples, for exam-
ple) (William et al., 2018). Sensitivity quantifies the
percentage of real positive cases (such as dysplastic
or malignant samples) that the model accurately de-
tects. In order to minimize false negatives, discover
cervical abnormalities early, and ensure that aber-
rant cases are not missed, high sensitivity is essen-
tial for cervical diagnostics (Sellamuthu Palanisamy
et al., 2022). The percentage of real negative cases
Table 1: Performance evaluation metrics for three state-of-
the-art model and the proposed model, BOVNet, obtained
for the DS1 data set.
% BOVNet ResNet AlexNet VGG
Sens. 98.81 96.39 96.05 96.3
Prec. 91.21 88.89 82.95 86.67
Acc. 94.86 92.49 89.02 91.23
AUC 93.18 94.44 94.3 93.33
(such as normal samples) that the model correctly
detects is known as specificity. When abnormalities
are absent, a low percentage of false positives is in-
dicated by high specificity, which is crucial for pre-
venting unnecessary treatments or interventions (Sell-
amuthu Palanisamy et al., 2022). The precision metric
quantifies the percentage of accurately identified pos-
itive cases among all cases that the model predicts to
be positive. It illustrates the model’s capacity to pre-
vent false positives and is especially crucial in situa-
tions when incorrectly classifying positive cases may
result in serious repercussions, including suggesting
needless follow-up procedures or treatments (Sompa-
wong et al., 2019).
For the first data set, DS1, when compared to other
models, BOVNet has the highest ROC-AUC score,
accuracy, sensitivity, and precision (Table 1). This
suggests that BOVNet distinguishes between cancer
BOVNet: Cervical Cells Classifications Using a Custom-Based Neural Network with Autoencoders
177
1 2 3 4
5 6
7 8 9 10 11 12 13 14
15
0.8
0.85
0.9
0.95
Data portion
Accuracy
BOVNet
ResNet-50
AlexNet
VGG-16
Figure 4: K-Fold cross-validation accuracy on 15 data portions, on DS1 data set.
100
200
300
82.44
86.18
114.3
92.41
56.7
92.41
75.2
53.45
223.4
332.4
236.8
252.3
DS1 DS2 DS3
Execution time (s)
BOVNet ResNet-50 AlexNet VGG-16
Figure 5: Execution times for models’ construction, training part, classification, and performance evaluation metrics for DS1,
DS2, and DS3 data sets.
in situ cells and normal columnar cells with high ac-
curacy. Given its great sensitivity and precision, it
appears to be able to minimize false positives while
efficiently identifying true positive cases. ResNet,
AlexNet, and VGG-16 nevertheless manage to ob-
tain respectable ROC-AUC scores and accuracy. All
things considered, BOVNet seems to be the model
that performs the best on this data set, suggesting that
it is capable of reliably identifying different types of
cervical cells.
For the second data set, DS2, BOVNet contin-
ues to outperform other models in terms of accuracy
(92%), sensitivity (90.9%), and precision (93.33%).
However, its ROC-AUC score is lower compared to
the previous data set (87.85%). VGG-16 achieves
similar accuracy to BOVNet but with slightly lower
sensitivity and precision. ResNet and AlexNet show
decreased performance compared to the previous
data set, indicating potential challenges in classify-
ing intermediate and superficial cell types accurately.
BOVNet still maintains its superiority in classifying
cervical cells on this data set, although the drop in
ROC-AUC suggests that it may struggle with dis-
tinguishing between intermediate and superficial cell
types.
BOVNet maintains its high accuracy (94.44%),
sensitivity (91.38%), and precision (98.15%), also for
the third data set, DS3, indicating its effectiveness in
distinguishing between different dysplastic cell types.
In this data set, BOVNet demonstrates its robustness
in classifying dysplastic cell types accurately, partic-
ularly with high precision, suggesting its potential
clinical utility in identifying severe dysplastic cells,
which are critical for early intervention and treatment.
