
PatSimBoosting: Enhancing Patient Representations for Disease
Prediction Through Similarity Analysis
Yuzheng Yan
a
, Ziyue Yu
b
and Wuman Luo
∗ c
Faculty of Applied Sciences, Macao Polytechnic University, China
Keywords:
Electronic Health Records, Similarity, Patient Representation Learning, Disease Prediction.
Abstract:
Patient representation learning based on electronic health records (EHR) is crucial for disease prediction.
So far, various deep learning-based methods have been proposed and have made great progress. In particular,
recent research has shown that trends and variations of dynamic features are of great importance in patient rep-
resentation learning. However, these methods ignored the similarity between the patients. Although a number
of similarity-based methods have been proposed for patient representation learning, they regarded each dy-
namic feature as a whole in similarity detection and failed to utilize the important fine-grained characteristics
of each feature. To address this issue, we propose a Patient Similarity-Based Representation Boosting frame-
work (PatSimBoost) to enhance patient representation for disease prediction based on EHR. Our proposed
framework consists of four modules: Frequency Extraction Module (FEM), Similarity Calculation Module
(SCM), Patient Representation Learning Module (PRLM), and Prediction Module (PM). FEM extracts trends
and variations of dynamic features, while SCM employs Dynamic Time Warping (DTW) to assess the similar-
ity between patients. PRLM learns patient representations, and the PM utilizes the representation of the most
similar patient, along with the current patient’s representation, to perform disease prediction. Experimental
results on two real-world public datasets demonstrate that PatSimBoost outperforms existing state-of-the-art
methods in terms of F1-score, AUROC, and AUPRC.
1 INTRODUCTION
Deep learning methods have been widely used for
disease prediction based on electronic health records
(EHR). Typically, EHR data is a set of visit records,
including static features (e.g., patient’s gender and
age), dynamic features (e.g., heart rate and oxygen
saturation ) and diagnosis results (e.g., COVID-19
and other viral pneumonia). The critical problem is
how to effectively learn patient representation based
on EHR datafor disease prediction (Shickel et al.,
2018).
So far, various patient representation learning
methods have been proposed for disease prediction.
For example, ConCare (Ma et al., 2020b) utilized the
attention mechanism to discern the relationships be-
tween different features and used an information de-
cay function to capture the importance variation of
time information. AdaCare (Ma et al., 2020a) adopted
a
https://orcid.org/0009-0009-6214-3203
b
https://orcid.org/0000-0002-1481-9362
c
https://orcid.org/0000-0002-2480-3997
∗
Denotes the corresponding author.
the multi-scale dilated convolutional operator to cap-
ture the variation patterns of historical visit records
and the correlations of different medical features.
StageNet (Gao et al., 2020) used stage-aware Long
Short-Term Memory (LSTM) to extract the long-term
and short-term disease progression patterns in patient
health status. These methods have made remarkable
progress in EHR-based disease prediction. However,
as discussed in MPRE (Yu et al., 2023), they ignored
two very important factors, i.e., trends and variations,
of dynamic features which are important to enhance
the patient representation. Specifically, trend repre-
sents the long-term development direction of the pa-
tient’s dynamic features, reflecting gradual changes in
the patient’s health status. And variation represents
the rapid change or abnormality of the patient’s dy-
namic features in a short period of time, reflecting
temporary or sudden changes in the patient’s health
status. In medical practices, a sustained upward trend
in amyloid beta levels in cerebrospinal fluid (CSF) is
typically associated with an increased risk of develop-
ing Alzheimer’s disease, indicating ongoing neurode-
generation (Nakamura et al., 1994). Besides, signifi-
48
Yan, Y., Yu, Z. and Luo, W.
PatSimBoosting: Enhancing Patient Representations for Disease Prediction Through Similarity Analysis.
DOI: 10.5220/0013224300003944
In Proceedings of the 10th International Conference on Internet of Things, Big Data and Security (IoTBDS 2025), pages 48-55
ISBN: 978-989-758-750-4; ISSN: 2184-4976
Copyright © 2025 by Paper published under CC license (CC BY-NC-ND 4.0)

cant and abrupt variations in procalcitonin levels may
suggest a severe bacterial infection or sepsis (Becker,
2001). MPRE achieved higher accuracy in disease de-
tection by adopting the trend and the variation infor-
mation of dynamic features.
