
Bioimages Synthesis and Detection Through Generative Adversarial
Network: A Multi-Case Study
Valeria Sorgente
1
, Ilenia Verrillo
1
, Mario Cesarelli
2
, Antonella Santone
1
, Fabio Martinelli
3
and
Francesco Mercaldo
1
1
Department of Medicine and Health Sciences “Vincenzo Tiberio”, University of Molise, Campobasso, Italy
2
Department of Engineering, University of Sannio, Benevento, Italy
3
Institute for High Performance Computing and Networking, National Research Council of Italy (CNR), Rende (CS), Italy
{valeria.sorgente, antonella.santone, francesco.mercaldo}@unimol.it, i.verrillo@studenti.unimol.it,
Keywords:
GAN, Generative Adversarial Networks, Bioimage, Deep Learning, Classification.
Abstract:
The rapid advancement of Generative Adversarial Networks technology raises ethical and security concerns,
emphasizing the need for guidelines and measures to prevent misuse. Strengthening systems to differentiate
real from synthetic images and ensuring responsible application in clinical settings could address data scarcity
in the biomedical field. For these reasons, considering the increasing popularity of the possibility to generate
synthetic images by exploiting artificial intelligence, we investigate the application of Generative Adversar-
ial Networks to generate realistic synthetic bioimages for common pathology representations. We propose a
method consisting of two steps: the first one is related to the training of a Deep Convolutional Generative
Adversarial Network, while the second step is represented by the evaluation of the bioimages quality using
classification-based metrics, comparing synthetic and real images. The model demonstrated promising results,
achieving visually realistic images for datasets such as PathMNIST and RetinaMNIST, with accuracy improv-
ing over training epochs. However, challenges arose with datasets like ChestMNIST and OCTMNIST, where
image quality was limited, showing poor detail and distinguishability from real samples.
1 INTRODUCTION AND
RELATED WORK
In recent years, deep learning (Mercaldo et al., 2022;
Zhou et al., 2023) has become one of the most popu-
lar techniques in the field of medical image analysis
(He et al., 2024; Huang et al., 2024), especially for
generative learning.
Among different models, Generative Adversarial
Network (GAN) is prominent for synthesizing realis-
tic medical image. This model is standout capabilities
in tasks such as generating images from textual de-
scriptions, upscaling of visual quality, and conversion
between different image styles. Because of their ver-
satility, GAN has found application in several areas
of medical imaging, which include digital pathology,
radiology and clinical neuroscience.
As a matter of fact, GAN can be used in the
biomedical field (Mercaldo et al., 2023) for appli-
cations such as medical image generation or clinical
data simulation. One of the major challenges in clin-
ical practice is the limited availability of high-quality
labeled biomedical images needed to train deep learn-
ing models for diagnostic applications. GAN offer a
solution to this problem by generating realistic syn-
thetic medical images, thereby expanding the avail-
able datasets and improving the robustness of models.
However, these technologies present risks of fraudu-
lent uses. In particular, GAN can be used to falsify
diagnostic images, such as X-ray or MRI, aimed to
manipulate diagnoses or research outcomes. Further-
more, the generation of altered synthetic data could
compromise the validity of clinical or epidemiologi-
cal studies. These possible scenarios raise ethical is-
sues and require control strategies to ensure the in-
tegrity and veracity of biomedical data. The use of
GAN is also complicated by some issues that limit
their potential. These include difficulties in training
and phenomena such as mode collapse, which reduce
model’s ability to generate different and accurate data.
Numerous studies have explored the application
of GANs in biomedical fields for various purposes
(Huang et al., 2022; Zhou et al., 2021; Huang et al.,
332
Sorgente, V., Verrillo, I., Cesarelli, M., Santone, A., Martinelli, F. and Mercaldo, F.
Bioimages Synthesis and Detection Through Generative Adversarial Network: A Multi-Case Study.
DOI: 10.5220/0013228900003911
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 18th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2025) - Volume 1, pages 332-339
ISBN: 978-989-758-731-3; ISSN: 2184-4305
Proceedings Copyright © 2025 by SCITEPRESS – Science and Technology Publications, Lda.

2023; Huang et al., 2021). For example, Orlando et
al. (Orlando et al., 2018) developed a method for
generating retinal fundus images with simulated le-
sions, aiming to enhance diagnostic models, while Fu
et al. (Fu et al., 2018) used GANs to augment reti-
nal fundus image data. While GANs have been ap-
plied in biomedical domains for applications like reti-
nal vessel segmentation and liver lesion classification,
this paper is focused on creating synthetic images that
closely resemble authentic ones and can evade detec-
tion by trained classifiers. We expected that with an
increase in training epochs, the quality of the syn-
thetic images improves, rendering them progressively
more realistic and increasingly challenging for clas-
sifiers to differentiate from real samples. This trend
should underscore the potential implications of GANs
in applications where realistic image synthesis is crit-
ical, as well as the challenges they may pose for cur-
rent diagnostic and classification systems.