ICT4AWE 2025 - 11th International Conference on Information and Communication Technologies for Ageing Well and e-Health
178

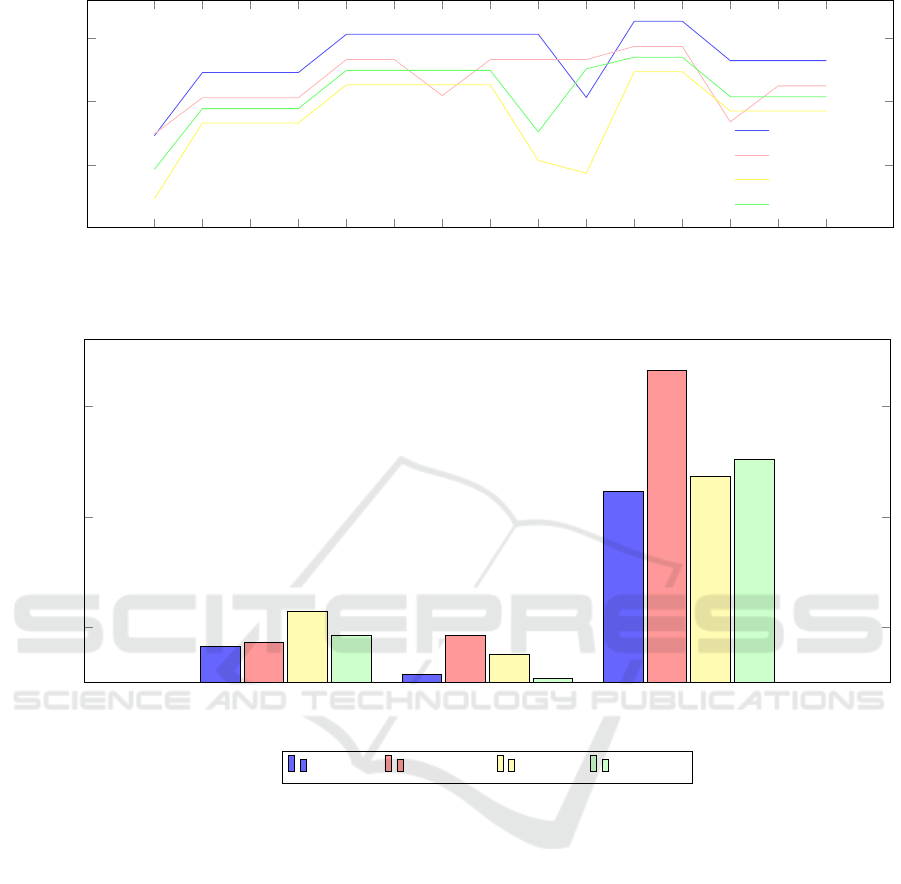
In the context of a 15-fold cross-validation system
applied to DS1, BOVNet demonstrated exceptional
performance across all but the first and tenth folds,
where ResNet-50 showed superior performance. No-
tably, BOVNet consistently achieved accuracy rates
ranging from 90% to 97% (see Figure 4).
Execution times were measured on a local ma-
chine equipped with an Intel Core i7, 12th generation
processor. All the models used the same setup. Com-
pared to alternative models, BOVNet often shows
shorter execution durations across all data sets (see
Figure 5). This suggests that, despite its complexity,
the architecture of BOVNet uses computing resources
rather effectively. ResNet and AlexNet consistently
exhibit higher execution times compared to BOVNet,
especially on the data sets containing a larger number
of classes. This suggests that their deeper architec-
tures and higher parameter counts result in longer in-
ference times. VGG-16 shows varied execution times
across different data sets.
The theoretical explanation for BOVNet’s effec-
tiveness lies in its ability to learn hierarchical features
through encoders, decoders, and convolutional lay-
ers, layers’ normalization and effectively fuse these
features for accurate classification, as evidenced by
its high sensitivity, precision, and ROC-AUC scores.
However, ResNet-50 excels at the best ROC-AUC re-
sult for the first data set, with 94.44%. ROC-AUC
offers a thorough assessment of the model’s perfor-
mance over all potential thresholds and is especially
helpful for evaluating the trade-off between sensitiv-
ity and specificity (Kanavati et al., 2022).
5 CONCLUSIONS
This research has provided significant insights into
the field of cervical cell classification. By leveraging a
novel approach that combines domain-specific knowl-
edge with advanced machine learning techniques, we
have demonstrated the potential for more accurate and
efficient classification of cervical cells.
From the model’s creation, procedures, and re-
sults, BOVNet emerges as a robust and efficient deep
learning architecture for the classification of cervi-
cal cell types. Across multiple data sets and eval-
uation metrics, BOVNet consistently outperforms or
matches the performance of other well-known archi-
tectures like ResNet-50, VGG-16, and AlexNet. This
highlights its suitability for medical image analysis
tasks, particularly in the context of cervical cell clas-
sification. One notable strength of BOVNet is its
efficiency, as evidenced by its relatively low execu-
tion times compared to other models. This efficiency
makes BOVNet an attractive choice for real-world ap-
plications where computational resources are limited
or real-time inference is crucial.