However, all the above works ignored the patient
similarity in representation learning. These days,
a number of similarity-based patient representation
learning methods have been proposed (Zhang et al.,
2021) (Yu et al., 2024) (Yu et al., 2022). However,
they regarded each dynamic feature as a whole in
similarity calculation and failed in detecting its fine-
grained characteristics. For example, Patient A, B and
C all have hyperglycemia. According to the tradi-
tional methods, they are similar patients. Thus, the
representations of Patient A and Patient B can be uti-
lized to enhance the representation of Patient C. How-
ever, while Patient A’s blood glucose levels are on a
declining trend, both Patient B and Patient C are expe-
riencing an upward trend in their blood glucose levels.
Consequently, compared with Patient A, Patient B is
more similar to Patient C (Goyal et al., 2009). Ignor-
ing this distinction may lead to erroneous assessments
of the patient’s health status, potentially resulting in
inappropriate treatment and management (Giannoula
et al., 2020).
To address this issue, in this paper, we propose
a Patient Similarity-Based Representation Boosting
framework (PatSimBoost) to enhance patient repre-
sentation for disease prediction. The objective is
to better detect the similar patients based on trends
and variations for representation learning. Specifi-
cally, the PatSimBoost consists of four components,
i.e., Frequency Extraction Module (FEM), Similarity
Calculation Module (SCM), Patient Representation
Learning Module (PRLM) and Prediction Module
(PM). We adopt symlets wavelet to obtain trend and
variation information of dynamic features in FEM.
Then, we adopted Dynamic Time Warping (DTW) to
calculate the similarity based on the extracted trend
and variation information in SCM. In addition, DTW
can handle differences in frequency by aligning visit
records that are similar but occur at different times.
After that, PRLM is used for learning the patient
representation. Based on SCM and PRLM results,
we identified the most similar patient representation
to enhance the current patient’s representation for
disease prediction. Finally, we predict the disease
through the PM.
In summary, the main contributions of this paper
are listed as follows:
1. We propose PatSimBoost to enhance the patient
representation for the disease prediction task.
The proposed framework consists of FEM, SCM,
PRLM and PM.
2. We adopt FEM to extract trends and variations in-
formation of dynamic features. SCM is employed
to calculate the similarity between patients.
3. We use PRLM for patient representation learning,
employing the most similar patient’s representa-
tion as auxiliary information in PM for prediction.
4. We evaluate the effectiveness of PatSimBoost on
two real-world public datasets. The experiment
results show that the proposed framework outper-
forms state-of-the-art baseline methods in terms
of F1-score, AUROC and AUPRC.
In the rest of this paper, we summarize the major
works in Section 2 and discuss the methodology of
PatSimBoost with its modules in Section 3. We elabo-
rate experiment results in Section 4 and conclude this
work in Section 5.
2 RELATED WORK
We summarize existing works into three categories,
namely correlation detection, variation pattern recog-
nition, and similar patient enhancement methods.
2.1 Correlation Detection
So far, various methods have been proposed to study
the correlation between different medical features of
patients. ConCare (Ma et al., 2020b) leverages the
attention mechanism to capture feature dependencies
and employs an information decay function to learn
patient representation. Retain (Choi et al., 2016) em-
ploys a two-level RNN with attention mechanisms to
learn the weights of each visit record. SAnD (Song
et al., 2018) employs a 1D convolutional layer to cap-
ture the correlation between features and utilizes an
attention mechanism to identify the important visit.
2.2 Variation Pattern Recognition
The dynamic features and health status of patients
often show irregular variations over time, posing a
challenge for accurate representation. Several studies
have been proposed to address this issue by exploring
variation patterns of historical dynamic information.
Specifically, T-LSTM (Baytas et al., 2017) addresses
the issue of irregular patient visit intervals by time de-
cay. MPRE (Yu et al., 2023) employs frequency con-
version to capture the variation and trends in patient
features, enabling the learning of time variation pat-
terns in dynamic features. AdaCare (Ma et al., 2020a)
PatSimBoosting: Enhancing Patient Representations for Disease Prediction Through Similarity Analysis
49

uses 1D dilated convolutional network to learn the
variations of patients’ health status at different scales.
StageNet(Gao et al., 2020) uses LSTM to extract dis-
ease progression based on different time intervals.
2.3 Similar Patients Enhancement
To address the issues of data sparsity and data
missing, a number of methods have been proposed
to leverage the data from similar patients for bet-
ter patient representation learning. Specifically,
GRASP (Zhang et al., 2021) clusters the health
status of patients, finds a group of patients with
similar health status to the current patient, and uses
GCN to enhance the current patient representation.
PPN (Yu et al., 2024) uses clustering to find a group
of ”prototype patients” closest to the cluster centroid.