As a matter of fact, this study aims to evaluate
the capability of GAN in producing two-dimensional
medical images from six different datasets represent-
ing common pathologies. In a nutshell, we introduce
an approach designed to assess the potential impact
of GAN-generated retinal fundus images on classifi-
cation tasks. Specifically, we employ a Deep Convo-
lutional GAN (DCGAN) to generate synthetic images
based on an existing dataset of retinal fundus images.
For this purpose, we exploit a set of of machine learn-
ing algorithms that, through image filters, evaluate the
quality of images generated by the GAN, providing
performance evaluation metrics, i.e., Precision, Re-
call, Accuracy and F-Measure.
The paper proceeds as follows: in the next sec-
tion we present the proposed method for bioimages
synthesis and detection, Section 3 presents the results
obtained from the experimental analysis and, finally,
in the last section conclusion and future research lines
are drawn.
2 THE METHOD
In this section we present the proposed method aimed
to understand whether it is possible by exploiting
machine learning to discriminate between real-world
bioimages and GAN-generated ones.
The proposed method is composed by two main
steps: the first one is related to bioimages genera-
tion (shown in Figure 1), the second one is related
to discrimination between real and fake bioimages
(shown in Figure 2). Thus, the first step of the pro-
posed method is the bioimage generation by means of
a GAN: this step is shown in Figure 1.
GAN is a machine learning model used to gener-
ate realistic data from random inputs. It consists of
two main components, a generator and a discrimina-
tor, that compete with each other, as shown in Figure
1. The idea is to train the generator to create data that
are indistinguishable from real data for the discrimi-
nator.
The generator will be completely trained when the
discriminator assigns a value of 0.5 to all images, de-
noting its inability to distinguish the inputs.
There are several variants of GAN, in this paper
we experiment with the DCGAN.
The developed DCGAN takes as input 28x28
pixel images, converted to grayscale and normalized
in the interval [-1,1], to align with the generator’s tanh
activation function. The generator receives a random
vector of size 100, which is transformed through a
series of layers: Dense, used to reorganize the noise
vector, BatchNormalization, to stabilize the training
process, Conv2DTranspose, to enlarge the image to a
format of 28x28x1.
The discriminator architecture includes, instead,
two sequences of Conv2D, BatchNormalization and
LeakyReLU to perform the downsampling, and the
sigmoid activation function in the last layer to obtain
a value suitable for binary classification. The train-
ing of the DCGAN is based on the competition be-
tween generator and discriminator. This dynamic is
managed using a loss function, different between the
two networks. The loss of the discriminator is com-
puted as the arithmetic mean between the loss of the
real images, with class 1 label, and that of the gen-
erated images, of class 0. The loss of the generator,
instead, is represented by the loss of the generated im-
ages. Its purpose is to maximize the discriminator’s
predictions, so that it classifies them with label 1.
The images generated during training were saved
for the next step, shown in Figure 2 where were com-
pared with the real ones.
The aim of the second step is to extract numeri-
cal features from the (real and fake) bioimages and to
build a set of machine learning models with the aim
to understand if it is possible to discriminate between
bioimages GAN-generated.
To obtain numerical features from real and gener-
ated bioimages, the images were subjected to a pre-
processing, applying image filters for the extraction
of features, as shown in Figure 2. This is essential to
optimize the classification accuracy, since it allows to
visualize salient features regarding the structure and
the chromatic variations of the images.