However, the analysis also underscores the im-
portance of considering data set-specific character-
istics. Although BOVNet generally performs well
across various data sets, there are instances where
other models, such as ResNet-50, exhibit superior
performance.
ACKNOWLEDGEMENT
The authors thank the West University of Timis¸oara
for the resources provided and Teodor-Florin Fortis¸
for suggestions.
REFERENCES
(2024). Pap smear data sets, last accessed 8 feb 2024, data
sets.
Abraham, N. and Khan, N. M. (2019). A novel focal tver-
sky loss function with improved attention u-net for le-
sion segmentation. In 2019 IEEE 16th international
symposium on biomedical imaging (ISBI 2019), pages
683–687. IEEE.
Adem, K., Kilic¸arslan, S., and C
¨
omert, O. (2019). Classi-
fication and diagnosis of cervical cancer with stacked
autoencoder and softmax classification. Expert Sys-
tems with Applications, 115:557–564.
AN, C. (2004). Liquid-based cytology and conventional
cervical smears: A comparison study in an asian
screening population. Cancer, 102:200–201.
Athinarayanan, S., Srinath, M., and Kavitha, R. (2016).
Computer aided diagnosis for detection and stage
identification of cervical cancer by using pap smear
screening test images. ICTACT Journal on Image &
Video Processing, 6(4).
Attallah, O. (2023). Cervical cancer diagnosis based on
multi-domain features using deep learning. Applied
Sciences, 13(3):1916.
Babuc, D., Ivascu, T., Ardelean, M., and Onchis, D.
(2024). Bionnica: A deep neural network architecture
for colorectal polyps’ premalignancy risk evaluation.
medRxiv, pages 2024–06.
Bedell, S. L., Goldstein, L. S., Goldstein, A. R., and Gold-
stein, A. T. (2020). Cervical cancer screening: past,
present, and future. Sexual medicine reviews, 8(1):28–
37.
Bodin, E., Malik, I., Ek, C. H., and Campbell, N. D.
(2017). Nonparametric inference for auto-encoding
variational bayes. arXiv preprint arXiv:1712.06536.
Breiman, L. (2001). Random forests. Machine learning,
45:5–32.
Chen, K., Wang, Q., and Ma, Y. (2022). Cervical optical
coherence tomography image classification based on
BOVNet: Cervical Cells Classifications Using a Custom-Based Neural Network with Autoencoders
179

contrastive self-supervised texture learning. Medical
Physics, 49(6):3638–3653.
Chen, T., He, T., Benesty, M., Khotilovich, V., Tang, Y.,
Cho, H., Chen, K., Mitchell, R., Cano, I., Zhou, T.,
et al. (2015). Xgboost: extreme gradient boosting. R
package version 0.4-2, 1(4):1–4.
Corlan, A.-S., Diogen, B., Flavia, C., and Darian, O. (2023).
Prediction and classification models for hashimoto. In
Endocrine Abstracts, volume 90. Bioscientifica.
Corlan, A.-S., Diogen, B., Flavia, C., Vlad, M., Balas, M.,
Golu, I., Amzar, D.-G., and Darian, O. (2024). An ar-
tificial intelligence system for estimating the improve-
ment of clinical and paraclinical parameters after ther-
apy in pituitary tumors. In Endocrine Abstracts, vol-
ume 99. Bioscientifica.
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J., and
Scholkopf, B. (1998). Support vector machines. IEEE
Intelligent Systems and their applications, 13(4):18–
28.
Hussain, E., Mahanta, L. B., Das, C. R., and Talukdar, R. K.
(2020). A comprehensive study on the multi-class
cervical cancer diagnostic prediction on pap smear
images using a fusion-based decision from ensemble
deep convolutional neural network. Tissue and Cell,
65:101347.
Kanavati, F., Hirose, N., Ishii, T., Fukuda, A., Ichihara,
S., and Tsuneki, M. (2022). A deep learning model
for cervical cancer screening on liquid-based cytol-
ogy specimens in whole slide images. Cancers,
14(5):1159.
Khamparia, A., Gupta, D., Rodrigues, J. J., and de Al-
buquerque, V. H. C. (2021). Dcavn: Cervical can-
cer prediction and classification using deep convolu-
tional and variational autoencoder network. Multime-
dia Tools and Applications, 80:30399–30415.
Kudva, V., Prasad, K., and Guruvare, S. (2020). Hybrid
transfer learning for classification of uterine cervix
images for cervical cancer screening. Journal of digi-
tal imaging, 33:619–631.
