
A Conceptual Model-Based Application for the Treatment and
Management of Data in Pediatric Oncology:
The Neuroblastoma Use Case
Jes
´
us Carre
˜
no-Bolufer
1 a
, Jos
´
e Fabi
´
an Reyes Rom
´
an
1 b
, Sergio P
´
erez Andr
´
es
1 c
,
D
´
esir
´
ee Ramal Pons
2
, V
´
ıctor Ju
´
arez Vidal
1
, Adela Ca
˜
nete Nieto
2 d
and
´
Oscar Pastor
1 e
1
Valencian Research Institute for Artificial Intelligence (VRAIN), Universitat Polit
`
ecnica de Val
`
encia, Valencia, Spain
2
Grupo de Investigaci
´
on Cl
´
ınica y Traslacional en C
´
ancer, Instituto de Investigaci
´
on Sanitaria La Fe (IIS La Fe),
Valencia, Spain
Keywords:
ClinGenNBL, Conceptual Modeling, Neuroblastoma, Pediatric Oncology, Data Management, CMN.
Abstract:
Neuroblastoma is one of the leading causes of death in childhood oncology. Current treatments for these
patients are general and not targeted, including radiotherapy, chemotherapy, and surgery. There is a need
for more efficient methods. Precision Medicine (PM) can help to overcome this challenge. PM incorporates
clinical, lifestyle, and genomic data, among others, into a standardized process to provide individualized
treatment. However, a large amount of data is needed to achieve PM, and the heterogeneity present in the
case of neuroblastoma poses a challenge for integration and, consequently, for knowledge generation. We
need a solid domain definition that provides a foundation for experts to work on, which implies generating
a conceptual model. Based on this model, any Information System (IS) can be developed. ISs play a vital
role in managing clinical data efficiently. Much of the clinical data has been captured and managed over the
years with inefficient tools such as spreadsheets. In this work, we first present the new Conceptual Model
of Neuroblastoma (CMN), with a special focus on genomics, and second, ClinGenNBL, a conceptual model-
based web application that implement the CMN with the goal of assisting clinicians in managing patients with
neuroblastoma through a user-friendly interface.
In Memoriam and in honor of the beloved Victoria
Castel S
´
anchez, who passed away during the research
and publication of this research work.
1 INTRODUCTION
The most frequent tumors in Spain (0-14 years, be-
tween 2010-2023) are leukemias 28.1%; lymphomas
12.1% and central nervous system tumors 24.8%
(Ca
˜
nete Nieto et al., 2023). The Neuroblastoma
(NBL), or secondary nervous system tumor, with
7.7% of cases, comes in fourth place in terms of inci-
dence. NBL is also the most frequent solid extracra-
nial tumor in childhood (Castleberry, 1997). In the
a
https://orcid.org/0009-0006-4720-4037
b
https://orcid.org/0000-0002-9598-1301
c
https://orcid.org/0000-0002-9316-210X
d
https://orcid.org/0000-0002-5669-5097
e
https://orcid.org/0000-0002-1320-8471
United States, it represents 6% of all pediatric can-
cers, with a survival rate of 82% for ages from birth
to 14 years old (Siegel et al., 2024).
In light of this problem, the Spanish NBL Group
of SEHOP
1
was created in 1987, led by the Pediatric
Oncology Unit of Hospital Universitari i Polit
`
ecnic
La Fe (HUP/IIS LaFe) (HUPLF). Since then, the
group has developed and fine-tuned therapeutic and
diagnostic protocols for NBLs and participated in in-
ternational groups with SIOPEN
2
and ANRA
3
. They
have also produced a wide variety of reports and dis-
seminated study results (Ca
˜
nete Nieto et al., 2023)
We encounter massive amounts of data from
which knowledge could be generated to advance Pre-
cision Medicine (PM) (McCabe et al., 2024) (Ca-
1
Sociedad Espa
˜
nola de Hematolog
´
ıa y Oncolog
´
ıa
Pedi
´
atricas. https://www.sehop.org/
2
International Society of Paediatric Oncology European
Neuroblastoma. https://www.siopen-r-net.org/
3
Advances in Neuroblastom Research Association.
https://www.anrmeeting.org/
Carreño-Bolufer, J., Reyes Román, J. F., Pérez Andrés, S., Ramal Pons, D., Juárez Vidal, V., Cañete Nieto, A. and Pastor, Ó.
A Conceptual Model-Based Application for the Treatment and Management of Data in Pediatric Oncology: The Neuroblastoma Use Case.
DOI: 10.5220/0013202600003928
In Proceedings of the 20th International Conference on Evaluation of Novel Approaches to Software Engineering (ENASE 2025), pages 15-26
ISBN: 978-989-758-742-9; ISSN: 2184-4895
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)
15

haney et al., 2022). This data can help us understand
patients who suffer from NBL. PM aims to provide
personalized treatment for each patient. Currently,
implementing PM in clinical data management re-
quires a high investment of resources, mainly due to
the complexity of the data and the lack of efficient In-
formation Systems (IS). Each disease is unique and
requires specialized tools.
Fortunately, technological advances in Software
Engineering (SE) in recent years have created many
ISs for better information management in different
contexts (Evans, 2016). In particular, in the context
of PM, these ISs play a fundamental role in manag-
ing patient follow-up and conducting studies on data
collected in the clinical context over the years.
However, when applied to NBL, one of the main
challenges in building an IS is the data heterogeneity
in this field. Consequently, there is a lack of inte-
grated information from a holistic perspective, which
hinders the development of automated pipelines for
identifying and analyzing relevant statistical data
(Digital Transition, ). A standard conceptual model
would facilitate the integration process from diverse
sources (Trujillo et al., 2018)(Ca
˜
nete et al., 2022).
In summary, data management in this context
presents the following problems: high heterogeneity
and dispersion of data and lack of efficient mecha-
nisms. To overcome these challenges, the following
goals are specified:
1. We aim to generate a Conceptual Model (CM) to
provide a common ontological framework for dis-
cussions with experts, increasing our understand-
ing of the neuroblastoma (NBL) domain. This
model will also serve as a foundation for de-
veloping an Information System (IS) to improve
data management and integration, encompassing
not only clinical data but also genomic data. To
achieve this, we establish a shared view of the
study domain, which must be supported by clin-
ical experts.
2. To design and develop an IS based on the de-
fined CM. This IS will improve the integration and
management of the existing data in the NBL do-
main. It will also help clinical experts to improve
their decision-making.