It then uses the similarity between the current patient
and this group of ”prototype patients” to enhance the
patient representation. SiaCo (Yu et al., 2022) finds
similar patients at the patient encounter level and
similar patients at the medical concept level, and uses
the similar patient information at these two levels to
enhance the representation of the current patient.
3 METHODOLOGY
In this section, we first give the problem formulation
of disease prediction tasks. Then, we show the overall
model framework of PatSimBoost. Finally, we will
introduce each module of proposed framework.
3.1 Problem Formulation
Let V = (r
1
, r
2
, ..., r
t
) ∈ R
|t|
be the vector of pa-
tient dynamic feature d. The dynamic features are
recorded |t| times. We denote the static feature as
C = (c
1
, c
2
, ..., c
|s|
) ∈ R
|s|
. In this work, we have
|n| patients, |d| dynamic features and |s| static fea-
tures. We formulate the disease prediction task with
the multi-class classification problem, in which each
patient has a corresponding label y. The prediction
function can be expressed as ˆy = F(V, s), where ˆy is
the predicted diagnosis result.
3.2 Framework Overview
Figure 1 presents an overview of the proposed Pat-
SimBoost.First, the Frequency Extraction Module
(FEM) extracts the frequency information for all dy-
namic features of each patient. High-frequency in-
formation is used as the variation information of the
corresponding feature, while low-frequency signals
are used as the trend information of the correspond-
ing feature. Then, the similarity based on the trend
and variation of each feature of all patients is calcu-
lated. Then the similarity of the corresponding fea-
tures of different patients can be obtained. Accord-
ing to the similarity between different features of all
patients, a similarity matrix can be finally obtained,
which records the similarity between all patients.The
backbone model learns and saves each patient’s repre-
sentation information in a representation list.The pre-
diction module receives input from both the current
patient’s representation and that of the most similar
patient. These combined inputs are used to perform
the final disease prediction.
3.3 Frequency Extraction Module
(FEM)
To fully capture the temporal information of each dy-
namic feature, we use symlets wavelet to decompose
dynamic feature data of patients.Decomposed low-
frequency information pertains to the trend of dy-
namic features, while high-frequency information re-
flects their variations. Extracting trend and variation
components enhances the original temporal informa-
tion, offering a more comprehensive understanding of
the data.
F
∗
=
T
∑
t=0
V
d
·
1
√
2
·Φ
n
(1)
H
∗
=
T
∑
t=0
V
d
·
1
√
2
·Ψ
n
(2)
where F = [F
1
, F
2
, ··· , F
d
] ∈R
|d|×m
refers to the low-
frequency information extracted by wavelet trans-
form of dynamic features, representing trend infor-
mation. H = [H
1
, H
2
, ··· , H
d
] ∈ R
|d|×m
refers to
the high-frequency information extracted by wavelet
transform of dynamic features, representing variation
information.∗ means the one specific extracted infor-
mation.
3.4 Similarity Calculation Module
(SCM)
We assess patient similarity in this module using the