Lehman, C. D., Wellman, R. D., Buist, D. S., Kerlikowske,
K., Tosteson, A. N., Miglioretti, D. L., Consor-
tium, B. C. S., et al. (2015). Diagnostic accuracy
of digital screening mammography with and without
computer-aided detection. JAMA internal medicine,
175(11):1828–1837.
Li, P., Liang, J., and Zhang, M. (2021). A degradation
model for simultaneous brightness and sharpness en-
hancement of low-light image. Signal Processing,
189:108298.
Nandy, A., Sathish, R., and Sheet, D. (2020). Identifica-
tion of cervical pathology using adversarial neural net-
works. arXiv preprint arXiv:2004.13406.
Pal, A., Xue, Z., Befano, B., Rodriguez, A. C., Long, L. R.,
Schiffman, M., and Antani, S. (2021). Deep metric
learning for cervical image classification. IEEE Ac-
cess, 9:53266–53275.
Park, Y. R., Kim, Y. J., Ju, W., Nam, K., Kim, S., and
Kim, K. G. (2021). Comparison of machine and
deep learning for the classification of cervical cancer
based on cervicography images. Scientific Reports,
11(1):16143.
Rahaman, M. M., Li, C., Yao, Y., Kulwa, F., Wu, X., Li, X.,
and Wang, Q. (2021). Deepcervix: A deep learning-
based framework for the classification of cervical cells
using hybrid deep feature fusion techniques. Comput-
ers in Biology and Medicine, 136:104649.
Ribeiro, E., Uhl, A., Wimmer, G., H
¨
afner, M., et al.
(2016). Exploring deep learning and transfer learning
for colonic polyp classification. Computational and
mathematical methods in medicine, 2016.
Sabottke, C. F. and Spieler, B. M. (2020). The effect of
image resolution on deep learning in radiography. Ra-
diology: Artificial Intelligence, 2(1):e190015.
Sellamuthu Palanisamy, V., Athiappan, R. K., and Na-
galingam, T. (2022). Pap smear based cervical cancer
detection using residual neural networks deep learning
architecture. Concurrency and Computation: Practice
and Experience, 34(4):e6608.
Sompawong, N., Mopan, J., Pooprasert, P., Himakhun, W.,
Suwannarurk, K., Ngamvirojcharoen, J., Vachiramon,
T., and Tantibundhit, C. (2019). Automated pap smear
cervical cancer screening using deep learning. In 2019
41st Annual International Conference of the IEEE En-
gineering in Medicine and Biology Society (EMBC),
pages 7044–7048. IEEE.
Tekchandani, H., Verma, S., Londhe, N. D., Jain, R. R.,
and Tiwari, A. (2022). Computer aided diagnosis sys-
tem for cervical lymph nodes in ct images using deep
learning. Biomedical Signal Processing and Control,
71:103158.
Tsikouras, P., Zervoudis, S., Manav, B., Tomara, E., Ia-
trakis, G., Romanidis, C., Bothou, A., and Galazios,
G. (2016). Cervical cancer: screening, diagnosis and
staging. J buon, 21(2):320–325.
Wang, Z., Chen, J., and Hoi, S. C. (2020). Deep learning
for image super-resolution: A survey. IEEE trans-
actions on pattern analysis and machine intelligence,
43(10):3365–3387.
William, W., Ware, A., Basaza-Ejiri, A. H., and Obun-
goloch, J. (2018). A review of image analysis and
machine learning techniques for automated cervical
cancer screening from pap-smear images. Computer
methods and programs in biomedicine, 164:15–22.
Xue, Z., Guo, P., Desai, K. T., Pal, A., Ajenifuja, K. O.,
Adepiti, C. A., Long, L. R., Schiffman, M., and
Antani, S. (2021). A deep clustering method for
analyzing uterine cervix images across imaging de-
vices. In 2021 IEEE 34th International Symposium
on Computer-Based Medical Systems (CBMS), pages
527–532. IEEE.
Zhang, L., Wang, X., Yang, D., Sanford, T., Harmon, S.,
Turkbey, B., Wood, B. J., Roth, H., Myronenko, A.,
Xu, D., et al. (2020). Generalizing deep learning for
medical image segmentation to unseen domains via
deep stacked transformation. IEEE transactions on
medical imaging, 39(7):2531–2540.
ICT4AWE 2025 - 11th International Conference on Information and Communication Technologies for Ageing Well and e-Health
180