In particular, the filters used are the following:
• AutoColorCorrelogramFilter: measures the spa-
tial correlation between the colors that compose
Bioimages Synthesis and Detection Through Generative Adversarial Network: A Multi-Case Study
333
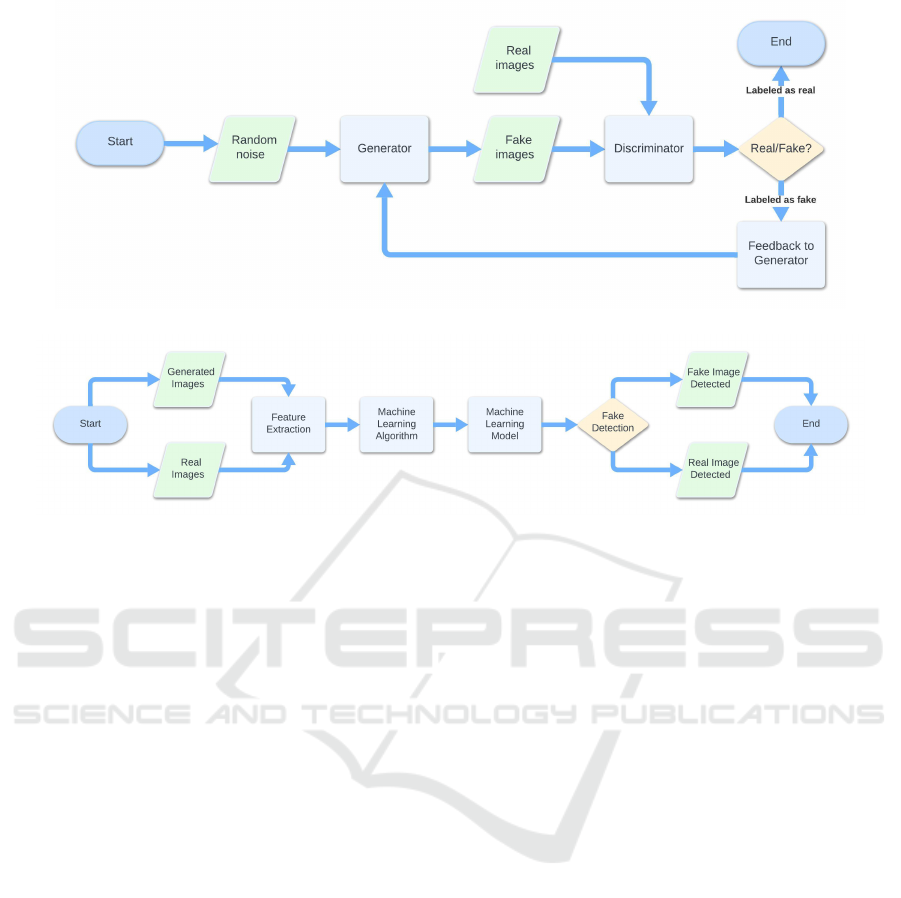
Figure 1: The first step of the proposed method related synthetic bioimages generation.
Figure 2: The second step of the proposed method related to synthetic bioimages detection.
the image, allowing to define the distribution of
the colors.
• BinaryPatternsPyramidFilter: extracts intensity
patterns around the points of the image, identify-
ing texture variations.
• ColorLayoutFilter: divides the image into a grid
of 64 blocks and calculates the average color for
each of them.
• FCTHFilter (Fuzzy Color and Texture Histogram
Filter): combines information on the color and on
the texture of the images in a single histogram, in
order to represent the main visual characteristics
of the image.
Once obtained a set of numerical features from
real and fake bioimages, as shown in Figure 2 we em-
ploy machine learning to perform classification. We
consider four classification algorithms, J48, Random
Forest, Random Tree and REPTree.
In the classification of medical images, algorithms
based on decision trees are particularly effective. In
particular, among the most used, J48 and Random
Tree (Dou and Meng, 2023) are relevant. The latter
acts by considering different random characteristics
for each node of the tree and does not perform prun-
ing operations, that is, unnecessary branches are not
removed. Among the models based on classification
trees, one of the most used is Random Forest, an al-
gorithm that combines several trees through Bagging,
based on the training of different models on subsets
of the dataset (Frank et al., 2016; Dou and Meng,
2023). This algorithm returns more stable models and
reduces variance. The aim of Random Forest is to im-
prove accuracy and reduce classification error. It has
proven to be particularly effective in several medical
contexts, in particular in the classification of images
depicting pathologies such as cancer (Dou and Meng,
2023).
Another classification algorithm is REPTree
which, unlike the models based on decision trees
mentioned above, prunes unnecessary branches. Fur-
thermore, it is designed to sort numerical values only
once, which is why it is faster (Frank et al., 2016).
3 EXPERIMENTAL ANALYSIS
In this section we present the experimental analysis
we performed aimed to demonstrate the effectiveness
of the proposed method.
3.1 Dataset
In order to collect real-world bioimages belonging to
different biomedical domains, we resort to MedM-
NIST (Yang et al., 2024), a collection of datasets
for biomedical image classification. The MedMNIST
collection includes 28x28 pixel bioimages, obtained
from real-world medical data and freely available for
research purposes.
In detail following datasets are exploited in the ex-
BIOIMAGING 2025 - 12th International Conference on Bioimaging
334

perimental analysis:
• PathMNIST, consisting of histological images of
colorectal cancer, stained with hematoxylin and
eosin;
• ChestMNIST, includes chest x-rays of fourteen
different diseases;
• DermaMNIST, it is based on dermatoscopic im-
ages of skin lesions, divided into seven diseases;
• OCTMNIST, it contains optical tomography im-
ages for the diagnosis of retinal diseases, divided
into four categories;
• PneumoniaMNIST, includes pediatric radio-
graphs for the classification of pneumonia
images;
• RetinaMNIST, it contains retinal images for as-
sessing the severity of diabetic retinopathy.
For each dataset 1000 bioimages are considered.
3.2 Experimental Settings
To understand whether DCGAN is able to generate
synthetic bioimages that are closer to the real ones,
following metrics are computed: Precision is a mea-
sure of the correctly classified instances compared to
all instances in the dataset. It is defined as:
Precision =
T P
T P + FP
(1)
with TP (true positive) the number of instances cor-
rectly identified as positive and FP (false positive) the
number of negative instances incorrectly classified as
positive.