extracted trend and variation data. Specifically, we
apply Dynamic Time Warping (DTW) (M
¨
uller, 2007)
to compute the similarity of this information. Below
IoTBDS 2025 - 10th International Conference on Internet of Things, Big Data and Security
50
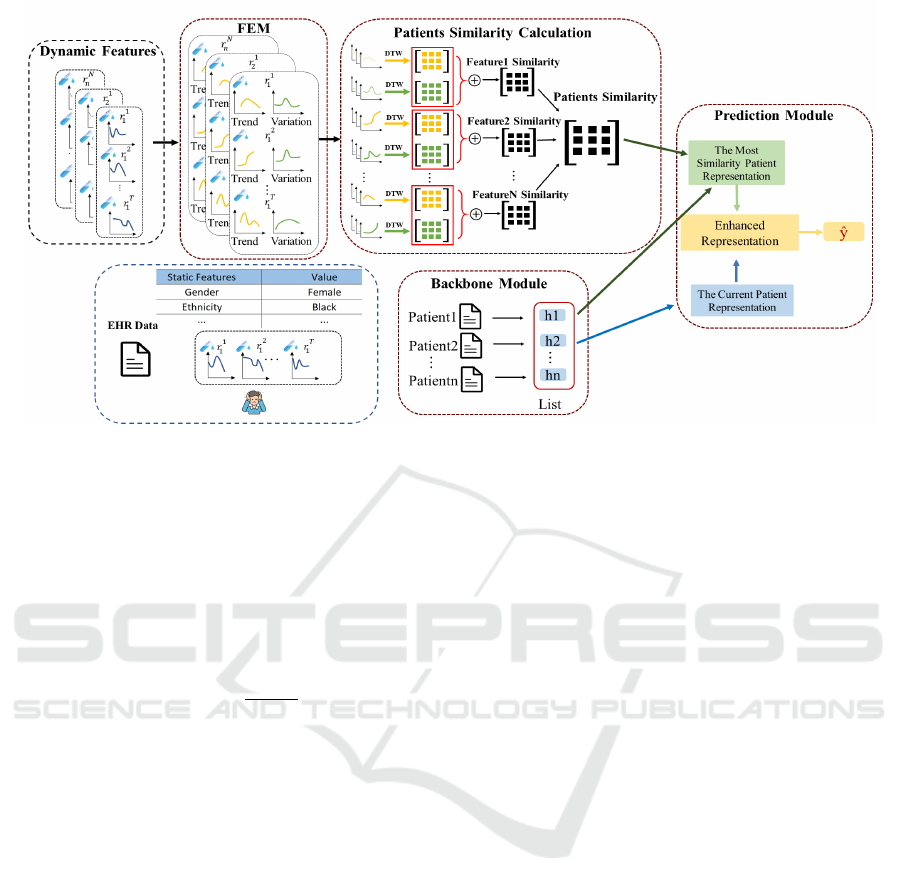
Figure 1: The overview of PatSimBoost. First, FEM conducts frequency decomposition on dynamic patient features to capture
trends and variations. Then, we analyze the patient similarity based on the trends and variations obtained from FEM. After
that, the patient representation module is used to capture original patient information. In the prediction module, we use the
information of the most similar patients to enhance the representation of the original patient and make the final prediction.
is the DTW calculation formula:
DTW (A, B) = dist(i, j) + min
DTW (i −1, j −1)
DTW (i −1, j)
DTW (i, j −1)
(3)
DTW (A
1
, B
1
) = dist(A
1
, B
1
) (4)
dist(i, j) =
p
i
2
+ j
2
(5)
where A and B represent two sequences to be calcu-
lated. The length of sequence A is i and the length of
sequence B is j. dist(i, j) refers to the distance calcu-
lation between i and j by Euclidean. The initial state
is equation(4). A
1
and B
1
refer to the values of the
first time point of sequences A and B respectively.
Here we consider using DTW to calculate the trend
and variation of patient features. The following for-
mula shows how we calculate the trend and variation
of a specific feature:
Similarity(F
∗
m
, F
∗
l
) = DTW (F
∗
m
, F
∗
l
) (6)
Similarity(H
∗
m
, H
∗
l
) = DTW (H
∗
m
, H
∗
l
) (7)
where m and l represent two different patients, F
∗
and
H
∗
refer to the trend and variation of a specific fea-
ture. The larger the value, the stronger the similar-
ity, and vice versa. We then calculate the similarity
of dynamic feature representation based on trend and
variation. The similarity of a patient special dynamic
feature is as follows:
Similarity(E
∗
m
, E
∗
l
) = W
α
·Similarity(F
∗
m
, F
∗
l
)+
W
β
·Similarity(H
∗
m
, H
∗
l
) (8)
where W
α
and W
β
refer to the weights of the simi-
larity between trend and variation. E
∗
represents a
specific dynamic feature of the patient. Each feature
represents a different aspect of the patient’s health sta-
tus. In order to fully reflect the similarity between
patients, we need to consider the similarity of all dy-
namic features, and finally we can get the similarity
between patients. The similarity between different
patients can be expressed as:
Similarity(m, l) =
|d|
∑
k=1
W ·Similarity(E
k
m
, E
k
l
) (9)
where|d|is the number of EHR dynamic features.
W ∈ R
|d|
refers to the weights of different specific
correspondences. Similarity(m, n) ∈ R
|n|×|n|
is a pa-
tient similarity matrix that contains the similarities be-
tween all patients. The closer the value is to 1, the
stronger the similarity between patients is, and 0 rep-
resents the weakest similarity between patients.
3.5 Patient Representation Learning
Module (PRLM)
Inspired by the work (Ma et al., 2020b), we adopt
ConCare to identify the important dynamic features,
which can be expressed as:
h
T
= ConCare(V
n
) (10)
where h
T
∈R
n×p
denotes all patients’ representation.