To achieve these goals, this work contributes to
version 2.0 (V2) of the Conceptual Model of Neu-
roblastoma (CMN) upon which a software platform,
ClinGenNBL, is implemented. ClinGenNBL, a con-
ceptual model-based web application, serves as a
work tool for clinicians, combining the management
and analysis of clinical data.
The rest of the article is organized as follows: in
Section 2, we provide context for the project and re-
view how information systems (IS) have been applied
to improve clinical and genomic data management.
Section 3, we detail the CMN, the conceptual model
that defines the NBL domain as the result of meet-
ings with experts in the field. Section 4 presents the
result of integrating the CMN with the Conceptual
Model of the Human Genome (CMHG) to expand the
genetic component and support the advancements in
the field of the PM. In Section 5, we introduce Clin-
GenNBL, an IS for clinical data management based
on the CMN. Finally, Section 6 concludes the article
with the contribution of this work and outlines future
lines of research.
2 PREVIOUS AND RELATED
WORK
This section provides context for understanding the
work carried out by presenting the background on
which this work is based in Section 2.1 and a sum-
mary of related works in Section 2.2.
2.1 Previous Work
The project of this work started in 2018, with
the first version (V1) of the CMN proposed by
Arevshatyan, Reyes Roman(co-author) and collabo-
rators (Arevshatyan et al., 2019), defining the domain.
Their work highlighted the importance of building
a Genomic Information System (GeIS) upon a con-
ceptual model. Later, in 2020, the CMN was im-
proved (V1.5), and the first version of the IS based on
this model was designed and developed (Arevshatyan
et al., 2020). It was adapted to the needs of the Pedi-
atric Oncology department of the HUPLF.
The clinical staff of the HUPLF used the prototype
to store patient data, diagnostic evaluations, and treat-
ments. However, this prototype has become obsolete
due to the lack of maintenance and the new needs of
the clinical staff. Most of the technologies used in
the prototype (jQuery, Bootstrap, Jasmine-Express)
are no longer maintained or have been replaced by
new ones. Moreover, the prototype does not have a
responsive design and has a lot of inconsistencies in
its user interfaces.
In recent years, new technologies have emerged
that facilitate development and maintenance. This
analysis, part of the work associated with this re-
search, culminated in a technical report that provides
a comprehensive analysis (Fern
´
andez Garc
´
ıa et al.,
2022). Rather than attempting to fix all the proto-
type’s flaws, a new version of the IS will be devel-
oped, incorporating best practices of modern tech-
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
16

nologies and based on an updated version of the
CMN.
Therefore, this work aims to generate the V2 of
the CMN using conceptual modeling techniques, as
they allow for an improved understanding of the do-
main and facilitate the treatment of complex data,
such as those about NBL. Based on this CMN, we
design and develop a new version of the IS, Clin-
GenNBL, that adapts to the new requirements de-
manded by the clinical staff. Finally, the resulting IS
must be implemented with novel but stable technolo-
gies to improve the development and maintenance
time.
2.2 Related Work
In order to develop a better solution, an analysis of
research related to this research work has been car-
ried out. This section presents several works that be-
long to the context of bioinformatics and/or aim to
design and develop an IS based on conceptual mod-
els. One of the areas where the use of conceptual
models has been most influential is in the human
genome. Since the contribution by Paton (Paton et al.,
2000), which introduced a collection of genomic (i.e.,
implementation-independent) conceptual data mod-
els for genomic data. These conceptual models are
amenable to (more or less direct) implementation on
different computing platforms.
Following the above work, numerous papers ap-
plying conceptual modeling techniques emerged. An
example of this is the work of Martin et. al., whose
goal is to design and develop an IS that integrates hu-
man genome variant data from different sources (Mar-
tin and Celma, 2011). To achieve this goal, the defini-
tion and categorization of variations is unified through
conceptualization. Once the conceptual model was
established, a database (Human Genome Data Base,
HGDB) was implemented.
In the Ph.D. thesis of Burriel Coll (Burriel Coll,
2017), the aim is to solve the problem of heterogene-
ity and dispersion of genomic data on breast cancer.
Within this domain, the biggest challenge is the treat-
ment of a high volume of data, as it faces a disease
with high prevalence. It aims to analyze the informa-
tion available in the databases and integrate it into an
IS, thanks to creating a CM.
Focusing the context on genomics, Bernasconi
et. al. present an article that proposes a conceptual
model of genomic metadata, whose purpose is to fa-
vor queries to experimental databases. Their work
can be divided into three phases. The first phase an-
alyzes the attributes of genomic metadata. The sec-
ond phase uses an up-bottom method to build the in-
tegrated schema. The last phase corresponds to vali-
dating the conceptual model with different databases
(Bernasconi et al., 2017).
The doctoral thesis of Reyes Rom
´
an (2018) de-
fines a framework focused on using CM. It proposes
to use an CMHG as a fundamental basis for generat-
ing Genomic Information Systems (GeIS), intending
to facilitate a conceptualization of the domain that al-
lows to reach accurate knowledge and to be able to
reach PM (Reyes Rom
´
an and Pastor, 2018). Within
the context of NBL is the work of Arevshatyan, which
analyzes and integrates clinical and genomic data. It
highlights that a Genomic IS based on a conceptual
model allows for improved adaptation to new domain
requirements. It also greatly simplifies the integra-
tion and management of heterogeneous and homo-
geneous data (Arevshatyan et al., 2019). Within a
broader clinical context (not only NBL), Arevshatyan
et. al. present a practical experience of data analysis
and decision-making process where a CM is designed
to develop an IS to manage clinical, pathological, and
molecular data in an integrated manner in the oncol-
ogy department of two hospitals (Arevshatyan et al.,
2020).
As a result of the COVID-19 disease, numerous
databases were created to mitigate the effects of the
pandemic. Within this context, Bernasconi et. al. re-
views the data integration efforts needed to access and
search SARSCoV2 genome sequences and metadata
deposited in the most important viral sequence repos-
itories (Bernasconi et al., 2020). The paper applies
conceptual modeling techniques to structure and or-
ganize the information.
Finally, the work of Garcia et. al. faces the chal-
lenge of integrating a huge amount of existing omic
data (Garc
´
ıa Sim
´
on et al., 2021). To this end, concep-
tual modeling techniques are applied to facilitate un-
derstanding, communication, and problem-solving in
the domain while establishing a common ontological
framework to stimulate the communication and evolu-
tion of complex domain knowledge. The work’s main
contribution is the presentation of the Genome Con-
ceptual Schema, independent of the species, which al-
lows the representation of proteomic data and facili-
tates their integration.