Recall instead measures the ability of a model to cor-
rectly identify all positive instances and is defined as
follows:
Recall =
T P
T P + FN
(2)
with FN (false negative) the number of instances in-
correctly classified as negative.
Accuracy represents the proportion of correctly clas-
sified instances compared to all those present in the
dataset. This measure is not very valid in the case
in which the classes within the dataset are not well
balanced. Accuracy is defined as:
Accuracy =
T P + TN
T P + T N + FP + FN
(3)
with TN (true negative) the number of instances cor-
rectly classified as negative.
F-Measure is another metric that is particularly useful
for measuring the model’s ability to recognize pat-
terns correctly. It is calculated as the harmonic mean
between Precision and Recall:
F − Measure = 2 ∗
Precision ∗ Recall
Precision + Recall
(4)
The classification of bioimages generated by DC-
GAN was carried out by comparing 1000 real images
with the same number of generated images, selected
at five different training epochs (i.e., 0, 24, 49, 74 and
99). The idea is to evaluate whether, as the epochs in-
crease, more and more realistic images are generated
by the DCGAN for all the analyzed datasets.
For feature extraction and model building we re-
sort to Weka, one of the most widespread data mining
tool suite, presenting several machine learning algo-
rithm implementation.
3.2.1 Experimental Results
In this section we show the results obtained from the
experimental analysis. In particular, we exploited:
• six different datasets (i.e., PathMNIST, ChestM-
NIST, DermaMNIST, OCTMNIST, Pneumoni-
aMNIST and RetinaMNIST);
• four different image filters for feature extrac-
tion (i.e., AutoColorCorrelogramFilter, Binary-
PatternsPyramidFilter, ColorLayoutFilter and FC-
THFilter);
• four different classification algorithms (i.e., J48,
Random Forest, Random Tree and REPTree);
• we trained a DCGAN for each dataset and we gen-
erated 1000 fake images for each epoch (for a to-
tal of 100 total epochs). Thus, for each dataset, we
generated 1000 images x 100 epochs = 100000 to-
tal images.
The aim of the experimental analysis is to explore
whether the images produced by the DCGAN as the
epochs increase are more similar to real bioimages.
For this reason, we train a series of binary classifiers,
where each classifier is trained on 1000 images gen-
erated at a given epoch and 1000 real images. We
then evaluate the trend of the accuracy to draw con-
clusions.
As previously stated, we use different feature ex-
traction and different classification algorithms for the
sake of generability of the proposed experimental
analysis, and we present the results obtained for five
epochs: the first epoch, epoch 25, epoch 50, epoch 75
and epoch 100 (i.e., the last one).
Thus, for each dataset we build following models:
4 machine learning algorithms x 4 feature extraction
techniques x 100 epochs = 1600 models. Consider-
ing that we consider 6 different datasets to understand
the trend of images generated by DCGAN, in total we
considered 1600 x 6 = 9600 models.
Bioimages Synthesis and Detection Through Generative Adversarial Network: A Multi-Case Study
335
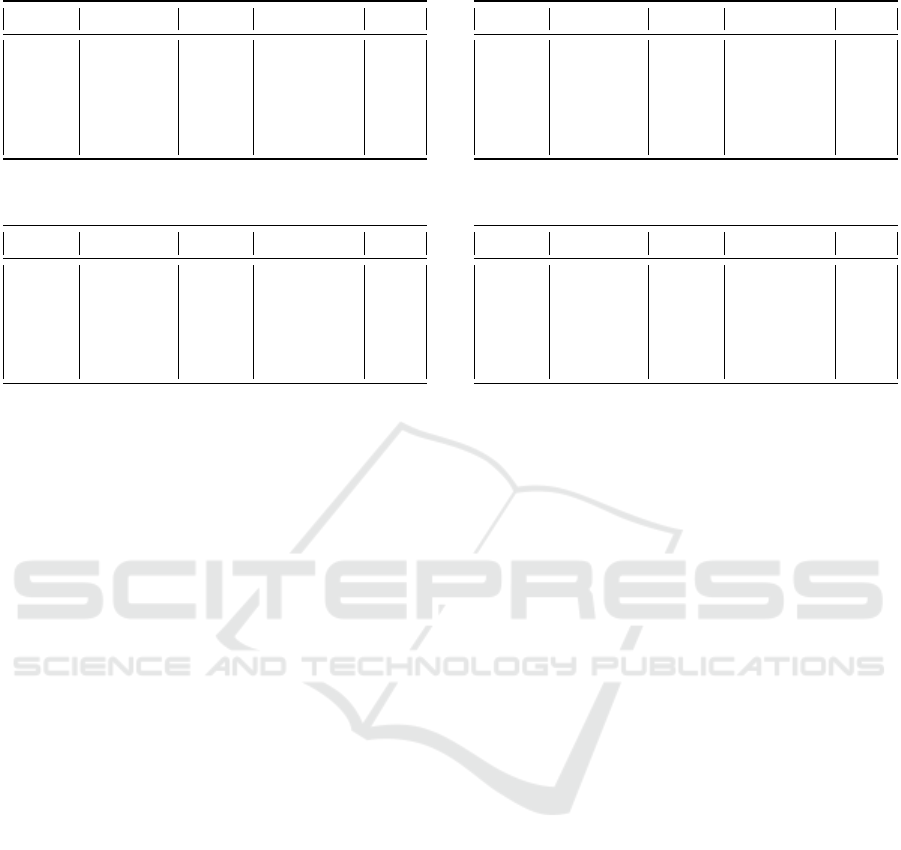
Table 1: PathMNIST Experimental analysis results.