Based on the most similar patient representation, we
PatSimBoosting: Enhancing Patient Representations for Disease Prediction Through Similarity Analysis
51

combine the representation with that as auxiliary in-
formation to the current patient.
s = W
T
×h
T
+W
c
×h
c
(11)
where W
T
, W
c
∈ R
n
refer to the learnable parameters.
s is the final representation information of the cur-
rent patient, h
T
represents the current patient repre-
sentation information learned by the ConCare, and h
c
refers to the representation information of the most
similar patient.
3.6 Prediction Module (PM)
Our objective is to predict diseases using the learned
patient representation. As we are dealing with multi-
classification tasks, the final prediction probability is
expressed as follows:
ˆy = So f tmax(W
s
×s) (12)
where W
s
∈ R
n
. Finally, the final loss is calculated by
the cross entropy loss function.
L(y, ˆy) = −
1
N
|n|
∑
n=1
y
n
|r|
∑
r=1
log( ˆy
r
) (13)
where |n|is the number of patients, ˆy
r
is the prediction
of patient n. |r|means the number of the disease class.
4 EXPERIMENTS
In this section, we experimentally study the perfor-
mance of the proposed PatSimBoost in two real-world
datasets. First, we introduce the two datasets. Then,
we describe the experimental settings, baseline mod-
els and the metrics used for performance evaluation.
Finally, we compare the PatimBoost with the base-
lines and analyze the experimental results.
4.1 Datasets
The SCRIPT CarpeDiem Dataset(Markov et al.,
2023) is a multi-classification dataset focusing on
pneumonia treatment in ICU patients. It contains clin-
ical data from 585 patients collected between June
2018 and March 2022, totaling 12,495 ICU patient
days. After feature engineering, 334 patients were se-
lected, each with an average of 23 visit records and
26 dynamic features per record. The dataset classi-
fies patients into four categories based on their diag-
nosis: COVID-19, respiratory viral pneumonia, bac-
terial pneumonia, and respiratory failure.
Besides, we use the Health Facts Database(Strack
et al., 2014) to predict whether diabetic patients will
suffer from circulatory system diseases in the future.
The visit records span 10 years (1999-2008) and
include 350 patients. We classify these diseases into
five categories based on the WHO’s ICD-9 codes (for
Disease Control et al., 2013). There are five labels
for these categories.
4.2 Baselines
We compare the proposed PatSimBoost with the
following baseline methods
• ConCare (Ma et al., 2020b) uses the attention
mechanism to learn the correlation between dif-
ferent dynamic features.The information decay
function simulates the gradual loss of information
importance.
• AdaCare (Ma et al., 2020a) employs a 1D dilated
convolutional network to analyze the variations
patterns in patients’ dynamic features across mul-
tiple scales time intervals.
• StageNet (Gao et al., 2020) utilizes LSTM to ex-
tract medical information over different time in-
tervals, enabling to capture changes of patient
health status.
• GRASP (Zhang et al., 2021) finds groups similar
to the current patient and uses GCN to enhance
the original patient representation.
• MPRE (Yu et al., 2023) employs frequency con-
version to capture the variations and trends of
each patient feature, allowing it to learn the tem-
poral patterns of dynamic features.
PPN (Yu et al., 2024) enhances patient represen-
tation by considering information of the given pa-
tients and prototypes while providing interpreta-
tion.
4.3 Evaluation Metrics
In this work, we use area under the curve (AUROC),
area under the precision-recall curve (AUPRC), and
F1 score as evaluation indicators to evaluate the
performance of PatSimBoost and Baseline methods.
AUROC is used to measure the overall classification
performance of the model, indicating the trade-off
between the true positive rate and the false positive
rate at different thresholds. AUPRC focuses on
the relationship between the precision and recall
of the model when dealing with imbalanced data.