3 CONCEPTUAL MODEL OF
NEUROBLASTOMA (CMN)
Conceptual Modeling (CM) is a technique used to un-
derstand a problem domain and communicate effec-
tively with users and subject matter experts. It has
been beneficial in homogenizing data in the clinical
A Conceptual Model-Based Application for the Treatment and Management of Data in Pediatric Oncology: The Neuroblastoma Use Case
17

context, as discussed in Section 2.2. In this section,
we present the V2 of the Conceptual Model of Neu-
roblastoma (CMN).
Figure 1 shows the CMN resulting from the do-
main study and collaborative meetings with clinical
experts at HUPLF. We conducted a preliminary lit-
erature review and did not find any other conceptual
models specifically addressing the neuroblastoma do-
main. Based on a previous CMN (Arevshatyan et al.,
2020), the analysis of each of the classes has been car-
ried out, structured in views, and extended to adapt it
to the new knowledge of the domain. Then, the model
is proposed to represent all the procedures performed
by the clinical staff throughout all the stages of patient
care.
The different views of the model shown in Figure
1 are discussed below:
• Patient View: It contains all demographic and
general information about the patient, including
their status throughout the course of the disease.
• Episode View: Encompasses all services provided
to the patient with the medical problem within
a specific period of time. This view includes
the protocol to which these services belong and
all information regarding symptoms, tumors, and
metastases.
• Treatment View: Represents all possible treat-
ments the patient can have. The subclasses rep-
resent the special types of treatments the patient
receives depending on the type of stage and how
it progresses.
• Test view: Represents the various diagnostic tests
physicians perform to detect and diagnose the dis-
ease.
• Genetic View: Contains all the genomic informa-
tion about the patient obtained through the perfor-
mance of genetic tests.
The Patient View is composed of all necessary pa-
tient data. It has been placed at the top as the rest of
the views depend on the existence of the patients. Be-
fore creating any patient, we must have at least one
”Hospital” in the system.
Once this requirement is fulfilled, it is possible
to start registering patients who belong to the class
”Patient”. This will occur on the first visit to the
hospital center, where the basic patient data, such as
name, date of birth, gender, weight, height, etc., will
be taken.
In addition to the basic data, every patient should
have a history of conditions over time. This history
is represented by the class ”Status”. A patient may
be alive, either with a tumor, ”AliveWithTumor” or
without it, class ”AliveFreeDisease”, and have local
relapses or metastases in the course of its evolution,
in which case it will be necessary to indicate its type,
included in the class ”Relapse”, or mark the disease
progression as death cause, represented by the class
”Deceased”.
The Episode View is the central pillar of the model
and around which all other views revolve. In the hier-
archy of views presented in Figure 1, it is in the center,
not just by chance, as the patient’s evolution is related
to the episodes. This is the one in charge of keeping
the history of diagnostic tests, treatments, symptoms,
etc.
Each time the patient is evaluated or receives any
treatment, an episode is created in the system. This
episode can be of a special type called a diagnosis,
manifested in the diagram with the class ”Diagnosis”,
which represents the event for which the patient first
goes to the hospital and the NBL diagnosis process
begins.
At the beginning of his assessment, the applica-
tion user will introduce the patient’s symptoms, repre-
sented in the CMN by the class ”Symptom”, through
a description and a date of the different symptoms of
the patient.
When diagnosing the patient, a primary tumor, de-
scribed by the class ”Tumor”, must be added. The ap-
plication user will specify the tumor’s type, location,
size, and date for each tumor. If a metastasis is de-
tected in a patient, it will be reflected through the en-
tity with that name and which contains the attributes
of type and date.
The episodes will be associated with a certain
phase, identified as ”Phase” in the model, correspond-
ing to a protocol called ”Protocol” in the model. A
medical protocol is a set of recommendations on the
diagnostic procedures to be used for any patient with
a given clinical condition. Or on the most appropriate
therapeutic approach to a clinical diagnosis or health
problem (UNITECO, 2019). Each patient receives
treatment based on a single protocol. These have a
duration and are renewed over the years.
Once the patient’s main data have been entered,
several diagnostic tests are performed to determine
the patient’s stage, making it possible to establish with
greater precision the treatment that the patient will re-
ceive.
Imaging methods, represented by the class ”Ra-
diological”, store all relevant data from tests such
as X-rays, ultrasound scans, and CT scans. Nuclear