Epoch Precision Recall F-Measure Acc.
0 0.90 0.90 0.89 0.90
24 0.81 0.81 0.81 0.81
49 0.79 0.79 0.79 0.79
74 0.81 0.81 0.81 0.81
99 0.81 0.81 0.80 0.80
Table 2: ChestMNIST Experimental analysis results.
Epoch Precision Recall F-Measure Acc.
0 0.99 0.99 0.99 0.99
24 0.99 0.99 0.99 0.99
49 0.99 0.99 0.99 0.99
74 1 1 0.99 0.99
99 0.99 0.99 0.99 0.99
We would expect that as the epochs increase, the
generated images are increasingly similar to the real
bioimages and therefore the classifier trained, for ex-
ample, with real images and images generated at
epoch 100 will have worse performances than a clas-
sifier trained with real images and images generated,
for example, at epoch 25. This is because we ex-
pect that as the epochs increase, since the generated
images are increasingly similar to the originals, the
classification algorithms will see them as increasingly
similar and therefore will not be able to discern them.
Tables 1, 2, 3, 4, 5, 6 shows the average Preci-
sion, Recall, F-Measure and Accuracy at five different
epochs.
For the PathMNIST dataset, which results are
shown in Table 1, the Accuracy trend was in line with
expectations. In the early training phases, the images
were of poor quality, while, as the epochs progressed,
the generator’s ability to produce realistic images im-
proved. The AutoColorCorrelogram image filter was
the most effective in distinguishing between the two
types of images, unlike BinaryPatternsPyramidFilter,
which led to a reduction in classification performance.
The Random Forest algorithm proved to be the most
suitable in classifying this type of histological images.
Other classifiers, such as REPTree and Random Tree,
showed lower performances in the last epochs, sug-
gesting a greater difficulty in distinguishing between
real and synthetic images.
In the case of the ChestMNIST dataset, which re-
sults are shown in Table 2, the Accuracy trend was
not in line with the predictions, showing a slope con-
trary to expectations. The generated images were
poorly characterized, especially in the last epochs,
suggesting a greater difficulty of the generator for this
Table 3: DermaMNIST Experimental analysis results.
Epoch Precision Recall F-Measure Acc.
0 0.99 0.99 0.99 0.99
24 0.96 0.96 0.95 0.95
49 0.94 0.94 0.94 0.94
74 0.94 0.94 0.94 0.94
99 0.94 0.94 0.94 0.94
Table 4: OCTMNIST Experimental analysis results.
Epoch Precision Recall F-Measure Acc.
0 0.99 0.99 0.99 0.99
24 0.99 0.99 0.99 0.99
49 0.99 0.99 0.99 0.99
74 0.99 0.99 0.99 0.99
99 0.99 0.99 0.99 0.99
dataset. In the classification, the combination of the
ColorLayoutFilter and the Random Tree algorithm
proved to be the most effective in distinguishing be-
tween the two types of images, although with worse
results than the previous dataset. In other filters, such
as the FCTHFilter, significant limitations were noted
due to the lack of sufficient information in the im-
age. Among the classification algorithms, Random
Forest was the one with the best performance, while
REPTree showed greater effectiveness in the most ad-
vanced epochs.
For the images of the DermaMNIST dataset,
which results are shown in Table 3, the generator
showed a constant progression, with a significant im-
provement of the images produced. In fact, in the first
epochs, images lacking details were found, which im-
proved significantly in the subsequent epochs. In this
context, AutoColorCorrelogramFilter and FCTHFil-
ter provided an evolution of the classification met-
rics according to the expectations. On the contrary,
BinaryPatternsPyramidFilter, although useful in the
texture analysis, showed a reduction in overall per-
formance compared to the other image filters used.
Random Forest proved to be the most effective classi-
fier, while Random Tree showed inferior results. The
trend of Accuracy confirmed the expectations, with a
progressive decrease during the advancement of the
epochs, suggesting a good ability of the generator to
generate increasingly realistic images.