The F1 Score is a balanced metric representing the
harmonic mean of precision and recall (Davis and
IoTBDS 2025 - 10th International Conference on Internet of Things, Big Data and Security
52
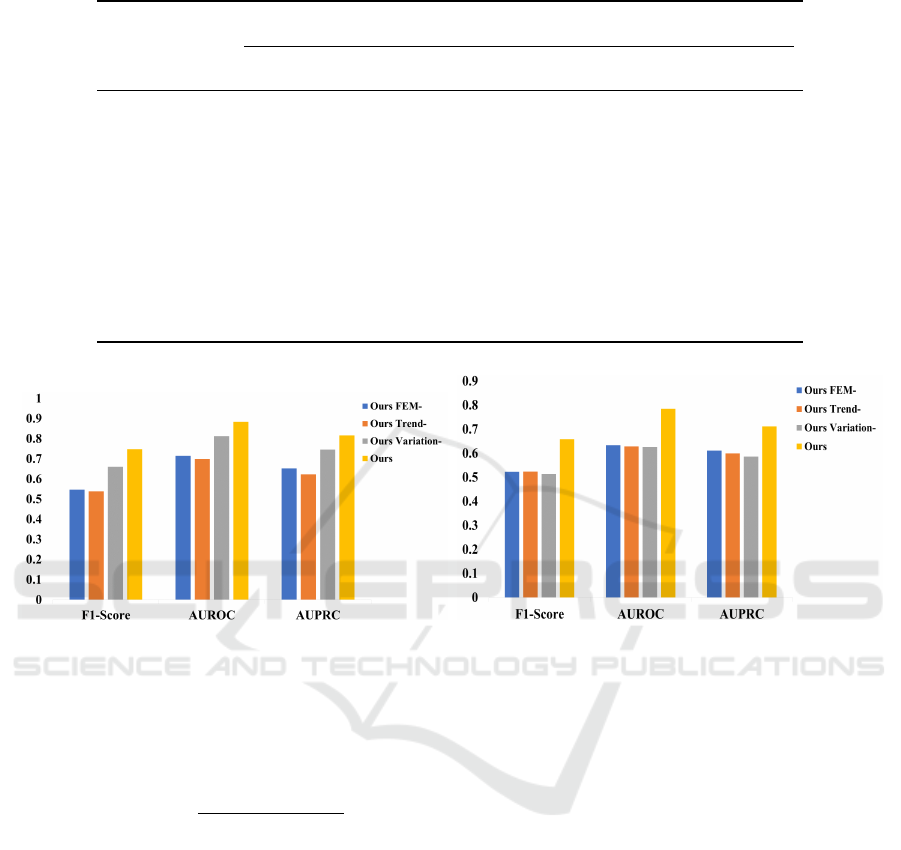
Table 1: Average performances of ours and baseline methods.
Models SCRIPT CarpeDiem Dataset Health Facts Database
F1-score AUROC AUPRC F1-score AUROC AUPRC
ConCare 0.3373 0.5851 0.5054 0.4417 0.5063 0.5180
AdaCare 0.4636 0.5995 0.5279 0.4421 0.5138 0.4911
StageNet 0.4270 0.6115 0.5330 0.4411 0.5194 0.476
GRASP 0.6129 0.8153 0.7534 0.6001 0.6796 0.6941
MPRE 0.6892 0.8785 0.7926 0.6403 0.7490 0.7121
PPN 0.5499 0.7074 0.6179 0.4407 0.5392 0.5199
OURS 0.7475 0.8836 0.8166 0.6576 0.7847 0.7115
(a) (b)
Figure 2: Ablation experiment results of F1-score, AUROC, and AUPRC. (a) shows the performance on the SCRIPT Car-
peDiem Dataset, and (b) shows the performance on the Health Facts Database.
Goadrich, 2006). The calculation formula of F1 score
is expressed as follows:
F1score = 2 ·
Precision ·Recall
Precision + Recall
(14)
4.4 Experimental Setup
Our proposed PatSimBoost and baseline methods are
based on Python 3.9 and Pytorch framework. We
use the Adam (Reddi et al., 2019) optimizer with a
learning rate of 10
−4
and the batch size is 64. We
use symlets-18 to perform feature decomposition on
the SCRIPT CarpeDiem Dataset and symlets-14 to
perform feature decomposition on the Health Facts
Database. We use 10-fold cross validation (Fushiki,
2011) and report the average performance in terms of
F1 score, AUROC and AUPRC.
4.5 Performance Analysis
The average performance results of PatSimBoost and
six baseline methods on F1 score, AUROC, and
AUPRC are shown in Table 1. We can see that Pat-
SimBoost outperforms the other models in most cases
in both datasets. In the SCRIPT CarpeDiem Dataset,
PatSimBoost outperforms the best method by 7.8% in
F1 score, 0.58% in AUROC, and 2.94% in AUPRC.
In the Health Facts Database, PatSimBoost surpasses
the best baseline method by 2.63% in F1 score and
4.55% in AUROC.
We find that MPRE outperforms other baseline
methods by effectively extracting trend and variation
information from dynamic features and capturing
the correlation between them, which other methods
do not consider this information. The GRASP and
PPN models perform well by utilizing auxiliary
information from similar patients to enhance the
representation of current patient. The reason why
PatSimBoost has better performance is that we also
PatSimBoosting: Enhancing Patient Representations for Disease Prediction Through Similarity Analysis
53

consider mining the trend and variation information
in dynamic patient features from the frequency
perspective. In addition, we use the similarity of
trend and variation to calculate the similarity between
patients. We adopt the most similar patient repre-
sentation to enhance the target patient representation
information. Our method can better help us find
similar patients and effectively enhance the original
patient representation information.