medicine encompasses information on two tests in
each entity and is represented by the class ”Nuclear”.
The different laboratory tests that can collect infor-
mation on the blood or urine tests performed on the
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
18
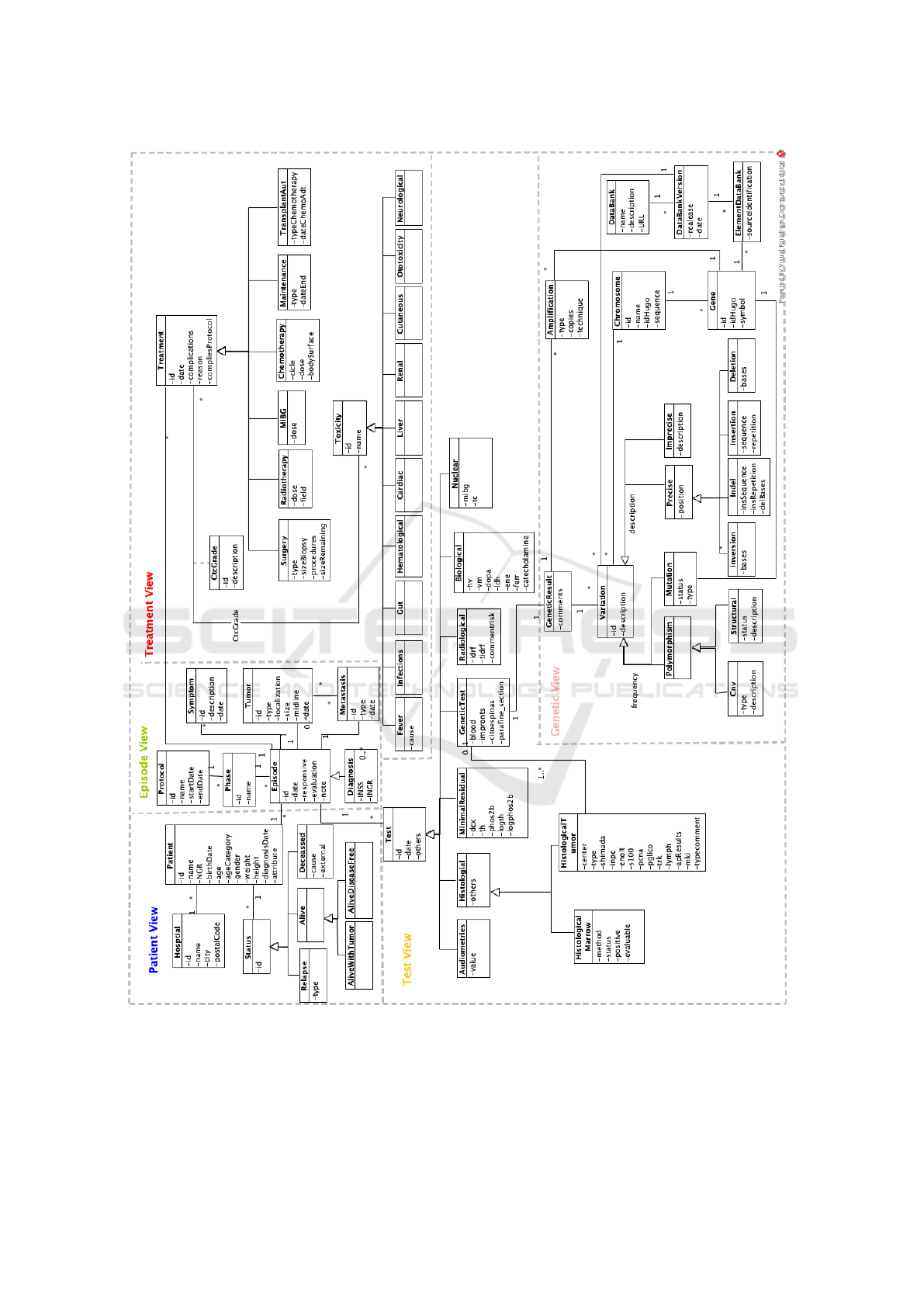
.
Figure 1: Conceptual Model of NBL.
patient are represented in the class Biological. The
study of audiometry tests, represented by the class
with the same name ”Audiometries” and the assess-
ment of minimal residual disease (class ”Minimal-
Residual”), which stores the result of different tests
performed on the patient.
Histological tests correspond to those tests that
use a histological technique. This is the series of or-
A Conceptual Model-Based Application for the Treatment and Management of Data in Pediatric Oncology: The Neuroblastoma Use Case
19

dered steps that prepare the tissue for observation un-
der the microscope (Guerrero Alquicira et al., 2017).
These tests include:
• Bone marrow evaluation, represented by the class
”HistologicalMarrow”, a test that records the
method or technique used, its status, whether it
has been positive or not, and whether it has been
evaluable or not.
• Tumoral evaluation, represented by the class
”HistologicalTumor”, it stores genetic informa-
tion following a study on genetic information after
a study has been performed on a tumor sample.
Such a study can generate instances of the class
”GeneticTest”, which will be accompanied by a
result represented by the class ”GeneticResult”.
Finally, clinicians will rely on genetic testing,
which studies the different variants of the patient.
These types of tests are studies on genes and chromo-
somes, represented as ”Gene” and ”Chromosome”,
which are divided into various tests, such as the study
of the amplifications of a gene, represented by ”Am-
plification”, its possible variants that appear in the
classes ”CNV” and ”Segmental” and, finally, the mu-
tations of the gene, represented by ”Mutation”.
Once the protocol to be followed for the patient
has been decided based on all the diagnostic tests, it
is the turn of the treatments, whose basic informa-
tion (date on which they were applied, the reason,
the protocol to which they belong, and whether or
not there were any complications, etc.) is available
in the class ”Treatments”. The treatments will always
be specific, i.e., they will always belong to surgery
(class ”Surgery”), chemotherapy (class ”Chemother-
apy”), autologous transplant (class ”TransplantAut”),
radiotherapy (class ”Radiotherapy”), or ”MIBG”.
Each treatment may cause different toxicity in
the patient (cardiac, nausea/vomiting, fever, infection,
etc.). For this reason, it is decided to represent the
different types of toxicities that may arise from each
treatment by the class ”Toxicity”, specifying the type
using its derivatives. In addition, toxicities can have a
degree and a specific description, represented by the
class ”CtcGrade”.
In summary, this section has described V2 of
CMN, supported by clinical experts at HUPLF. It
includes several views for patients, episodes, treat-
ments, tests, and genetics, providing a base to man-
age clinical data for neuroblastoma in common terms.
However, the genetic component can still be extended
with other conceptual models. This task is done in
Section 4. Numerous model-based applications can
be built upon the CMN, for example, ClinGenNBL
(Section 5).
4 INTEGRATION OF THE
CONCEPTUAL MODEL OF
NEUROBLASTOMA (CMN)
AND THE CONCEPTUAL
MODEL OF HUMAN GENOME
(CMHG)
As exposed in Section 1, PM is the pathway to effec-
tively and efficiently treat individuals suffering from
NBL. It consists of applying special treatments de-
pending on the characteristics of the patient. In this
context, DNA is what defines humans the most. To
this end, we focus on the genetic component of the
CMN by integrating the CMN with the Conceptual
Model of Human Genome (CMHG), which we detail
in this section.
The CMHG used is version 2 from the thesis
of Reyes Rom
´
an, which features a chromosome-
centered view (Reyes Rom
´
an and Pastor, 2018). Be-
low is a summary of the objectives of each view cor-
responding to the CMHG, which will be incorporated
into the CMN presented previously in this work (Sec-
tion 3) and that will give rise to the new model, CMN-
CMHG.
• Structural View: As its name indicates, it de-
scribes the structure of the genome.
• Transcript View: Shows the components and con-
cepts related to protein synthesis.
• Variation View: Models the knowledge related to
the differences found in the DNA sequences of
different individuals.
• Phenotype View: Represents the phenotypes as-
sociated with one or several DNA variations. The
phenotype is the set of physical, biochemical, and
behavioral characteristics that can be observed.
• Bibliographic View: View provides information
about the data sources from which the data to be
stored in the model has been extracted.
• Pathway View: Composition of processes com-
posed of chemical reactions that take place inside
a cell.
The CMN-CMHG integration can be found here
4
.