As for results obtained with the ChestMNIST
dataset, the results related to OCTMNIST dataset,
shown in Table 4, did not produce the expected re-
sults. During training, less complex images than ex-
pected emerged, denoting an almost ascending curve
in the epoch-Accuracy graph. Among the image fil-
BIOIMAGING 2025 - 12th International Conference on Bioimaging
336

Table 5: PneumoniaMNIST Experimental analysis results.
Epoch Precision Recall F-Measure Acc.
0 0.99 0.99 0.99 0.99
24 0.99 0.99 0.99 0.99
49 0.99 0.99 0.99 0.99
74 0.99 0.99 0.99 0.99
99 0.99 0.99 0.99 0.99
Table 6: RetinaMNIST Experimental analysis results.
Epoch Precision Recall F-Measure Acc.
0 0.99 0.99 0.99 0.99
24 0.99 0.99 0.99 0.99
49 0.99 0.99 0.99 0.99
74 0.99 0.99 0.99 0.99
99 0.99 0.99 0.99 0.99
ters used, ColorLayoutFilter allowed a greater distinc-
tion between real and generated images, in particular
in combination with J48 and REPTree. BinaryPat-
ternsPyramidFilter showed better results in terms of
Accuracy, although not in line with the predictions.
The Accuracy trend was not in line with expecta-
tions, with an increase of this value in the last epochs.
This indicates a greater difficulty in generating im-
ages, which are not sufficiently realistic.
The PneumoniaMNIST dataset, which results are
shown in Table 5, showed particular results. In the
first epochs, the generator produced images with little
detail, which progressively improved in quality until
epoch 49. Subsequently, a slight worsening in the per-
formance of the classifier was observed, with a conse-
quent increase in Accuracy. BinaryPatternsPyramid-
Filter proved to be the best filter for the classifica-
tion of pneumonia images, unlike the others, which
showed different results. In fact, it is noted how the
Accuracy values are very high during all the training
epochs, with the exception of epochs 49 or 74, de-
pending on the image filters and classification algo-
rithms used. Among the latter, Random Forest was
the one that provided the best results. Overall, Ac-
curacy followed a trend similar to that expected, al-
though the slight increase in the last training epochs.
For RetinaMNIST dataset results, shown in Table
6, similar results were found to the previous dataset.
As a matter of fact, in the first epochs, the gener-
ated images were lacking in detail, while in the fol-
lowing ones an increase in their quality was noted,
with a slight change in trend at epoch 74. The Auto-
ColorCorrelgoramFilter and BinaryPatternsPyramid-
Filter filters showed good results. Despite this, the
combination of ColorLayoutFilter with the Random
Tree algorithm was the one that produced a result
more consistent with expectations. The Accuracy
trend is in line with expectations, showing a general
improvement in the generator’s capabilities, despite a
slight drop in performance in the second half of train-
ing.
With the aim to provide a full overview of the ex-
perimental analysis results, Figure 3 shows the aver-
age accuracy for each epoch (from 0 to 100) for the
datasets involved in the experimental analysis.
In each plot shown in Figure 3 (one plot for each
dataset), the x-axis indicates the epochs, while the y-
axis is related to the average accuracy for each model
trained with the original bioimage dataset and the fake
images generated for a certain epoch. We note that
for some datasets like ChestMNIST and OCTMNIST
there is a consistent improvement in accuracy when
images obtained from higher epochs are considered.
From the other side, when are considered images gen-
erated from the PathMNIST, DermaMNIST, Pneumo-
niaMNIST, and RetinaMNIST exhibit a decline or
fluctuation in accuracy over epochs, indicating that in
these cases the DCGAN is able to generated images
more similar to the real ones, as thee number of epoch
increases.
4 CONCLUSION AND FUTURE
WORK
In this paper we explored the possibility to exploit
GANS to generate realistic synthetic images, partic-
ularly in contexts such as the representation of com-
mon pathologies. In several cases, such as for PathM-
NIST and RetinaMNIST, the results confirmed that
the model is able to generate visually realistic im-
ages. In the classification process, the Accuracy trend
showed a progressive improvement over the training
epochs, indicating an increasing verisimilitude of the
synthetic images. However, for some datasets, includ-
ing ChestMNIST and OCTMNIST, the generated im-
ages did not reach the expected quality, resulting in
poor detail and easily distinguishable from real im-
ages. The limitations found, such as the non-uniform
quality of the images generated for some datasets, in-
dicate that there is still room for improvement in the
use of GANs for biomedical image generation. In par-
allel, the evolution of GANs raises ethical and secu-
rity issues. It will be necessary to implement mea-
sures to ensure the responsible use of these technolo-
gies, ensuring that synthetic images are not misused.