Table 2: Ablation experiment comparison study.
Models FEM Trend Variation
Ours
FEM−
× × ×
Ours
Trend−
✓ × ✓
Ours
Variation−
✓ ✓ ×
OURS ✓ ✓ ✓
4.6 Ablation Study
We conduct ablation experiments to verify the effec-
tiveness of each module in PatSimBoost. Table 2
shows the configuration of this ablation study.
• Ours
FEM−
directly calculates the similarity be-
tween patients based on feature similarity, without
converting dynamic features into the trends and
variations using FEM.
• Ours
Trend−
calculates only the similarity of vari-
ation in dynamic features, not the similarity of
trends after frequency extraction.
• Ours
Variation−
uses the FEM on dynamic features,
but we only calculate the similarity of the trend
information, not the similarity of the variation.
• Ours employs FEM to extract and analyze the
trends and variations of dynamic features, calcu-
lating their similarity accordingly.
As shown in Figure 2, the comparison results in
the SCRIPT CarpeDiem Dataset and the Health Facts
Database indicate that the performance becomes sub-
optimal when relying solely on trend or variation.
The reason is that considering only trends will ignore
short-term fluctuations or emergencies in the patient’s
features. If only variation is considered, the long-term
trend of variations in the patient’s features will be lost.
Therefore, by considering both trends and variations
of patients’ dynamic features, we can better extract
the patient’s detailed important information and ef-
fectively learn the patient representation.
5 CONCLUSION
In this paper, we propose the framework called Pat-
SimBoost to enhance patient representation based on
similar patients. PatSimBoost uses frequency-based
feature extraction and similarity analysis to analyze
patient data. Our Frequency Extraction Module ef-
fectively distinguishes high-frequency variation from
low-frequency trend information in patient features.
By calculating the similarity of these features across
patients, we constructed a comprehensive similarity
matrix. This matrix not only facilitates the identifi-
cation of the most similar patients but also enhances
the predictive accuracy of our model. By integrat-
ing representation information from both the target
patient and their most similar counterpart, our pre-
diction module delivers improved disease prediction
outcomes. This methodology offers a robust frame-
work for personalized healthcare, enabling more ac-
curate and tailored treatment strategies. Future work
will focus on refining the model and exploring its ap-
plication to diverse clinical datasets.
ACKNOWLEDGEMENTS
This work was supported in part by the Macao
Polytechnic University – Research on Representation
Learning in Decision Support for Medical Diagnosis
(RP/FCA-11/2022).
REFERENCES
Baytas, I. M., Xiao, C., Zhang, X., Wang, F., Jain, A. K.,
and Zhou, J. (2017). Patient subtyping via time-
aware lstm networks. In Proceedings of the 23rd ACM
SIGKDD international conference on knowledge dis-
covery and data mining, pages 65–74.
Becker, K. (2001). Procalcitonin: how a hormone became a
marker and mediator of sepsis. Swiss medical weekly,
131(4142):595–602.
Choi, E., Bahadori, M. T., Sun, J., Kulas, J., Schuetz, A.,
and Stewart, W. (2016). Retain: An interpretable pre-
dictive model for healthcare using reverse time atten-
tion mechanism. Advances in neural information pro-
cessing systems, 29.
Davis, J. and Goadrich, M. (2006). The relationship be-
tween precision-recall and roc curves. In Proceed-
ings of the 23rd international conference on Machine
learning, pages 233–240.
for Disease Control, C., Prevention, for Disease Control, C.,
Prevention, et al. (2013). International classification
of diseases, ninth revision, clinical modification (icd-
9-cm).
IoTBDS 2025 - 10th International Conference on Internet of Things, Big Data and Security
54

Fushiki, T. (2011). Estimation of prediction error by us-
ing k-fold cross-validation. Statistics and Computing,
21:137–146.
Gao, J., Xiao, C., Wang, Y., Tang, W., Glass, L. M., and
Sun, J. (2020). Stagenet: Stage-aware neural networks
for health risk prediction. In Proceedings of The Web
Conference 2020, pages 530–540.
Giannoula, A., Centeno, E., Mayer, M.-A., Sanz, F., and
Furlong, L. I. (2020). A system-level analysis of
patient disease trajectories based on clinical, phe-
notypic and molecular similarities. Bioinformatics,
37(10):1435–1443.