In the model presented, we find three types of colors
that allow us to quickly and efficiently distinguish the
different parts where integration occurs. Blue marks
the existing classes in both models. Purple represents
the existing classes in the CMN that the inclusion of
the new model has modified. Finally, green represents
4
https://doi.org/10.5281/zenodo.13152470
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
20

the new connection classes, i.e. the first point of con-
tact between the CMN and the CMHG.
The following subsections provide a detailed ex-
planation of how the integration took place. Some
views of the CMHG presented intersect with the Ge-
netic View in the CMN, leading to segregation of this
view (Subsection 4.1), while others introduce new
knowledge to the CMN (Subsection 4.2).
4.1 Extension of Knowledge:
Segregation of the Genetic View
All genetic information is grouped under the Genetic
View in the CMN. When this view is compared with
the CMHG for the first time, it is divided into four
new views: the Variation View, Structural View, Tran-
scription View, and Bibliographic View, using the
same names as in the CMHG.
The first new view comes from the variations, rep-
resented by the ”Variation” class in both models. This
class becomes the main class of the new Variation
View by adding the attributes provided by the CMHG
(clinical importance, private, and version of creation).
In addition to this class, the inheritance hierarchy of
the classes is maintained in both models.
The contribution on the Variation View with a
higher knowledge value is the extension of structural
polymorphic variants. Represented by the ”Struc-
tural” class in the CMN, is replaced by the class
”SNP” (Single Nucleotide Polymorphism) of the
CMHG. This renaming is closely related to data stor-
age, e.g. the use of dbSNP (Sherry et al., 2001).
Continuing with the segregation of the Genetic
View, the second new view is the Structural View, de-
rived from chromosomes. The ”Chromosome” class
is the core around which the CMHG revolves. The
”Chromosome” class now includes the sequence at-
tribute. Additionally, the ”Species” class has been in-
cluded to determine the family to which each chro-
mosome belongs; the points in the sequence where
recombinations occur, represented by the ”Hotspot”
class; and the cytogenetic bands, represented by the
”Cytoban” class, which provide characteristics of the
chromosome and are, in turn, related to the imprecise
subtype of the variant, in the ”Imprecise” class.
In the Structural View, special attention is given
to the elements of a chromosome. Where there was
previously a direct connection between chromosomes
and genes, the model is now enriched with transcrib-
able elements, conserved regions, and regulatory ele-
ments. All information related to the regions is con-
centrated in the Transcription View, the third new
view.
Of the three types of elements, special attention
is given to the transceptors, represented by the class
”TranscriptableElment” whose purpose is to represent
a region of DNA that can be transcribed. In turn, these
regions can be specialized into two types:
1. Gene: a concept that we find in the CMN enriched
with new attributes, and more specifically, biotype
determines the specialization of the various types
of genes. of genes.
2. Exon: each of the transcribable elements that
form part of the gene.
The relationship between these two classes is made
through the transcripts, i.e. the ”Transcript” class rep-
resents the different transcripts present in a gene and
comprises a series of exons.
Finally, we find the last new view, the Bib-
liographic View. Similarly to what occurred in
the CMN, this view provides information about the
source of the data, meaning where the information to
be stored in the model has been obtained. It retains
the data source, represented by the ”DataBank” class,
its version (”DataBankVersion” class), and the rela-
tionship between genes and the data source, shown
in the ”ElementDataBank” class. Notably, this ele-
ment’s relationship has changed from the Gene class
to its predecessor, ”ChromosomeElement”.
4.2 New Knowledge: Phenotype and
Pathway
During the integration process, entirely new views
were identified, meaning any element of the CMN
did not represent them. Therefore, the introduction of
these new views will provide previously unavailable
knowledge.
One of the most important contributions made in
the model’s V2 extension was the inclusion of the
Phenotype View. The phenotype results from genes
that can be expressed and the external factors that af-
fect their expression: environmental, nutritional, and
chemical factors.
This contribution, introduced in the context of
NBL, will endow the model with a combined
genotype-phenotype perspective within diagnostic
evaluations. This means that the model can represent
phenotypic expressions for a patient.
Just as in the CMN we find parts unaffected by
integration, there is the list of metabolic pathways. A
metabolic pathway (”Pathway” class) is a succession
of chemical reactions that take place inside the cell.
In summary, in this section, we have presented
the integration of the CMN and the CMHG into a
unified framework (CMN-CMHG), with an expanded
genetic view that can serve as a common language
A Conceptual Model-Based Application for the Treatment and Management of Data in Pediatric Oncology: The Neuroblastoma Use Case
21

for communication in the domain of genetic Preci-
sion Medicine (PM) applied to neuroblastoma (NBL).
It includes information on chromosomes, variations,
phenotypes, bibliographic data, and pathways, among
other elements. This integration also facilitates inter-
operability between different PM and NBL tools, of-
fering deeper insights into the field.
5 CLINGENNBL: A CMN-BASED
APPLICATION
This section introduces ClinGenNBL, the solution
generated based on the conceptual model defined in
Section 3. On the one hand, this CM has helped im-
prove communication between the various multidisci-
plinary experts during the requirements definition. On
the other hand, it has served as ontological support in
creating software solutions developed with innovative
technologies and great support from the SE commu-
nity. All this is to provide an IS that responds to the
needs of clinical experts in NBL and has an appro-
priate degree of maturity for its application in medi-
cal practice. This section describes the solution de-
signed, developed, and validated. It also presents sev-
eral screenshots from extensive use involving more
than 800 real patients.
5.1 Requirements
The first phase of the solution design consisted of
gathering the functional needs from the requirements
analysis sessions conducted with the HUPLF clinical
experts. The functional and non-functional require-
ments were analyzed and converted to small pieces of
information using the use case analysis. ClinGenNBL
allows doctors to manage and analyze the clinical data
associated with NBL with a user-friendly interface.
As introduced in this section, it is based on the CMN;
however, some concepts are not part of the require-
ments as they are not needed by the experts at the time
of the writing of this article.
Among its features, the data analytics section
should be noted, which includes the following ca-
pabilities: i) Generate a patient state report in CSV
format, ii) Search neuroblastoma hospitals by allow-
ing the user to search for hospitals, iii) Filter neurob-
lastoma protocols and hospitals by any property, iv)
Show the analysis of distributions in which patients
are found (INRG
5
or INSS
6
) and the number of pa-
tients associated with each status, v) Display the chro-
5
International Neuroblastoma Risk Group.
6
International Neuroblastoma Staging System.
mosome alteration analysis in a table composed of pa-
tient names, applied protocol, date of birth and diag-
nosis, INSS, as well as Del1p
7
, vi) Show gene ampli-
fication analysis composed of patient names, protocol
applied, date of birth and diagnosis, INSS, as well as
copies of the MYCN gene.