Strengthening systems that can accurately distinguish
between real and generated images, along with the
definition of guidelines for the use of GANs in the
Bioimages Synthesis and Detection Through Generative Adversarial Network: A Multi-Case Study
337
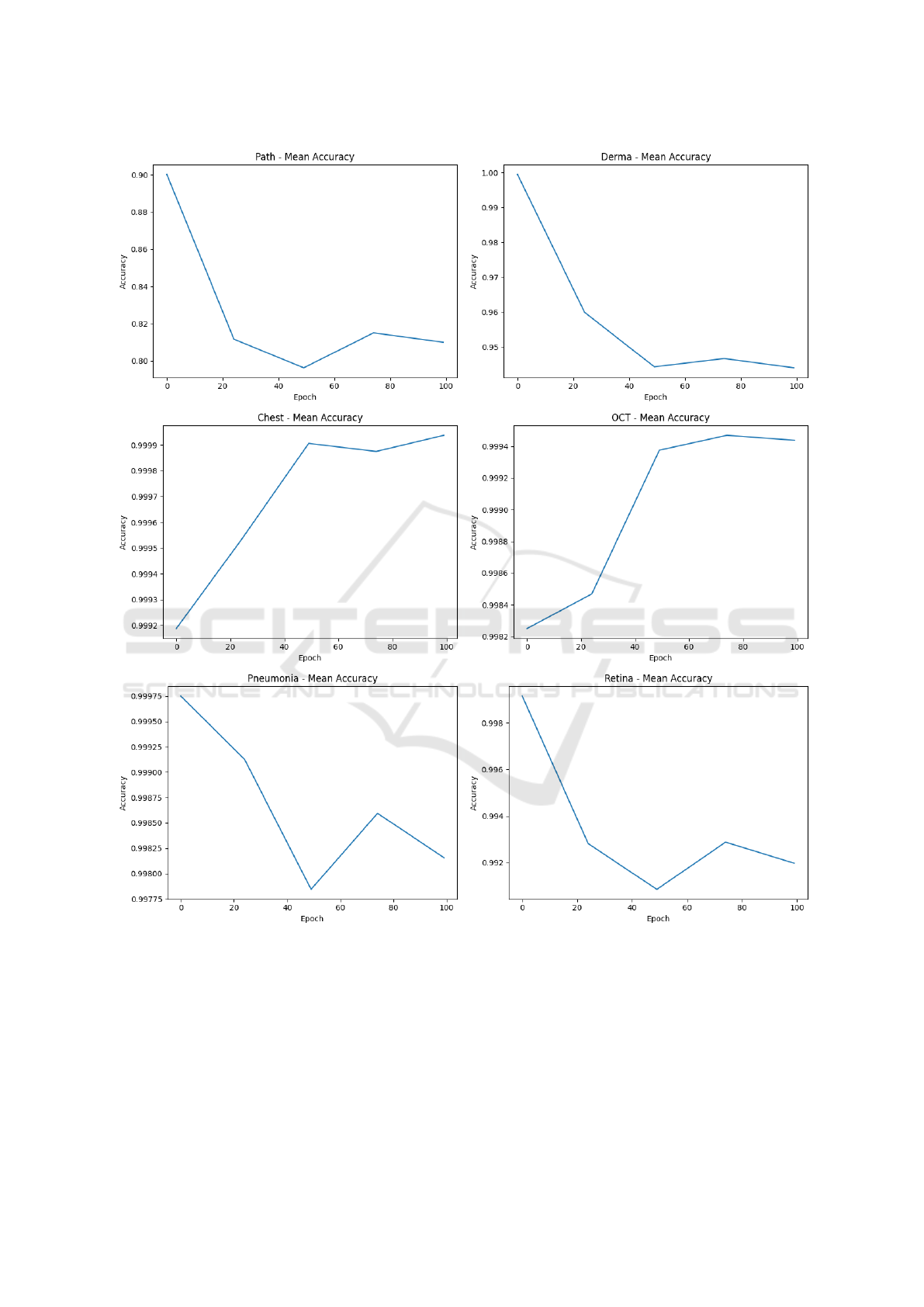
Figure 3: Average Accuracy while epochs are increasing for the multi-case study.
medical field, will be crucial to ensure transparency
in the use of these technologies.
ACKNOWLEDGEMENTS
This work has been partially supported by EU DUCA,
EU CyberSecPro, SYNAPSE, PTR 22-24 P2.01 (Cy-
bersecurity) and SERICS (PE00000014) under the
MUR National Recovery and Resilience Plan funded
by the EU - NextGenerationEU projects, by MUR -
REASONING: foRmal mEthods for computAtional
analySis for diagnOsis and progNosis in imagING -
PRIN, e-DAI (Digital ecosystem for integrated anal-
ysis of heterogeneous health data related to high-
impact diseases: innovative model of care and re-
BIOIMAGING 2025 - 12th International Conference on Bioimaging
338

search), Health Operational Plan, FSC 2014-2020,
PRIN-MUR-Ministry of Health, the National Plan for
NRRP Complementary Investments D
∧
3 4 Health:
Digital Driven Diagnostics, prognostics and therapeu-
tics for sustainable Health care, Progetto MolisCTe,
Ministero delle Imprese e del Made in Italy, Italy,
CUP: D33B22000060001, FORESEEN: FORmal
mEthodS for attack dEtEction in autonomous driv-
iNg systems CUP N.P2022WYAEW and ALOHA: a
framework for monitoring the physical and psycho-
logical health status of the Worker through Object de-
tection and federated machine learning, Call for Col-
laborative Research BRiC -2024, INAIL.