Goyal, A., Mehta, S. R., Gerstein, H. C., D
´
ıaz, R., Afzal,
R., Xavier, D., Zhu, J., Pais, P., Lisheng, L., Kazmi,
K. A., Zubaid, M., Piegas, L. S., Widimsky, P., Budaj,
A., Avezum, A., and Yusuf, S. (2009). Glucose levels
compared with diabetes history in the risk assessment
of patients with acute myocardial infarction. Ameri-
can Heart Journal, 157(4):763–770.
Ma, L., Gao, J., Wang, Y., Zhang, C., Wang, J., Ruan, W.,
Tang, W., Gao, X., and Ma, X. (2020a). Adacare: Ex-
plainable clinical health status representation learning
via scale-adaptive feature extraction and recalibration.
In Proceedings of the AAAI Conference on Artificial
Intelligence, volume 34, pages 825–832.
Ma, L., Zhang, C., Wang, Y., Ruan, W., Wang, J., Tang, W.,
Ma, X., Gao, X., and Gao, J. (2020b). Concare: Per-
sonalized clinical feature embedding via capturing the
healthcare context. In Proceedings of the AAAI Con-
ference on Artificial Intelligence, volume 34, pages
833–840.
Markov, N., Gao, C. A., Stoeger, T., Pawlowski, A.,
Kang, M., Nannapaneni, P., Grant, R., Rasmussen, L.,
Schneider, D., Starren, J., Wunderink, R., Budinger,
G. S., Misharin, A., Singer, B., and Investigators, N.
S. S. (2023). Script carpediem dataset: demographics,
outcomes, and per-day clinical parameters for criti-
cally ill patients with suspected pneumonia (version
1.1.0).
M
¨
uller, M. (2007). Dynamic time warping. Information
retrieval for music and motion, pages 69–84.
Nakamura, T., Shoji, M., Harigaya, Y., Watanabe, M.,
Hosoda, K., Cheung, T. T., Shaffer, L. M., Golde,
T. E., Younkin, L. H., Younkin, S. G., et al. (1994).
Amyloid β protein levels in cerebrospinal fluid are el-
evated in early-onset alzheimer’s disease. Annals of
Neurology: Official Journal of the American Neuro-
logical Association and the Child Neurology Society,
36(6):903–911.
Reddi, S. J., Kale, S., and Kumar, S. (2019). On the
convergence of adam and beyond. arXiv preprint
arXiv:1904.09237.
Shickel, B., Tighe, P. J., Bihorac, A., and Rashidi, P. (2018).
Deep ehr: A survey of recent advances in deep learn-
ing techniques for electronic health record (ehr) anal-
ysis. IEEE Journal of Biomedical and Health Infor-
matics, 22(5):1589–1604.
Song, H., Rajan, D., Thiagarajan, J., and Spanias, A.
(2018). Attend and diagnose: Clinical time series
analysis using attention models. In Proceedings of the
AAAI conference on artificial intelligence, volume 32.
Strack, B., DeShazo, J. P., Gennings, C., Olmo, J. L., Ven-
tura, S., Cios, K. J., and Clore, J. N. (2014). Impact
of hba1c measurement on hospital readmission rates:
analysis of 70,000 clinical database patient records.
BioMed research international, 2014(1):781670.
Yu, F., Cui, L., Cao, Y., Liu, N., Huang, W., and Xu, Y.
(2022). Similarity-aware collaborative learning for
patient outcome prediction. In International Confer-
ence on Database Systems for Advanced Applications,
pages 407–422. Springer.
Yu, Z., Wang, J., Luo, W., Tse, R., and Pau, G. (2023).
Mpre: Multi-perspective patient representation ex-
tractor for disease prediction. In 2023 IEEE Inter-
national Conference on Data Mining (ICDM), pages
758–767. IEEE.
Yu, Z., Zhang, C., Wang, Y., Tang, W., Wang, J., and Ma,
L. (2024). Predict and interpret health risk using ehr
through typical patients. In ICASSP 2024-2024 IEEE
International Conference on Acoustics, Speech and
Signal Processing (ICASSP), pages 1506–1510. IEEE.
Zhang, C., Gao, X., Ma, L., Wang, Y., Wang, J., and Tang,
W. (2021). Grasp: generic framework for health status
representation learning based on incorporating knowl-
edge from similar patients. In Proceedings of the AAAI
conference on artificial intelligence, volume 35, pages
715–723.
PatSimBoosting: Enhancing Patient Representations for Disease Prediction Through Similarity Analysis
55