More information about the requirements
can be found in the following technical report
(Fern
´
andez Garc
´
ıa et al., 2022) (Spanish), which
includes comprehensive documentation covering all
the information, parameters, and options managed
by the developed system. During the design of an
application interface, mockups are common, which
allow us to present a possible approach to the final
design.
5.2 Architecture
To ensure that an IS meets the needs of stakeholders
and has the necessary level of maturity for a medical
application, technologies that are widely used in the
software development industry and that receive the
support and backing of very important companies at a
professional level have been used.
ClinGenNBL consists of three components: fron-
tend, backend, and database. By decoupling the logic
and model (backend) from the view (frontend), we
achieve greater flexibility and increased productivity
as we get a greater separation or composition of the
application. Figure 2 shows the architecture repre-
sented in a UML component diagram. Below, the
components are detailed:
1. Backend: NodeJS has been used as the JavaScript
execution platform. This level of abstraction aims
to provide an interface, usually an API, which al-
lows easier interaction with the database or other
external services.
2. Frontend: It consists of the user interface defined
in ReactJS, coded in a proprietary language called
JSX, an extension of the JavaScript programming
language. Other libraries, such as i18n, have been
used as well. It was decided to use React as the
main tool for client-side development since it is
one of the most widely used web development li-
braries today. This component interacts with the
backend through a REST API, which uses the
HTTP protocol and exchanges messages in JSON
format.
3. Database: Object-Relational Mapping (ORM) has
been used, a technique that allows the manipu-
lation of database data using an object-oriented
7
Genetic deletion affecting the short arm of chromo-
some 1.
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
22

paradigm. In this case, it was decided to use
Sequelize
8
, a library written in TypeScript for
Node.js, which allows the definition of models
and relationships, the database query and the han-
dling of these models in the form of objects. In
addition, this library is compatible with many
DBMSs, including MySQL8
9
, which has been
chosen to store all the data related to the applica-
tion since it is a very mature system used through-
out the history of software application develop-
ment.
5.3 Validation
Validating the solution means justifying that it would
contribute to the stakeholders’ objectives if imple-
mented. In the Engineering Cycle (EC), validation is
performed before implementation. It involves investi-
gating the effects of the interaction between an artifact
prototype and the problem model, always compar-
ing them with the solution requirements (Wieringa,
2014).
Expert opinion is one of the simplest ways to val-
idate the solution (Wieringa, 2014). The design of
the solution is submitted to a committee of experts,
in this case, different physicians and doctors from the
Clinical and Translational Research Group in Cancer
(GICT-Cancer) of the HUPLF/IIS La Fe, who imag-
ine how it will interact with the context problem (NBL
data management) and try to predict the effects they
think the solution will have.
The main objectives of these meetings are: i) to
obtain the maximum knowledge of the domain and
the research problem and ii) to consolidate and vali-
date the progress on developing the solution. The first
meetings held by the HUPLF team of experts focused
on defining the requirements and studying the prob-
lems they were facing daily. Once analyzed, priori-
tized, and classified, they were shared along with the
conceptual model with the team to validate them. The
rest of the meetings that were agreed upon with the
group focused on reviewing the application’s progress
through demonstrations. Each of them had a deliver-
able associated with the progress of the functionalities
implemented since the previous meeting. Finally, to
test the final developed software, the expert opinion
exercise was divided into two stages: i) User observa-
tion and ii) User interviews.
In the user observation phase, the experts of the
HUPLF were asked to perform a set of predefined
tasks within the developed IS. The tasks that were
8
Sequelize. https://sequelize.org/
9
MySQL. https://www.mysql.com/
defined following the application requirements in-
cluded.
• Hospital registration: The user must register a
new hospital, display the list of existing hospi-
tals, and search for the new hospital by two cri-
teria ”Name” and ”City”.
• Registration and protocol query: The user must
register a new protocol, display the list of existing
protocols, and add the different phases that make
up the protocol.
• Patient registration: The user must register a new
patient, display the list of existing patients, and
search for the new patient by MRN
10
.
• Symptom, tumor, metastasis registration: The
user must register a new symptom, tumor, and
metastasis on the previously created patient.
• Diagnostic evaluation registration: The user must
register at least once for each of the different types
of diagnostic evaluations.
• Treatment Discharge: The user must discharge at
least once each treatment, including all types of
toxicity.
• Consultation of patient information: The user
must display the data entered in the previous
tasks.
• Correction of patient errors: The user will have to
edit one of the attributes of each entity and finally
delete them.
The objective of the user interview is to know the
user’s opinion regarding the use of the application af-
ter performing the previously defined tasks. The com-
ments can be summarized in four relevant points:
• Improvement of daily work: Using the IS dur-
ing patient diagnosis and treatment allows them to
offer better care and achieve Precision Medicine
or Personalized Medicine. In addition, the IS al-
lows them to perform more efficient and effective
queries on patient data, speeding up the data ex-
ploitation process.
• The tool is easy to use: Team members found the
application easy to use, as they could recognize
and locate most of the tasks they were asked to
perform without assistance.
• Stakeholders’ objectives are met: The team of ex-
perts expressed that they were very satisfied with
the application. They indicated that the data man-
agement would help improve the treatment pro-
vided to patients, as they would be able to analyze
and extract all the data.
10
Medical Record Number.