REFERENCES
Dou, Y. and Meng, W. (2023). Comparative analysis
of weka-based classification algorithms on medical
diagnosis datasets. Technology and Health Care,
31(S1):397–408. Available from: PMC10200164.
Frank, E., Hall, M. A., and Witten, I. H. (2016). The WEKA
Workbench. Morgan Kaufmann, 4th edition. On-
line Appendix for ”Data Mining: Practical Machine
Learning Tools and Techniques”.
Fu, H., Cheng, J., Xu, Y., Wong, D. W. K., Liu, J., and Cao,
X. (2018). Joint optic disc and cup segmentation based
on multi-label deep network and polar transformation.
IEEE transactions on medical imaging, 37(7):1597–
1605.
He, H., Yang, H., Mercaldo, F., Santone, A., and Huang, P.
(2024). Isolation forest-voting fusion-multioutput: A
stroke risk classification method based on the multidi-
mensional output of abnormal sample detection. Com-
puter Methods and Programs in Biomedicine, page
108255.
Huang, P., He, P., Tian, S., Ma, M., Feng, P., Xiao, H.,
Mercaldo, F., Santone, A., and Qin, J. (2022). A vit-
amc network with adaptive model fusion and multiob-
jective optimization for interpretable laryngeal tumor
grading from histopathological images. IEEE Trans-
actions on Medical Imaging, 42(1):15–28.
Huang, P., Li, C., He, P., Xiao, H., Ping, Y., Feng, P., Tian,
S., Chen, H., Mercaldo, F., Santone, A., et al. (2024).
Mamlformer: Priori-experience guiding transformer
network via manifold adversarial multi-modal learn-
ing for laryngeal histopathological grading. Informa-
tion Fusion, 108:102333.
Huang, P., Tan, X., Zhou, X., Liu, S., Mercaldo, F., and
Santone, A. (2021). Fabnet: fusion attention block
and transfer learning for laryngeal cancer tumor grad-
ing in p63 ihc histopathology images. IEEE Journal
of Biomedical and Health Informatics, 26(4):1696–
1707.
Huang, P., Zhou, X., He, P., Feng, P., Tian, S., Sun, Y., Mer-
caldo, F., Santone, A., Qin, J., and Xiao, H. (2023).
Interpretable laryngeal tumor grading of histopatho-
logical images via depth domain adaptive network
with integration gradient cam and priori experience-
guided attention. Computers in Biology and Medicine,
154:106447.
Mercaldo, F., Brunese, L., Martinelli, F., Santone, A., and
Cesarelli, M. (2023). Generative adversarial net-
works in retinal image classification. Applied Sci-
ences, 13(18):10433.
Mercaldo, F., Zhou, X., Huang, P., Martinelli, F., and San-
tone, A. (2022). Machine learning for uterine cervix
screening. In 2022 IEEE 22nd International Confer-
ence on Bioinformatics and Bioengineering (BIBE),
pages 71–74. IEEE.
Orlando, J. I., Barbosa Breda, J., Van Keer, K., Blaschko,
M. B., Blanco, P. J., and Bulant, C. A. (2018).
Towards a glaucoma risk index based on sim-
ulated hemodynamics from fundus images. In
Medical Image Computing and Computer Assisted
Intervention–MICCAI 2018: 21st International Con-
ference, Granada, Spain, September 16-20, 2018,
Proceedings, Part II 11, pages 65–73. Springer.
Yang, J., Shi, R., Wei, D., Liu, Z., Zhao, L., Ke, B., Pfister,
H., and Ni, B. (2024). Medmnist+ 18x standardized
datasets for 2d and 3d biomedical image classification
with multiple size options: 28 (mnist-like), 64, 128,
and 224. Dataset, Version 3.0, Published on January
16, 2024. Accessed: 26 September 2024.
Zhou, X., Tang, C., Huang, P., Mercaldo, F., Santone, A.,
and Shao, Y. (2021). Lpcanet: classification of laryn-
geal cancer histopathological images using a cnn with
position attention and channel attention mechanisms.
Interdisciplinary Sciences: Computational Life Sci-
ences, 13(4):666–682.
Zhou, X., Tang, C., Huang, P., Tian, S., Mercaldo, F., and
Santone, A. (2023). Asi-dbnet: an adaptive sparse
interactive resnet-vision transformer dual-branch net-
work for the grading of brain cancer histopathological
images. Interdisciplinary Sciences: Computational
Life Sciences, 15(1):15–31.
Bioimages Synthesis and Detection Through Generative Adversarial Network: A Multi-Case Study
339