A Conceptual Model-Based Application for the Treatment and Management of Data in Pediatric Oncology: The Neuroblastoma Use Case
23
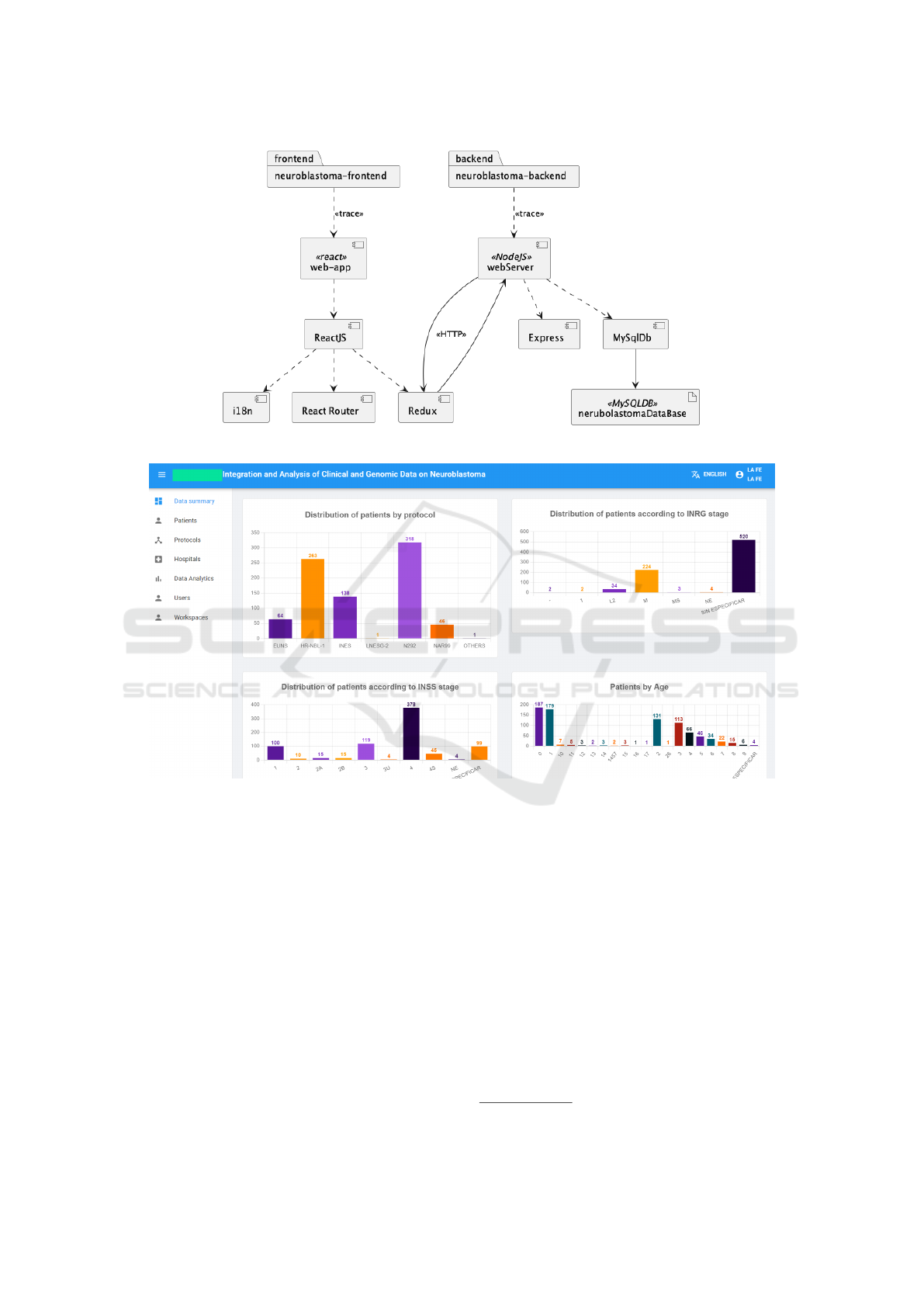
Figure 2: Solution architecture.
Figure 3: Screenshot of ClinGenNBL: Sumamry.
• Possible improvements to the application to facil-
itate its use were identified: The team of clinical
experts suggested possible visual improvements
that would facilitate the process when entering
data: the grouping of certain toxicities and the cre-
ation of shortcuts in the creation of lookup tables.
In general, stakeholders reported that the use of
the application was beneficial. The tool allows them
to achieve their goals and improve the treatment of
NBL easily and intuitively. They also mentioned that
adopting the tool would take little time and could be
extended to more hospitals.
In conclusion, homogenizing data related to NBL
treatment and its access through a common IS pro-
duces greater satisfaction and benefits than manual
and dispersed treatment (through different applica-
tions). The validation has provided encouraging find-
ings, but the results should be studied further.
5.4 Using ClinGenNBL
Thanks to the extensive knowledge of experts in var-
ious areas relevant to this work, such as Information
Systems Engineering (ISE), Bioinformatics, and Web
Engineering, among others, it has been possible to
achieve a high degree of maturity (TRL5 (European
Comission, 2014)) in both the initial design and the
final result of the application. It is ready to be trans-
ferred to HUPLF. We present several screenshots of
the platform (Figures 3 and 4) and a demonstration
video, where 831 real patients with relevant data were
introduced. The demonstration can be found here
11
.
11
https://doi.org/10.5281/zenodo.13138339
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
24

Figure 4: Screenshot of ClinGenNBL: Manage Patient.
All the clinical data was entered in compliance with
GDRP
12
legislation (GDRP, 2016).
6 CONCLUSIONS
Finally, to conclude, Section 6.1 summarizes the con-
tribution of this work to the defined goals.
6.1 Contribution
This research aimed to generate a conceptual model
for the neuroblastoma domain and to implement an
information system (IS) based on it, using conceptual
modeling techniques to address the identified prob-
lems. This work has contributed to the development
of the V2 of the CMN and the implementation of ro-
bust software based on it (ClinGenNBL). Clinical ex-
perts continuously validate this solution at a technical
and functional level. Additionally, it has highlighted
the genetic aspects of the domain to advance preci-
sion medicine through the integration of CMN and
CMHG.
Firstly, the CMN improves the understanding of
the domain and establishes a solid foundation on
which to build the solution design. The knowledge
obtained from the domain study can be integrated
and centralized. It also improves stakeholder com-
munication, which is very useful during the require-
ments gathering and validation phase. Secondly, Clin-
GenNBL serves as an information system instance of
12
General Data Protection Regulation.
the CMN and provides greater efficiency to the clin-
icians at HUPLF in managing clinical data and gen-
erating knowledge about neuroblastoma. It comprises
two independent components implemented with mod-
ern and consolidated technologies such as NodeJS,
React, and MySQL. Experts have successfully vali-
dated and tested it with more than 800 real patient
cases.
6.2 Future Work
CMN has been developed to a stable version (V2),
and ClinGenNBL has reached a high degree of matu-
rity (TRL5). However, there is still more work to be
done. Future work includes:
• Maintenance and evolution of the application:
The initial application has been validated satisfac-
torily with the expert opinion of the HUPLF doc-
tors. Based on the needs detected and the possible
improvements identified, future developments are
proposed to address new functionalities for the ap-
plication.
• Implementation of the integration of the CMN-
CMHG: Given the knowledge gathered through-
out the work, the integration between both models
(CMN and CMHG) is proposed in Section 4 but
not implemented in the application. However, the
integration design and development could be part
of a future engineering cycle.
• Implementation of the tool in the real working en-
vironment: The next step for a successful evolu-
tion is using the application in a realistic environ-
ment. To this end, implementing the HUPLF is
A Conceptual Model-Based Application for the Treatment and Management of Data in Pediatric Oncology: The Neuroblastoma Use Case
25

only the first step, with the possibility of taking
the IS to the rest of the national hospitals and ex-
panding internationally. This must be done in col-
laboration with the technical service of each de-
partment in order to follow the protocols estab-
lished by the healthcare organization.
ACKNOWLEDGEMENTS
The authors would like to thank the PROS Re-
search Center Genome group for their valuable
discussions on the application of CM in medicine,
as well as the staff of the Medical and Pediatric
Oncology Service at Hospital La Fe for their sup-
port and commitment to facilitating our work and
enhancing our understanding of the domain. This
work was supported by the Generalitat Valenciana
through the CoMoDiD project (CIPROM/2021/023),
and the Spanish State Research Agency through
the SREC project (PID2021-123824OB-I00),
MICIN/AEI/10.13039/501 100011033 and cofi-
nanced with ERDF and the European Union Next
Generation EU/PRT.
REFERENCES
Arevshatyan, S. et al. (2019). Integration and analysis of
clinical and genomic data of neuroblastoma applying
conceptual modeling. IEEE Journal of Biomedical
and Health Informatics.
Arevshatyan, S. et al. (2020). An application of an ehr based
on conceptual modeling to integrate clinical and ge-
nomic data and guide therapeutic strategy. ANALES.
Bernasconi, A., Canakoglu, A., Masseroli, M., Pinoli, P.,
and Ceri, S. (2020). A review on viral data sources and
search systems for perspective mitigation of covid-19.
Briefings in Bioinformatics, 22(2):664–675.
Bernasconi, A. et al. (2017). Conceptual modeling for
genomics: Building an integrated repository of open
data. In Conceptual Modeling, pages 325–339, Cham.
Springer International Publishing.
Burriel Coll, V. (2017). Dise
˜
no y Desarrollo de un Sistema
de Informaci
´
on para la Gesti
´
on de Informaci
´
on sobre
C
´
ancer de Mama. Universitat Polit
`
ecnica de Val
`
encia.
Cahaney, C., Dhir, A., and Ghosh, T. (2022). Role of preci-
sion medicine in pediatric oncology. Pediatric Annals,
51(1):e8–e14.
Castleberry, R. P. (1997). Neuroblastoma. Eur J Cancer,
33(9):1430–7; discussion 1437–8.
Ca
˜
nete, A. et al. (2022). Neuroblastoma in spain: Link-
ing the national clinical database and epidemiological
registries – a study by the joint action on rare cancers.
Cancer Epidemiology, 78:102145.
Ca
˜
nete Nieto, A. et al. (2023). C
´
ancer infantil en Espa
˜
na.
Estad
´
ısticas 1980-2023. Registro Espa
˜
nol de Tumores
Infantiles (RETI-SEHOP). Universitat de Val
`
encia,
Valencia.
Digital Transition. The digital transition in health-
care: An urgent need. En l
´
ınea. Available
at: https://digitalforeurope.eu/the-digital-transition-
in-healthcare-an-urgent-need?lang=en. Last access:
28 of October 2024.
European Comission (2014). Technology readdiness levels.
European Commission Decision C (2014)4995 of 22
July 2014). Online. Available at: https://ec.europa.eu.
Evans, R. S. (2016). Electronic health records: Then, now,
and in the future. Yearbook of Medical Informatics,
25(S 01):S48–S61.
Fern
´
andez Garc
´
ıa, F., Reyes Rom
´
an, J. F., Baghiu, C.-
B., P
´
erez, S., and Pastor L
´
opez, O. (2022). Clin-
gennbl (v2): Integration and analysis of neurob-
lastoma clinical and genomic data. Available at:
http://hdl.handle.net.
Garc
´
ıa Sim
´
on, A., Palacio Le
´
on, A., Reyes Rom
´
an, J. F.,
Casamayor R
´
odenas, J. C., and Pastor, O. (2021). A
conceptual model-based approach to improve the rep-
resentation and management of omics data in preci-
sion medicine. IEEE Access, 9:154071–154085.
GDRP (2016). Regulation (EU) 2016/679 of the European
Parliament and of the Council of 27 April 2016 on the
protection of natural persons with regard to the pro-
cessing of personal data and on the free movement of
such data, and repealing Directive 95/46/EC (General
Data Protection Regulation). Online.
Guerrero Alquicira, R., Rojas Lemus, M., and Fortoul
van der Goes, T. (2017). Histolog
´
ıa y biolog
´
ıa celular.
MCGRAW-HILL.
Martin, A. and Celma, M. (2011). Integrating human
genome variation data: An information system ap-
proach. In 22nd International Workshop on Database
and Expert Systems Applications, pages 65–69.
McCabe, M. G. et al. (2024). Precision medicine for
childhood cancer: Current limitations and future per-
spectives. JCO Precision Oncology, 8(8):e2300117.
PMID: 38207228.
Paton, N. et al. (2000). Conceptual modelling of genomic
information. Bioinformatics, 16(6):458–557.
Reyes Rom
´
an, J. F. and Pastor, O. (2018). Dise
˜
no y desar-
rollo de un sistema de informaci
´
on gen
´
omica basado
en un modelo conceptual hol
´
ıstico del genoma hu-
mano. PhD thesis, Universitat Polit
`
ecnica de Val
`
encia.
Sherry, S. T., Ward, M.-H., Kholodov, M., Baker, J., Phan,
L., Smigielski, E. M., and Sirotkin, K. (2001). dbSNP:
the NCBI database of genetic variation. Nucleic Acids
Research, 29(1):308–311.
Siegel, R. L., Giaquinto, A. N., and Jemal, A. (2024). Can-
cer statistics, 2024. CA: A Cancer Journal for Clini-
cians, 74(1):12–49.
Trujillo, J. C. et al., editors (2018). Conceptual Model-
ing: 37th International Conference, ER 2018, Pro-
ceedings. Springer International Publishing.
UNITECO (2019). ¿Qu
´
e son los protocolos m
´
edicsos?
Available at: https://www.unitecoprofesional.es.
Wieringa, R. J. (2014). Design Science Methodology
for Information Systems and Software Engineering.
Springer.
ENASE 2025 - 20th International Conference on Evaluation of Novel Approaches to Software Engineering
26
